Abstract
The prpR1 of Porphyromonas gingivalis codes for three distinct enzymes with specificity for arginyl peptide bonds termed RI, RIA, and RIB. These three isoforms comprise the majority of the extracellular, arginine-specific protease activity in P. gingivalis W50. RI is a heterodimer in which the catalytic α chain is noncovalently associated with a second chain involved in adherence phenomena. RIA and RIB are both monomeric species. RIA represents the free α chain, and RIB is a highly posttranslationally modified form of the α chain which is exclusively vesicle or membrane associated and migrates as a diffuse band on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. In previous studies, insertional inactivation of the prpR1 demonstrated that arginine-specific protease activity can also arise from a closely related second gene, prR2. In the present work, the prR2 was insertionally inactivated in P. gingivalis W50 in order to establish the contribution of this locus to the arginine-specific protease activity of this periodontal bacterium. Loss of prR2 function had several effects on prpR1-derived enzymes. First, the total Arg-X activity was reduced by approximately 50% relative to that of the parent strain. The reduction in total activity was a consequence of decreased concentrations of the monomeric enzymes derived from the prpR1, while the heterodimeric enzyme, RI, was unaffected by this mutation. Second, the chromatographic behavior of both the soluble and vesicle- or membrane-associated monomeric enzymes was radically different from the behavior of RIA and RIB from the parent strain. Finally, the vesicle- or membrane-associated enzyme in the prR2 mutant strain lacked the extensive posttranslational additions which are found on RIB in P. gingivalis W50. These data suggest that the product(s) of the prR2 plays a significant role in the maturation pathway of prpR1-derived enzymes, and this may contribute to the coconservation of these two genes in P. gingivalis.
Porphyromonas gingivalis is a highly proteolytic, gram-negative, anaerobic bacterium which is a frequent component of the microbial community in the subgingival plaque of patients with adult periodontal disease. The proteolytic activities of P. gingivalis are thought to present a significant challenge to the colonized host through their deregulatory effects on the inflammatory response and host defenses (8, 24, 26) in addition to direct action on the structural components of the host tissues (9). Proteases with specificity for peptide bonds containing arginine in the P1 position comprise a major proportion of the total extracellular activity in vitro (4, 20), account for several of the deregulatory effects on host systems observed by using whole bacterial cells and culture supernatants (16, 25), and are therefore considered to be important virulence determinants (6).
Gene inactivation studies have demonstrated that insertions at two chromosomal loci are required to abolish all the Arg-X protease activity of P. gingivalis (14), and the two genes have now been cloned and sequenced (1, 11, 17, 18) and their products characterized (21, 22). The prpR1 (protease polyprotein RI) gives rise to three separate enzyme species referred to as RI, RIA, and RIB. RI is a heterodimer (Mr, 110,000) composed of a catalytically active α chain in noncovalent association with a β chain which has adhesin or hemagglutinin properties (5, 10). These two chains are contiguous on the initial translation product and are flanked by long N- and C-terminal extensions. RIA (54K) and RIB (70 to 80K) are both monomers of the α chain which are differentially posttranslationally modified with components common to the lipopolysaccharide (LPS) of this organism. The individual steps in the maturation pathway of the three isoenzymes derived from the prpR1 have not been established but may involve proteolytic processing of a full-length translation product to give rise to the heterodimeric RI, truncated transcription of the prpR1 to generate the monomeric enzymes (21), and differential posttranslational modifications to these forms to give rise to RIA and RIB (22).
Two additional arginine-specific proteases, termed RIIA and RIIB, are produced by the second gene, prR2 (protease RII), and these proteases are very similar to RIA and RIB, respectively, with respect to structure and enzymatic properties. However, since RIIA and RIIB have only been detected in the culture supernatants of a prpR1 isogenic mutant of P. gingivalis W50, they may not represent true extracellular enzymes of the wild-type strain (21). In the present study we wished to determine the contribution of prR2-derived enzymes to the overall arginine-specific protease activity of P. gingivalis W50. Given the low levels of these enzymes in the wild-type culture supernatant we predicted that loss of prR2 expression would have only a minimal effect. However, the data demonstrate that the products of the prR2 play an important role in the maturation pathway of the prpR1-derived enzymes via an effect(s) at the level of posttranslational modification.
MATERIALS AND METHODS
Bacteria, growth conditions, and plasmids.
P. gingivalis W50 and the isogenic mutants, W501 (prpR1) and W50D7 (prR2), were cultured anaerobically on blood agar or in brain heart infusion media (Oxoid, Basingstoke, United Kingdom) supplemented to 5 μg ml−1 with hemin as described previously (1, 2, 22). Escherichia coli XL-1 Blue MRF′ (Stratagene) was grown at 37°C in Luria-Bertani (tryptone, 10 g liter−1; yeast extract, 5 g liter−1; NaCl, 5 g liter−1 [23]) medium containing 20 μg of tetracycline per ml. For selection of P. gingivalis insertion mutants, clindamycin chloride was added to 5 μg ml−1, and for maintenance of pUC-derived plasmids in E. coli, the medium was supplemented with ampicillin to 50 μg ml−1. Luria-Bertani medium containing 300 μg of erythromycin per ml was used to select for E. coli containing Erm cassette. Ultrapure plasmids were prepared with the ion-exchange columns of Qiagen Inc.
Generation of prR2 mutant P. gingivalis strain.
prR2 in P. gingivalis W50 was insertionally inactivated with an ermF-ermAM tandem macrolide-lincosamide cassette (Erm) by allelic exchange following electrotransformation with a 3.8-kb amplicon derived from pKE1 as described previously (21).
Southern blotting.
P. gingivalis chromosomal DNA was purified, restricted, transferred onto membranes, and probed under stringent conditions as previously described (1, 2). A 270-bp EcoRV-SphI fragment from the pKpL insert (Fig. 1) and a 2.1-kb SstI-PstI fragment of pVA2198 (7) containing the Erm cassette were excised out of preparative agarose gels following electrophoresis and purified by using Qiaquick (Qiagen). DNA labelling was performed by the random priming method (Ready-To-Go; Pharmacia) and by using Redivue [α-32P]dCTP (3,000 mCi/mmol; Amersham).
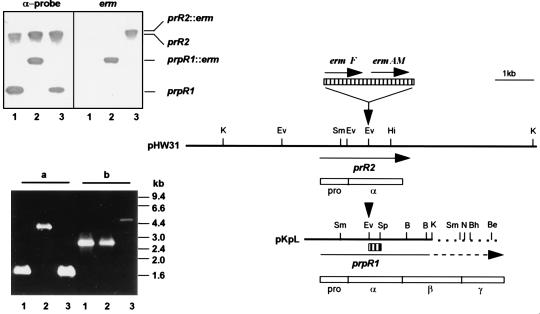
FIG. 1.
Genomic organization of the major arginine-specific protease genes prR2 and prpR1 and Southern and PCR analyses of P. gingivalis W50 strains. The two bold lines represent an ∼8.5-kb-HindIII (pHW31) or Sau3AI-KpnI (pKpL, nucleotide 1 to 3471 of the sequence with accession no. X82680) fragment of P. gingivalis W50 genomic DNA bearing prR2 and truncated prpR1, respectively. Unique restriction sites are shown as bars above the fragments. Note that both pKpL and prpR1 have been extended for comparative purposes, and this is shown as broken lines. Insertion of the Erm cassette, encoding ermF and ermAM (hatched box above restriction maps), is at the EcoRV site (indicated by down-pointing arrows) for generation of P. gingivalis W501 or W50D7 via electrotransformation. Positions and direction of genes are indicated by arrows under the DNA fragments. Empty boxes are schematic representations of translated sequences (PrRII and PrpRI) with their domain characteristics (pro, α, β, and γ); pro and α domains are common to the two gene products. The α-probe, used in the Southern hybridization experiment, is shown as a small hatched box under pKpL. In the Southern hybridization (upper inserted panel), P. gingivalis chromosomal DNA was restricted with SmaI and membranes were probed with either the 270-bp α-probe or the 2.1-kb Erm cassette. The positions of the 3.2-kb prpR1 and ≥12-kb prR2 together with the corresponding erm inserted loci are shown. Lanes: 1, W50 (wt); 2, W501 (prpR1 mutant); 3, W50D7 (prR2 mutant). The PCR panel (lower insert) shows the agarose electrophoresis of amplicons generated with primers specific for prpR1 (a) or prR2 (b) by using P. gingivalis chromosomal DNA as templates. The lane numbers are the same as for the Southern hybridization experiment. B, BamHI; Be, BstEII; Bh, BssHII; Ev, EcoRV; Hi, HincII; K, KpnI; N, NcoI; Sm, SmaI; Sp, SphI.
Characterization of P. gingivalis protease mutants by PCR.
Chromosomal DNAs from P. gingivalis strains were used as templates in PCR by using primer pairs specific for either the α-coding region of prpR1 (21) or the prR2 locus. The former reaction mixture has been described previously. The latter reaction mixture contained chromosomal DNA, 100 ng; deoxynucleoside triphosphates, 250 μM; MgCl2, 2.7 mM; primer TwoF4 (5′-ATATATggtaccAATGATGCTCGGTTTGGG-3′), 0.5 μg; primer TwoR2 (5′-ATATATaagcttGGATTCCTCGGCACAGCC-3′), 0.5 μg; Thermus icelandicus DNA polymerase (Advanced Biotechnologies), 2.5 U. The mixture was subjected to 25 cycles of 94, 60, and 72°C for 1, 1, and 5 min, respectively. Control reaction mixtures did not contain any DNA or one of the two primers.
Protease purifications.
Purification of P. gingivalis RI, RIA, and RIB was performed by using a combination of gel filtration, affinity, and ion-exchange chromatographies as described previously (22).
Enzyme assays.
Arg-X protease activity was measured routinely in 0.1 M Tris-HCl (10 mM l-cysteine, 10 mM CaCl2, pH 8.1, 30°C) with N-benzoyl-dl-arginine p-nitroanilide (dl-BApNA) (500 μM) as the substrate. The reaction was monitored at 405 nm, and enzyme activity was expressed as increase in absorbance/min at 30°C. Lys-X protease activity was measured with N-α-acetyl-l-lysine-p-nitroanilide (AcLyspNA) (250 μM) as substrate in the same reaction buffer and under the same conditions as described above.
Gel electrophoresis.
Polyacrylamide gel electrophoresis (PAGE) in the presence of sodium dodecyl sulfate (SDS) (12) was carried out at 5°C in 12.5% (wt/vol) polyacrylamide slab gels (10 by 7 by 0.15 cm). Samples of protease (10 to 20 μg) were first treated with 50 μl of leupeptin (1 mM) at 25°C for 20 min, heated at 100°C for 5 min, and dried in vacuo. Fluorescently labelled protease samples were prepared as described below. PAGE in the presence of 8 M urea at pH 8.8 in 7.5% (wt/vol) slab gels was carried out according to the method of Marshall and Inglis (13).
Fluorescence labelling of proteases by using DNS-EGR-CK.
Protease solutions were treated with an equal volume of 0.2 M Tris-HCl (20 mM CaCl2, 20 mM 2-mercaptoethanol [pH 8.4]) at 4°C for 10 min. Dansyl-glutamyl-glycyl-arginyl chloromethyl ketone (DNS-EGR-CK) (0.25 mg; Calbiochem) was dissolved in 600 μl of 95% (vol/vol) aqueous ethanol just before use. Fifty microliters was added to the reduced protease solution, and the reaction was allowed to proceed at 4°C until the enzyme activity (monitored by dl-BApNA hydrolysis) was abolished. Samples of protein solution were then dried in vacuo. Labelling was also performed in the presence of 50 μM leupeptin to unambiguously identify the protease band after SDS-PAGE. The dried fluorescently labelled proteases were then treated with either SDS-PAGE or 8 M urea sample buffer and subjected to electrophoresis.
Following 8 M urea-PAGE it was necessary to transfer the proteins to polyvinyl difluoride membranes (Millipore) before the fluorescent protein bands could be visualized under UV light. This procedure removed the excess DNS-EGR-CH2OH, the byproduct of the labelling reaction which migrated to approximately the same position as the protease bands on urea gels.
LPS preparation.
Cells from 48-h brain heart infusion broth cultures of each strain were pelleted by centrifugation at 4,000 × g for 30 min at 4°C, washed twice in phosphate-buffered saline (pH 7.2), and resuspended in the same buffer to an optical density at 525 nm of 1.0. Cells from the 1-ml cell suspension were then pelleted and resuspended in 75 μl of reducing SDS-PAGE sample buffer and heated at 100°C for 10 min. Proteinase K (50 μg) was then added, and the mixtures were incubated at 50°C for 18 h. Twenty-five microliters of each preparation was then subjected to SDS-PAGE (12.5% polyacrylamide gel) and the LPS was visualized by silver staining by the method of Wray et al. (27).
RESULTS
Generation of prR2 mutant strain P. gingivalis W50D7.
Southern hybridization analyses of DNA from a number of laboratory (1) and clinical isolates (3) of P. gingivalis have confirmed the existence of a gene (prR2) highly homologous to the α-encoding domain of prpR1. Therefore, an α::erm fragment was constructed by insertion of a tandem macrolide-lincosamide resistance cassette (Erm) into the α-coding region of prpR1 in pKpL (21) for allelic exchange via electrotransformation of P. gingivalis W50. Chromosomal integration of the Erm cassette into either the prpR1 or prR2 locus on the P. gingivalis W50 genome was initially examined by Southern blot hybridization and then confirmed by PCR by using locus-specific primers (Fig. 1). The prpR1-specific primer pair amplified the 1.7-kb α-coding domain of prpR1. Insertion of a 2.1-kb Erm cassette in P. gingivalis W501 shifted the PCR product to 3.8 kb, while the unaltered product size was found in a prR2 mutant strain (W50D7). Similarly, the prR2-specific primers TwoF4 and TwoR2 amplified a 2.8-kb amplicon in P. gingivalis W50 corresponding to the entire prR2 gene and 600 bp of the 5′ sequence. The insertion of the 2.1-kb Erm cassette in P. gingivalis W50D7 increased the amplicon size to 4.9 kb. These data confirmed the Southern blot information (Fig. 1) that chromosomal integration of the Erm cassette into the prR2 had been achieved in P. gingivalis W50D7, and this mutant was chosen for analysis of the effects on protease expression.
Inactivation of prR2 results in 50% reduction in total BApNA activity.
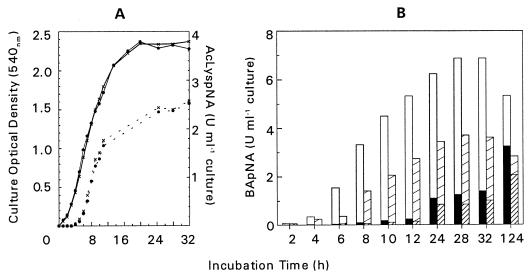
Insertional inactivation of prR2 had no measurable effect on either the growth rate or final cell yield of the mutant compared to the parent strain in brain heart infusion broth culture. Similarly, no differences were found with respect to the level of AcLyspNA hydrolysis (Fig. 2A). However, the total BApNA activity of the prR2 mutant strain was reduced by approximately 50% throughout all stages of growth. This reduction was more pronounced in the cell-associated BApNA activity. For example, at the end of log-phase growth (t = 32 h) the cell-bound activity of the mutant was reduced by 52.5% while the activity in the supernatant was still 72.5% of that in the parent strain (Fig. 2B). Exactly the converse was obtained when the distribution of activity in a prpR1 mutant strain was compared to that of the parent (21). In that instance, BApNA activity in the extracellular compartment was selectively reduced. These data may indicate that in P. gingivalis W50 the products of prR2 represent mainly cell-bound forms of Arg-X proteases, whereas the prpR1 products are predominantly extracellular and support previous observations that the prR2 products are not readily detectable in the culture supernatant of the wild-type P. gingivalis W50.
FIG. 2.
(A) Growth curves and Lys-X protease activity of P. gingivalis W50 (x - x) and W50D7 (prR2 mutant) (• - •). A 10% inoculum of overnight cultures of both strains was used to inoculate brain heart infusion broth supplemented with hemin (5 mg liter−1). Samples were withdrawn throughout growth for absorbance measurements (solid lines), and total Lys-X protease activity (dotted lines) was measured following sonication of whole culture mixed with an equal volume of 25 mM MOPS (morpholinepropanesulfonic acid) pH 6.0 buffer (10 mM CaCl2, 0.2 M NaCl). Units of enzyme activity are expressed as increase in absorbance at 405 nm/min. (B) Arg-X activity in culture supernatant and in sonicated cultures was measured as described in the text. Open bars indicate cell-bound activity, and solid bars indicate supernatant of P. gingivalis W50. Broad hatched bars indicate cell-bound activity, and fine hatched bars indicate supernatant in W50D7 (prR2 mutant). Units of enzyme activity are expressed as increase in absorbance at 405 nm/min.
RIA and RIB are significantly reduced in the prR2 mutant strain and have altered chromatographic behavior relative to wild-type enzymes.
In order to confirm that the residual Arg-X protease activity in the prR2 mutant strain was derived from prpR1 expression and also to examine the relative proportions of the isoforms of the RI proteases in this mutant, the enzymes in the 6-day culture supernatant were fractionated by using procedures developed for the purification of RI, RIA, and RIB from the parent strain (22). All the activity was precipitated by saturating the culture supernatant with 85% ammonium sulfate. Only a small proportion (15%) of the precipitate could be resolubilized with low-detergent (0.0055% Zwittergent) buffer (S-fraction). The remainder (P-fraction) required buffer containing 0.05% detergent for solubilization, indicating that the vast majority (85%) of the extracellular Arg-X protease activity in the prR2 mutant strain is associated with vesicles or membrane fragments. In the parent strain this activity is routinely of the order of 60% of the total (Table 1).
TABLE 1.
Distribution of Arg-X enzyme activity in culture supernatants of P. gingivalis W50 and W50D7 (prR2 mutant)
| Fraction | Extracellular Arg-X enzyme activity (%)
|
|
|---|---|---|
| W50 | W50D7 (prR2 mutant) | |
| Culture supernatant | 100 | 100a |
| 85% Ammonium sulfate | ||
| Supernatant | 0 | 0 |
| Pellet | 100 | 100 |
| Solubilized with 0.0055% detergent | ||
| S-fraction (soluble) | 40 | 15 |
| ↓ | ||
| RI | 15 | 6.8 |
| RIA | 25 | 8.2 |
| Solubilized with 0.05% detergent | ||
| P-fraction (vesicle or membrane bound) | 60 | 85 |
| ↓ | ||
| RI | 10 | 51 |
| RIB | 50 | 34 |
Total extracellular Arg-X activity of P. gingivalis W50D7 (prR2 mutant) is approximately 50% of that of the parent strain W50.
The sum of RI purified by affinity chromatography from the S- and P-fractions represented approximately 58% of the total Arg-X activity in the culture supernatant of the prR2 mutant strain. In the parent strain, RI routinely comprises approximately 25% of the total activity. However, as the total extracellular activity in the prR2 mutant strain is half that of the parent W50, approximately equal concentrations of RI are present in the culture supernatants of the two strains, and hence inactivation of prR2 has no effect on the amount of this isoform.
In contrast to the situation for RI, which displayed chromatographic behavior identical to that of the parent strain enzyme, there were significant alterations to the physical properties of the enzymes which were not retained by affinity chromatography of the S- and P-fractions of W50D7 compared to the parent strain RIA and RIB. All attempts to purify these activities by using chromatographic procedures which are reproducible for the parent strain enzymes were unsuccessful (see below). Furthermore, separations based on cationexchange hydrophobic or hydroxyapatite chromatography failed to generate homogeneous preparations. In addition to their aberrant chromatographic behavior and consistent with their decreased stability, the proportions and total levels of these monomeric Arg-X enzymes in the prR2 mutant strain were markedly different to those of the parent strain RIA and RIB. This was particularly significant in the case of the monomeric enzyme present in the S-fraction, which comprised only 8.2% of the total extracellular activity in the prR2 mutant strain (versus 25% in the parent), corresponding to a greater than sixfold reduction in its absolute concentration. Similarly, a threefold reduction in the concentration of monomeric enzyme associated with the vesicle or membrane fraction in the prR2 mutant strain was observed.
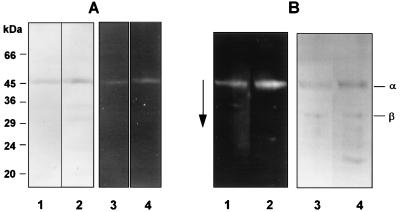
The SDS-PAGE and urea-PAGE profiles of RI from the parent strain P. gingivalis W50 and the prR2 mutant are shown in Fig. 3. RI from both strains gives rise to a single band (Mr, ∼54K) which contains both the α and β components of the heterodimer (Fig. 3A). Several minor lower-molecular-weight protein bands were also present in RI from the prR2 mutant. N-terminal sequence analysis of all of these components gave the sequence SGQAE…, demonstrating that they represent C-terminally truncated products of the β component which we have shown previously to be highly susceptible to autolytic degradation (22). The rates of migration of the α and β chains of RI from the prR2 mutant strain were indistinguishable from rates for the parent RI on urea-PAGE, indicating that there are no significant alterations to the charges of these molecules (Fig. 3B). RI from both W50 and W50D7 showed several faster moving minor bands on 8 M urea-PAGE, which again correspond to peptides arising from breakdown of the RI β chain. Hence, on the basis of size and chromatographic and charge characteristics, RI from the prR2 mutant strain was identical to the parent strain enzyme.
FIG. 3.
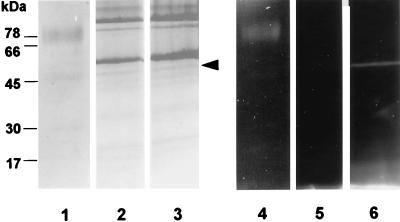
Gel electrophoresis of DNS-EGR-CK-labelled RI from P. gingivalis W50 and W50D7 (prR2 mutant). Enzymes from strains W50 and W50D7 were labelled with the fluorescent irreversible inhibitor DNS-EGR-CK as described in the text and subjected to SDS-PAGE on 12.5% gels (A) or 8 M urea-PAGE on 7.5% gels (B) followed by blotting onto polyvinylidene difluoride membranes. Gels and blots were viewed under UV light to visualize labelled proteins and then stained for total protein with Coomassie brilliant blue. (A) Lanes: 1 and 2, stained with Coomassie blue; 3 and 4, viewed under UV light; 1 and 3, RI-W50; 2 and 4, RI-W50D7. The molecular masses of marker proteins are indicated alongside the gel. (B) Lanes: 1 and 2, viewed under UV light; 3 and 4, stained with Coomassie blue; 1 and 3, RI-W50; 2 and 4, RI-W50D7 (prR2 mutant). The arrow indicates the direction of migration. α and β indicate the α-catalytic chain and the β-adhesin chain of RI.
The residual enzyme activities in the soluble and vesicle or membrane fractions of the prR2 mutant strain were subjected to ion-exchange chromatography. In the parent strain this procedure yields homogeneous preparations of RIA and RIB, respectively. However, there were still multiple proteins in the molecular weight range of 15 to 100K on SDS-PAGE in both fractions from W50D7. In order to determine which of these proteins represented the catalytically active moieties, a protease active-site labelling technique was employed by using a fluorescent peptide chloromethyl ketone protease inhibitor.
Insertional inactivation of prR2 affects the maturation pathway of prpR1-derived enzymes.
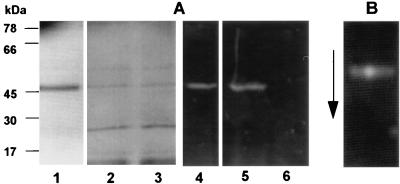
DNS-EGR-CK binds irreversibly to the active site of the arginine-specific proteases of P. gingivalis. The labelled proteases can then be detected by UV illumination following electrophoresis. Thus, labelling of RI from the parent strain and the prR2 mutant with DNS-EGR-CK generates fluorescent bands in the expected positions on SDS-PAGE and urea-PAGE (Fig. 3). Treatment of the residual enzyme activity in the S-fraction from the prR2 mutant strain with DNS-EGR-CK specifically labelled a minor protein (Mr, ∼54K) which was indistinguishable from the parent strain RIA on the basis of migration on SDS-PAGE and urea-PAGE (Fig. 4). Throughout the remainder of the paper this enzyme is therefore referred to as RIAs-W50D7. However, labelling of the residual enzyme activity in the vesicle or membrane fraction from the mutant with DNS-EGR-CK failed to demonstrate the presence of a high-molecular-weight, modified form of the α chain which is characteristic of RIB from P. gingivalis W50 (Fig. 5). Instead, the catalytic moiety in this preparation appeared identical in size to the α chain of RI and RIA. This enzyme is therefore referred to as RIAv-W50D7. The migration of this enzyme on urea-PAGE was not examined since RIB from the parent strain barely enters the resolving gel in this system. In the case of both RIAs-W50D7 and RIAv-W50D7 the labelling with DNS-EGR-CK was inhibited by leupeptin, thereby confirming the specificity of the reaction to protease active sites.
FIG. 4.
Gel electrophoresis of DNS-EGR-CK-labelled RIA from P. gingivalis W50 and monomeric enzyme from the S-fraction of W50D7 (RIAs-W50D7). Enzymes from strains W50 and W50D7 were labelled with DNS-EGR-CK as described in the text and subjected to SDS-PAGE (A) or 8 M urea-PAGE (B) and detected as described in the legend to Fig. 3. (A) Lanes: 1 to 3, stained with Coomassie blue; 4 to 6, viewed under UV light; 1 and 4, RIA-W50; 2 and 5, RIAs-W50D7; 3 and 6, RIAs-W50D7 labelled in the presence of 50 μM leupeptin. (B) A mixture of RIA-W50 and RIAs-W50D7 labelled with DNS-EGR-CK and run on an 8 M urea-PAGE gel.
FIG. 5.
Gel electrophoresis of DNS-EGR-CK-labelled RIB from P. gingivalis W50 and monomeric enzyme from the P-fraction of W50D7 (RIAv-W50D7). Enzymes from strains W50 and W50D7 were labelled as described in the legend to Fig. 3 and subjected to SDS-PAGE on 12.5% gels. Lanes: 1 to 3, stained with Coomassie blue; 4 to 6, viewed under UV light; 1 and 4, RIB-W50; 2 and 5, RIAv-W50D7 labelled in the presence of 50 μM leupeptin; 3 and 6, RIAv-W50D7. The arrowhead next to lane 3 indicates the protein which is labelled with DNS-EGR-CK.
These data suggest that insertional inactivation of the prR2 in P. gingivalis W50 affects the maturation of the monomeric enzymes derived from the prpR1, leading to altered chromatographic behavior of these proteases and a failure to generate the highly posttranslationally modified RIB.
LPS synthesis is unaltered in P. gingivalis W50D7 (prR2 mutant).
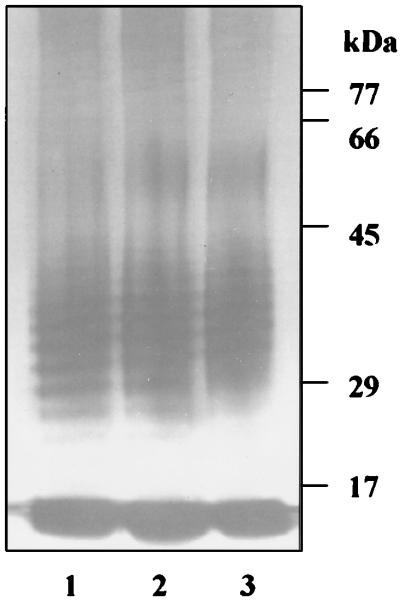
The posttranslational modifications to RIB in P. gingivalis W50 involve the addition of components which are common to the LPS of this organism. The altered posttranslational modifications to RIB from the vesicle fraction of the prR2 mutant (Fig. 5) led us to compare the gross structures of the LPSs in the prR2 mutant and the parent strain. However, on the basis of silver-stained SDS-PAGE we were unable to detect any significant differences between the LPS purified from W50 and that of the prR2 mutant, suggesting that the observed alterations to RIA and RIB are not due to a gross defect in LPS synthesis (Fig. 6).
FIG. 6.
Silver-stained SDS-PAGE of LPS preparations from P. gingivalis W50 (lane 1), W501 (prpR1 mutant) (lane 2), and W50D7 (prR2 mutant) (lane 3). Proteinase K digests of stationary phase cells were subjected to SDS-PAGE on 12.5% gels followed by silver staining. No Coomassie blue staining material was present.
DISCUSSION
The study over the last decade of proteases of P. gingivalis with specificity for arginyl peptide bonds has led to conflicting data regarding the number of these enzymes, their molecular weights, and genomic origin (19). However, these differences are beginning to be rationalized by gene inactivation studies and careful analysis of the residual enzyme activity. In brief, although some of the conflicting data may have arisen through analysis of different strains of this genomically heterogeneous species, it is likely that the production of different enzyme isoforms from the same gene has added to the confusion. The production of three separate biochemical entities, each of different molecular weight and posttranslational modification, from the prpR1 and two from the prR2 suggests that a dedicated and multicomponent maturation pathway(s) is involved in the production of these arginine-specific proteases (21). The biological significance of this pathway(s) and the resulting biochemical species is unclear, although it presumably reflects a requirement by this organism for different functional activities to facilitate colonization and survival at inflamed sites in the periodontal tissues of humans.
The data presented in this communication demonstrate that insertional inactivation of the prR2 of P. gingivalis W50 results in a significant reduction in the total Arg-X protease activity, particularly in the cell-associated fraction. This could have been predicted from our earlier observations (21) that inactivation of the prpR1 only results in an approximately 50% reduction in the total Arg-X protease activity which selectively affects the extracellular enzyme fraction. Furthermore, significant levels of prR2 mRNA expression are detectable in both the parent strain and a P. gingivalis prpR1 mutant strain (21).
Of greater significance, however, particularly with respect to the maturation pathway of the arginine-specific enzymes, was the finding that inactivation of the prR2 had significant effects on the biochemical properties of two of the prpR1-derived enzymes: RIA and RIB. The strategies which have been developed for the purification of RIA and RIB and which are consistently effective for these enzymes from the parent strain (22) were unable to yield homogeneous preparations from the prR2 mutant strain. The altered chromatographic behavior was also reflected in the molecular size of the vesicle-associated enzyme on SDS-PAGE, which suggested that the normal posttranslational additions were not present.
These observations suggest that insertional inactivation of prR2 leads to different processing of monomeric proteases from the prpR1 in the mutant strain compared to the wild type and a failure to generate the highly posttranslationally modified RIB. Previous biochemical and immunochemical analyses have demonstrated that RIB stains intensely with the periodic acid-Schiff reagent (22) and contains periodate-sensitive epitopes which are recognized by monoclonal antibodies immunoreactive with the LPS of P. gingivalis (5). Hence, RIB from P. gingivalis W50 appears to be a highly glycosylated protein, which probably explains its migration as a 70- to 80-kDa species on SDS-PAGE. Recent investigations using monoclonal antibodies raised to RIA suggest that this isoform also carries covalently attached carbohydrate residues which in this case are insufficient to influence significantly the molecular weight (unpublished observations). It is noteworthy that these investigations have failed to detect any glycosylation to the α chain of RI. Therefore, it is possible that the different behavior of the monomeric enzymes from the prR2 mutant strain compared to the parent W50 may result from an interruption in the normal process of glycosylation.
In contrast to the situation for RIA and RIB, insertional inactivation of prR2 had no effect on either the total levels or chromatographic behavior of RI from the mutant. Furthermore, the electrophoretic characteristics of the α and β chains of this heterodimer from the prR2 mutant strain were exactly the same as those of their wild-type counterparts on both SDS-PAGE and 8 M urea-PAGE. Analysis of the sequence of the full-length PrpRI precursor suggests that proteolytic processing is required at three Arg-X peptide bonds in order to remove the propeptide and C-terminal γ region and release the α and β chains of RI. The data in the present report indicate that this processing takes place in the absence of prR2 expression, perhaps via an autolytic mechanism.
We have considered a number of possible mechanisms by which insertional inactivation of prR2 could lead to aberrant maturation of RIA and RIB. First, loss of PrRII itself may affect the proteolytic processing of the RIA and RIB precursor(s), and this may have consequences for the availability of sites on the mature protein for posttranslational additions. This seems unlikely, however, since the proteolytic processing pathway which gives rise to RI is unaffected by loss of PrRII. Furthermore, in the case of RIA, aberrant proteolytic processing of the precursor molecule would be expected to lead to a change in the size and/or charge properties of the prR2 mutant form, and neither of these parameters appeared different. Second, PrRII may be required for proteolytic activation of another P. gingivalis cell protein which is essential to the RIA and RIB maturation pathway. Some support for this proposal comes from the studies of Nakayama et al. (15), which suggest that the arginine-specific proteases of P. gingivalis are involved in the normal proteolytic processing of at least two, unrelated, cell surface components of this organism: fimbrillin and a 75-kDa outer membrane protein. Third, insertional inactivation of the prR2 may affect transcription of another gene, the product of which is necessary for RIA and RIB maturation. Although mRNA analysis suggests that the prR2 is not cotranscribed with an upstream or downstream gene on a polycistronic message and hence is unlikely to form part of an operon (21), we cannot exclude the possibility that loss of the PrRII has consequences on transcription at another locus.
While the precise nature of the effect of insertional inactivation of the prR2 on RIA and RIB maturation is uncertain, the phenotypic properties of this mutant do reveal some characteristics of the maturation pathway of prpR1-derived enzymes. First, maturation of RI from the polyprotein precursor occurs independently of the steps in the maturation process which give rise to the differently modified monomeric forms RIA and RIB. Second, although the highly modified RIB isoform is exclusively located in the vesicles in the 6-day supernatant of the parent P. gingivalis W50, the posttranslational additions are not essential for the targeting and retention of monomeric protease in the extracellular membranous compartment. Finally the data suggest that the PrRII plays an important role in the processing of prpR1-derived enzymes, and this functional link between the two genes may contribute to their coconservation in laboratory and clinical isolates of this species (3).
ACKNOWLEDGMENT
This study was supported by the Medical Research Council (PG9318173).
REFERENCES
- 1.Aduse-Opoku J, Muir J, Slaney J M, Rangarajan M, Curtis M A. Characterization, genetic analysis, and expression of a protease antigen (PrpRI) of Porphyromonas gingivalis W50. Infect Immun. 1995;63:4744–4754. doi: 10.1128/iai.63.12.4744-4754.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aduse-Opoku J, Slaney J M, Rangarajan M, Muir J, Young K A, Curtis M A. The Tla protein of Porphyromonas gingivalis W50: a homolog of the RI protease precursor (PrpRI) is an outer membrane receptor required for growth on low levels of hemin. J Bacteriol. 1997;179:4778–4788. doi: 10.1128/jb.179.15.4778-4788.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allaker R P, Aduse-Opoku J, Batten J E, Curtis M A. Natural variation within the principal arginine-specific protease gene, prpR1, of P. gingivalis. Oral Microbiol Immunol. 1997;12:298–302. doi: 10.1111/j.1399-302x.1997.tb00394.x. [DOI] [PubMed] [Google Scholar]
- 4.Curtis M A, Ramakrishnan M, Slaney J M. Characterisation of the trypsin-like enzymes of Porphyromonas gingivalis W83 using a radiolabelled active-site-directed inhibitor. J Gen Microbiol. 1993;139:949–955. doi: 10.1099/00221287-139-5-949. [DOI] [PubMed] [Google Scholar]
- 5.Curtis M A, Aduse-Opoku J, Slaney J M, Rangarajan M, Booth V, Cridland J, Shepherd P. Characterization of an adherence and antigenic determinant of the ArgI protease of Porphyromonas gingivalis which is present on multiple gene products. Infect Immun. 1996;64:2532–2539. doi: 10.1128/iai.64.7.2532-2539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curtis M A. Analysis of the protease and adhesin domains of the PrpRI of Porphyromonas gingivalis. J Periodont Res. 1997;32:133–139. doi: 10.1111/j.1600-0765.1997.tb01394.x. [DOI] [PubMed] [Google Scholar]
- 7.Fletcher H M, Schenkein H A, Morgan R M, Bailey K A, Berry C R, Macrina F L. Virulence of Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect Immun. 1995;63:1521–1528. doi: 10.1128/iai.63.4.1521-1528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imamura T, Pike R N, Potempa J, Travis J. Pathogenesis of periodontitis: a major arginine-specific cysteine proteinase from Porphyromonas gingivalis induces vascular permeability enhancement through activation of the Kallikrein/Kinin pathway. J Clin Invest. 1994;94:361–367. doi: 10.1172/JCI117330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kadowaki T, Yoneda M, Okamoto K, Maeda K, Yamamoto K. Purification and characterization of a novel arginine-specific cysteine proteinase (Argingipain) involved in the pathogenesis of periodontal disease from the culture supernatant of Porphyromonas gingivalis. J Biol Chem. 1994;269:21371–21378. [PubMed] [Google Scholar]
- 10.Kelly C G, Booth V, Kendal H, Slaney J M, Curtis M A, Lehner T. The relationship between colonization and hemagglutination inhibiting and B cell epitopes of Porphyromonas gingivalis. Clin Exp Immunol. 1997;110:285–291. doi: 10.1111/j.1365-2249.1997.tb08329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirszbaum L, Sotiropoulos C, Jackson C, Cleal S, Slakeski N, Reynolds E. Complete nucleotide sequence of a gene prtR of Porphyromonas gingivalis W50 encoding a 132kDa protein that contains an arginine specific thiol endopeptidase domain and a haemagglutinin domain. Biochem Biophys Res Commun. 1995;207:424–431. doi: 10.1006/bbrc.1995.1205. [DOI] [PubMed] [Google Scholar]
- 12.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 13.Marshall R C, Inglis A S. Protein oligomer composition, preparation of monomers and constituent chains. In: Darbre A, editor. Practical protein chemistry. Chichester, England: John Wiley and Sons Ltd; 1987. pp. 1–66. [Google Scholar]
- 14.Nakayama K, Kadowaki T, Okamoto K, Yamamoto K. Construction and characterisation of arginine-specific cysteine proteinase (Arg-gingipain)-deficient mutants of Porphyromonas gingivalis. J Biol Chem. 1995;270:23619–23626. doi: 10.1074/jbc.270.40.23619. [DOI] [PubMed] [Google Scholar]
- 15.Nakayama K, Yoshimura F, Kadowaki T, Yamamoto K. Involvement of arginine-specific cysteine protease (Arg-gingipain) in fimbriation of Porphyromonas gingivalis. J Bacteriol. 1996;178:2818–2824. doi: 10.1128/jb.178.10.2818-2824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nilsson T, Carlsson J, Sundqvist G. Inactivation of key factors of the plasma proteinase cascade systems by Bacteroides gingivalis. Infect Immun. 1985;50:467–471. doi: 10.1128/iai.50.2.467-471.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okamoto K, Misumi Y, Kadowaki T, Yoneda M, Yamamoto K, Ikehara Y. Structural characterisation of argingipain, a novel arginine-specific cysteine protease as a major periodontal pathogenic factor from Porphyromonas gingivalis. Arch Biochem Biophys. 1995;316:917–925. doi: 10.1006/abbi.1995.1123. [DOI] [PubMed] [Google Scholar]
- 18.Pavloff N, Potempa J, Pike R N, Prochazka V, Kiefer M C, Travis J, Barr P J. Molecular cloning and structural characterization of the Arg-gingipain proteinase of Porphyromonas gingivalis. J Biol Chem. 1995;270:1007–1010. doi: 10.1074/jbc.270.3.1007. [DOI] [PubMed] [Google Scholar]
- 19.Potempa J, Pavloff N, Travis J. Porphyromonas gingivalis: a proteinase/gene accounting audit. Trends Microbiol. 1995;3:430–434. doi: 10.1016/s0966-842x(00)88996-9. [DOI] [PubMed] [Google Scholar]
- 20.Potempa J, Pike R, Travis J. The multiple forms of trypsin-like activity present in various strains of Porphyromonas gingivalis are due to the presence of either Arg-gingipain or Lys-gingipain. Infect Immun. 1995;63:1176–1182. doi: 10.1128/iai.63.4.1176-1182.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rangarajan M, Aduse-Opoku J, Slaney J M, Young K A, Curtis M A. The prpR1 and prR2 arginine-specific protease genes of Porphyromonas gingivalis W50 produce five biochemically distinct enzymes. Mol Microbiol. 1997;23:955–965. doi: 10.1046/j.1365-2958.1997.2831647.x. [DOI] [PubMed] [Google Scholar]
- 22.Rangarajan M, Smith S J M, U S, Curtis M A. Biochemical characterisation of the arginine-specific proteases of Porphyromonas gingivalis W50 suggests a common precursor. Biochem J. 1997;323:701–709. doi: 10.1042/bj3230701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 24.Scott C F, Whitaker E J, Hammond B F, Colman R W. Purification and characterization of a potent 70-kDa thiol lysyl-proteinase (Lys-gingivain) from Porphyromonas gingivalis that cleaves kininogens and fibrinogen. J Biol Chem. 1993;268:7935–7942. [PubMed] [Google Scholar]
- 25.Sundqvist G, Carlsson J, Herrmann B, Tarnvik A. Degradation of human immunoglobulins G and M and complement factors C3 and C5 by black-pigmented Bacteroides. J Med Microbiol. 1985;19:85–94. doi: 10.1099/00222615-19-1-85. [DOI] [PubMed] [Google Scholar]
- 26.Wingrove J A, DiScipio R G, Chen Z, Potempa J, Travis J, Hugli T E. Activation of complement components C3 and C5 by a cysteine proteinase (gingipain-1) from Porphyromonas (Bacteroides) gingivalis. J Biol Chem. 1992;267:18902–18907. [PubMed] [Google Scholar]
- 27.Wray W, Boulikas T, Wray V P, Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem. 1981;118:197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]