Abstract
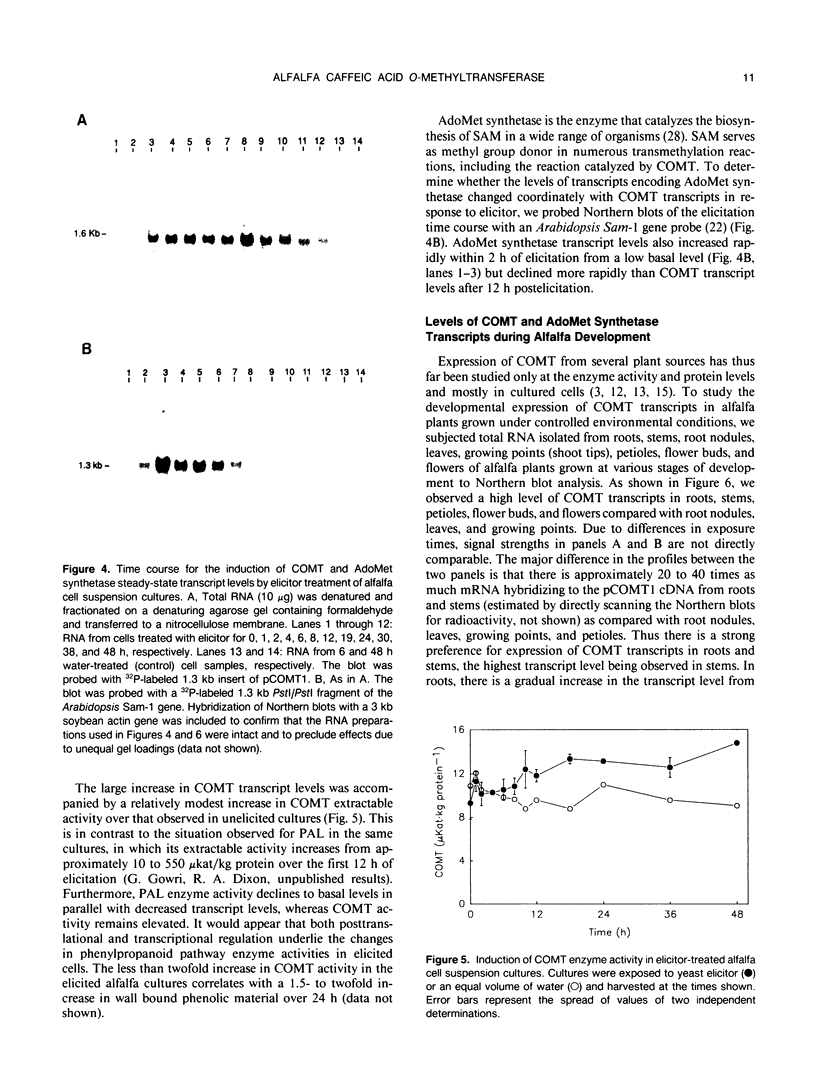
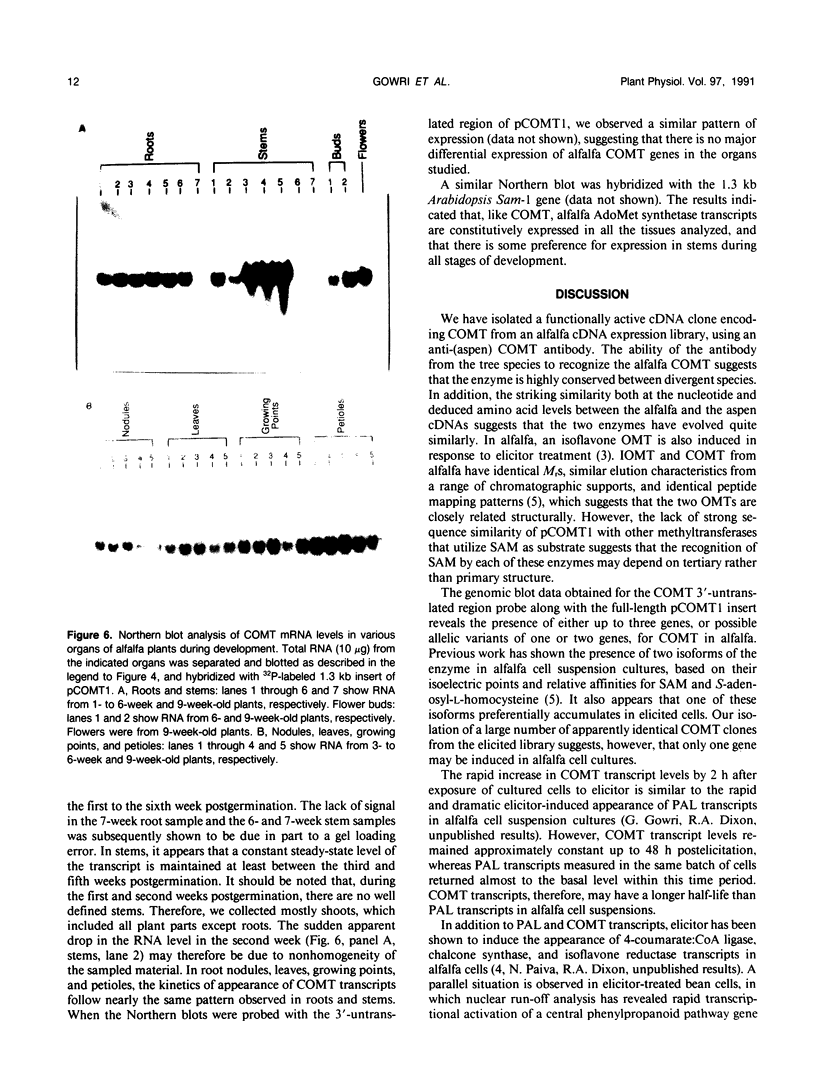
S-Adenosyl-l-methionine:caffeic acid 3-O-methyltransferase (COMT, EC 2.1.1.6) catalyzes the conversion of caffeic acid to ferulic acid, a key step in the biosynthesis of lignin monomers. We have isolated a functionally active cDNA clone (pCOMT1) encoding alfalfa (Medicago sativa L.) COMT by immunoscreening a λZAPII cDNA expression library with anti-(aspen COMT) antibodies. The derived amino acid sequence of pCOMT1 is 86% identical to that of COMT from aspen. Southern blot analysis indicates that COMT in alfalfa is encoded by at least two genes. Addition of an elicitor preparation from bakers' yeast to alfalfa cell suspension cultures resulted in a rapid accumulation of COMT transcripts, which reached a maximum level around 19 hours postelicitation. Northern blot analysis of total RNA from different organs of alfalfa plants at various developmental stages showed that COMT transcripts are most abundant in roots and stems. Transcripts encoding ATP: i-methionine-S-adenosyl transferase (AdoMet synthetase, EC 2.5.1.6), the enzyme responsible for the synthesis of the methyl donor for the COMT reaction, were coinduced with COMT transcripts in elicitor-treated cells and exhibited a similar pattern of expression to that of COMT in different organs of alfalfa plants at various stages of development.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Binns A. N., Chen R. H., Wood H. N., Lynn D. G. Cell division promoting activity of naturally occurring dehydrodiconiferyl glucosides: do cell wall components control cell division? Proc Natl Acad Sci U S A. 1987 Feb;84(4):980–984. doi: 10.1073/pnas.84.4.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalkin K., Edwards R., Edington B., Dixon R. A. Stress Responses in Alfalfa (Medicago sativa L.): I. Induction of Phenylpropanoid Biosynthesis and Hydrolytic Enzymes in Elicitor-Treated Cell Suspension Cultures. Plant Physiol. 1990 Feb;92(2):440–446. doi: 10.1104/pp.92.2.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R., Dixon R. A. Purification and characterization of S-adenosyl-L-methionine: caffeic acid 3-O-methyltransferase from suspension cultures of alfalfa (Medicago sativa L.). Arch Biochem Biophys. 1991 Jun;287(2):372–379. doi: 10.1016/0003-9861(91)90492-2. [DOI] [PubMed] [Google Scholar]
- Egli M. A., Griffith S. M., Miller S. S., Anderson M. P., Vance C. P. Nitrogen Assimilating Enzyme Activities and Enzyme Protein during Development and Senescence of Effective and Plant Gene-Controlled Ineffective Alfalfa Nodules. Plant Physiol. 1989 Nov;91(3):898–904. doi: 10.1104/pp.91.3.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer E. E. Effects of fungal elicitor on lignin biosynthesis in cell suspension cultures of soybean. Plant Physiol. 1985 Jun;78(2):338–342. doi: 10.1104/pp.78.2.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Grand C., Sarni F., Lamb C. J. Rapid induction by fungal elicitor of the synthesis of cinnamyl-alcohol dehydrogenase, a specific enzyme of lignin synthesis. Eur J Biochem. 1987 Nov 16;169(1):73–77. doi: 10.1111/j.1432-1033.1987.tb13582.x. [DOI] [PubMed] [Google Scholar]
- Hermann C., Legrand M., Geoffroy P., Fritig B. Enzymatic synthesis of lignin: purification to homogeneity of the three O-methyltransferases of tobacco and production of specific antibodies. Arch Biochem Biophys. 1987 Mar;253(2):367–376. doi: 10.1016/0003-9861(87)90190-1. [DOI] [PubMed] [Google Scholar]
- Khouri H. E., Tahara S., Ibrahim R. K. Partial purification, characterization, and kinetic analysis of isoflavone 5-O-methyltransferase from yellow lupin roots. Arch Biochem Biophys. 1988 May 1;262(2):592–598. doi: 10.1016/0003-9861(88)90410-9. [DOI] [PubMed] [Google Scholar]
- Lawton M. A., Lamb C. J. Transcriptional activation of plant defense genes by fungal elicitor, wounding, and infection. Mol Cell Biol. 1987 Jan;7(1):335–341. doi: 10.1128/mcb.7.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann J., Schell J., Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal Biochem. 1987 May 15;163(1):16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- Lütcke H. A., Chow K. C., Mickel F. S., Moss K. A., Kern H. F., Scheele G. A. Selection of AUG initiation codons differs in plants and animals. EMBO J. 1987 Jan;6(1):43–48. doi: 10.1002/j.1460-2075.1987.tb04716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleman J., Boerjan W., Engler G., Seurinck J., Botterman J., Alliotte T., Van Montagu M., Inzé D. Strong cellular preference in the expression of a housekeeping gene of Arabidopsis thaliana encoding S-adenosylmethionine synthetase. Plant Cell. 1989 Jan;1(1):81–93. doi: 10.1105/tpc.1.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen M., Lundström K., Tilgmann C., Savolainen R., Kalkkinen N., Ulmanen I. Molecular cloning and characterization of rat liver catechol-O-methyltransferase. Gene. 1990 Sep 14;93(2):241–247. doi: 10.1016/0378-1119(90)90231-f. [DOI] [PubMed] [Google Scholar]
- Tabor C. W., Tabor H. Methionine adenosyltransferase (S-adenosylmethionine synthetase) and S-adenosylmethionine decarboxylase. Adv Enzymol Relat Areas Mol Biol. 1984;56:251–282. doi: 10.1002/9780470123027.ch4. [DOI] [PubMed] [Google Scholar]
- Walter M. H., Grima-Pettenati J., Grand C., Boudet A. M., Lamb C. J. Cinnamyl-alcohol dehydrogenase, a molecular marker specific for lignin synthesis: cDNA cloning and mRNA induction by fungal elicitor. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5546–5550. doi: 10.1073/pnas.85.15.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M. H., Grima-Pettenati J., Grand C., Boudet A. M., Lamb C. J. Extensive sequence similarity of the bean CAD4 (cinnamyl-alcohol dehydrogenase) to a maize malic enzyme. Plant Mol Biol. 1990 Sep;15(3):525–526. doi: 10.1007/BF00019173. [DOI] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Yeast RNA polymerase II genes: isolation with antibody probes. Science. 1983 Nov 18;222(4625):778–782. doi: 10.1126/science.6356359. [DOI] [PubMed] [Google Scholar]