Abstract
Despite recent advances in nucleic acid delivery systems with the success of LNP vehicles, adeno-associated virus (AAV) remains the leading platform for targeted gene delivery due to its low immunogenicity to humans, high transduction efficiency, and range of serotypes with varying tropisms. Depending on the therapeutic goals and serotype used, different production conditions may be more amenable, generating an ever-growing need for rapid yet robust analytical techniques to support the high-quality manufacturing of AAV. A critical bottleneck exists for assessing full capsids where rapid, high-throughput techniques capable of analyzing a range of serotypes are needed. Here, we present a rapid, high-throughput analytical technique, microfluidic electrophoresis, for the assessment of full capsids compatible with AAV1, AAV2, AAV6, AAV8, and AAV9 without the need for assay modifications or optimizations, and AAV5 with some constraints. The method presented in this study uses a mathematical formulation we developed previously with a reference standard to combine the independently obtained capsid protein and single-stranded DNA (ssDNA) profiles to estimate the percentage of full capsids in a sample of unknown concentration. We assessed the ability to use a single serotype (AAV8) as the reference standard regardless of the serotype of the sample being analyzed so long as the melting temperature (Tm) of the capsids is within 12 °C from the Tm of AAV8. Using this method, we are able to characterize samples ±6.1% with an average analytical turnaround time of <5 min/sample, using only 10 μL/sample at a concentration of 2.5 × 1012 VG/mL.
Introduction
Over 30 years after the first gene therapy, there has been a significant increase in the arsenal of therapies and modalities, such as viral (i.e., lentivirus) and nonviral (i.e., lipid nanoparticle), available. Recombinant adeno-associated viruses (rAAV) have become one of the leading platforms for gene therapy. With a diameter of roughly 25 nm1 and a single-stranded DNA (ssDNA) genome of 4.7 kb,2 the characteristics that set them apart are their long-term gene expression,3 lack of pathogenicity in humans,3 and selective tropisms.3−5 Consequently, AAV characterization remains high on the bioanalytical research agenda, and significant resources have been dedicated to developing methods to assess critical quality attributes (CQAs) to meet the standards and regulations of the FDA. Currently, the most common characterization methods are digital droplet PCR (ddPCR) for the genome titer,6−8 enzyme-linked immunosorbent assay (ELISA) for the capsid titer,9−11 and transmission electron microscopy (TEM) for the content ratio in terms of full and empty capsids.6,12,13 Notwithstanding, analytical ultracentrifugation (AUC) remains the golden standard, particularly in industry, for content ratio assessment due to its increased accuracy and ability to quantify partially filled capsids.12,14−16 Some other notable techniques include charge-detection mass spectrometry (CDMS),17 size exclusion chromatography multi-angle light scattering (SEC-MALS),18 anion-exchange chromatography (AEC),19 and mass photometry20 among others.16
While many techniques have been developed and optimized to characterize AAV samples, most require high sample volumes, are low throughput, or have high turnaround times. Therefore, a need remains to support the streamlined analysis of the numerous samples synthesized during the production of AAV and large-scale batch-to-batch analysis. One alternative solution to this need are miniaturized, high-throughput compatible platforms, such as microfluidics. Microfluidics is a particularly attractive technology due to its precise control of fluids, high-throughput capabilities, and rapid sample processing, making it capable of outperforming traditional technologies.21 Additionally, the use of high-resolution miniaturized platforms may lower sample and reagent volumes and costs.22 As a result, depending on their intended use, they can replace or complement many traditional techniques used to characterize AAVs. For instance, this type of platform (micron or submicron) has been used to assess the genomic content,23 capsid proteins,23,24 and content ratio of AAV samples.23,25 However, rather than focusing on the development of a new analytical method, the focus of this study is to increase the fundamental understanding of how different serotypes and sample treatments affect AAV behavior in electrokinetic microfluidic systems and to categorize optimized methods based on serotype stability and overall response.
In a previous study, we successfully determined the percentage of full capsids in AAV8 samples of unknown concentration using an AAV8 standard.23 However, despite the great potential shown by the method, it was limited by (1) using a standard that matched the serotype of the sample, (2) the limited translation to other serotypes due to serotype-specific capsid thermal stability, and (3) the requirement for a time-consuming sample treatment step for nucleic acid extraction. These limitations are important since 10–13 naturally occurring AAV serotypes and over 100 variants have been purified and studied for gene delivery.3,26,27 The wide range of serotypes and variants makes AAV highly versatile, as they may express different tropisms, or tissue targets, based on their binding receptors (Table 1).3,26,27 To further increase their therapeutic robustness, it has been demonstrated that AAV vectors could be equipped with the desired high-affinity ligands, resulting in increased efficiency targeting the desired cells.28 Having such an extensive repertoire of AAV vehicles is of great clinical significance as it has been reported that over 90% of humans have been infected with AAV, approximately 50% of which may have neutralizing antibodies against the virus.3,26,29,30 As such, serotype or variant selection may significantly impact therapy efficiency based on its ability to evade the immune system. Therefore, it is essential to make a versatile analytical platform that enables the purity analysis of different AAV serotypes for maximal applicability, particularly in a rapid, high-throughput, low-volume format that can support manufacturing optimization and downstream purity assessment.
Table 1. Description of the Melting Temperature, Tm, of the Capsids of Each Serotype,31−33 the Organs They Target,34,35 and the Number of Clinical Trials Using That Serotype36,a.
| serotype | capsid Tm (°C)31−33 | organs targeted34,35 | number of clinical trials36 |
|---|---|---|---|
| AAV1 | 82–84.5 | eye, muscle, CNS, heart | 12 |
| AAV2 | 66.33–71.6 | eye, brain, lung, liver, muscle, joint, CNS, kidney | 59 |
| AAV3 | 67.3–71.7 | 0 | |
| AAV4 | 74.0–75.0 | eye, lung, CNS | 0 |
| AAV5 | 88.07–90.5 | eye, lung, liver, CNS | 19 |
| AAV6 | 77.31–78.5 | lung, muscle | 2 |
| AAV7 | 76.5–76.7 | liver, muscle | 0 |
| AAV8 | 70.59–73 | eye, liver, muscle, CNS, heart, pancreas | 30 |
| AAV9 | 76.17–77.0 | lung, liver, muscle, CNS, heart | 40 |
The capsid Tm was determined elsewhere using differential scanning fluorescence (DSF),31−33 and the values are reported here as a range combining full and empty capsid values. The number of clinical trials using each serotype was based on queries using the keywords “adeno-associated virus” and “AAVX”, where “X” represents the serotype number on the clinicaltrials.gov website. The search using “adeno-associated virus” yielded 180 trials, and other studies have reported as many as 331 trials,37 whereas the sum of the clinical trials included below is 162. This discrepancy is due to the lack of specificity in the description or use of different variants in the trials.
Our initial method was developed and validated using AAV8,23 and prior to this study, further assay optimizations were conducted using AAV8 as well; therefore, it was decided to retain AAV8 as the reference standard in this study due to the extent of the prior body of work and the reference standard serotype comparison conducted in this study. Based on the body of scientific literature available and ongoing clinical trials (Table 1) for each serotype, it was determined that AAV2, 8, and 9 were the most commonly used in AAV research. After investigating the differences and similarities across the different serotypes, we found that as long as the capsid melting temperature of the serotypes (Table 1) was within 12 °C of AAV8, they responded similarly to the chemical, biochemical, and physical (heating) treatment used to characterize the samples in this study. This range should include AAV1, 2, 3, 4, 6, 7, and 9 (Table 1). Based on the organs targeted and the number of clinical trials, we validated the method with serotypes 1, 2, 6, 8, and 9, which cover the expected operational temperature range. We also attempted to analyze AAV5 due to its prevalence and number of clinical trials. However, the results were beyond our accepted threshold for prediction accuracy due to the significantly higher capsid Tm, which prevents its proper denaturing and digestion using the current protocol. Notwithstanding, based on its clinical relevance, we evaluated the performance of the assay to predict the percentage of full capsids in AAV5 samples and highlighted the current limitations and a potential solution to overcome them.
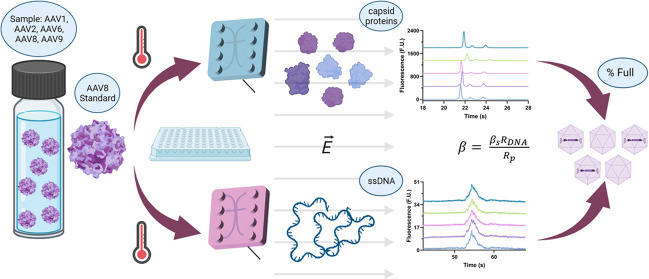
Upon narrowing down the functional melting temperature (Tm) range for the assay, we evaluated how different serotypes (1, 2, 6, 8, and 9) performed as the reference standard for the estimation of full and empty capsids. Despite the ranging degrees of success, there was no statistical difference between their efficiency, with AAV1 and AAV9 showing the highest variability. Ultimately, AAV8 was selected, as its capsid Tm is roughly in the middle of the most commonly used serotypes (AAV2, 8, and 9). By reducing the time required to extract the nucleic acid from the capsid and optimizing the nucleic acid and capsid protein assays, we were able to develop a more robust method that enables the rapid characterization of an unknown AAV sample of serotypes 1, 2, 6, 8, and 9 (Figure 1) using low volumes (10 μL) within our desired threshold, and AAV5 with a larger error. In addition, the insights provided by the study regarding the relationship between sample Tm and treatment response can provide valuable information to expand the method into a platform method including proprietary AAV variants for development laboratories. Being able to assess the purity of AAV samples rapidly serves a 2-fold purpose: upstream, it can help optimize the AAV manufacturing conditions (postpurification), while downstream, it can help characterize the final product.
Figure 1.
Workflow for the microfluidic electrophoresis characterization of AAV1, AAV2, AAV6, AAV8, and AAV9 was calculated by analyzing the capsid and genomic composition separately and integrating the information using a reference standard and a mathematical formulation to estimate the percentage of full capsids in the sample.
Materials and Methods
Materials
AAV (1, 2, 5, 6, 8, and 9) reference standards were purchased/obtained from Revvity (Waltham, MA) at a concentration of 1 × 1013 GC/mL and were diluted using PBS + 0.001% Pluronic to 2.5 × 1012 GC/mL. Different serotypes either came or were diluted with empty capsids to achieve different percentages of full, which will be specified throughout the study. The LabChip GXII Touch platform, custom AAV protein microchip and chip reagents, and custom AAV ssDNA microchip and chip reagents were obtained from Revvity. The UV/vis percentage of full capsids was estimated by Revvity and provided as a single value, which is why there are no error bars associated with those values.
Methods
The AAV characterization experiments discussed in this study were conducted on the analytical platform LabChip GXII Touch (Revvity) for optics, robotic motion, pressure, and electrokinetic control. Two separate assays were developed or optimized for the purity assessment of the AAV samples: a capsid protein and an ssDNA assay. The samples were treated, heated on a 96- or 384-well plate for both assays, and transferred onto the LabChip platform. For each assay, an assay-specific (AAV protein or AAV ssDNA) microfluidic chip was loaded with a gel or a gel-dye and a marker for calibration purposes. The chip was placed on a LabChip platform. Once the well plate and chip are loaded onto the platform, the system moves the desired sample well under a metal sipper attached to the microfluidic chip. By vacuum pressure, 20 nL of each sample was loaded onto the chip individually for analysis. Additional information can be found under Methods for Capsid Protein and ssDNA Characterization in the Supporting Information. The statistical analysis for this study was conducted using GraphPad Prism 9, and the figures were made using GraphPad and/or BioRender.com.
Theory
In a previous study, we proposed a mathematical formulation that enabled us to estimate the percentage of full capsids in a sample of unknown concentration and percentage of full capsids using a microfluidic electrophoresis method that resulted from the combination of a protein and ssDNA assay that we developed for AAV purity assessment.23 To achieve this, a key innovation was integrating a reference standard of the known percentage of full capsids and concentration into the workflow to normalize the raw fluorescent data obtained from the electropherograms. For this, we described the total number of particles in a sample, N, and standard, Ns, in terms of the full, f, and empty, e, particles present in the sample23
| 1 |
| 2 |
Full and empty capsids are composed of the same protein subunits (VP1–3); therefore, regardless of the genetic content, each capsid should contain the same amount of protein α. Therefore, the relation between the concentration, c, of proteins in the samples and the standard can be represented by23
| 3 |
while that of the ssDNA insert present in the sample and standard can be expressed as23
| 4 |
We then defined the known percentage of full capsids in the standard, βs, as23
| 5 |
which can be used to estimate the percentage of full capsids in the sample through the relation23
| 6 |
By independently analyzing both the protein and ssDNA profiles of the unknown AAV sample(s) in tandem with a reference standard, we can mitigate the error introduced by the use of two independent assays by helping normalize the concentrations yielded by each assay to the standard, allowing us to combine the assays. Using this approach, we were able to estimate the percentage of full capsids in an AAV8 sample using an AAV8 standard ±4%.23 Since the basis for this mathematical formulation is independent of serotype and showed a high degree of accuracy in the past, it will also be used to estimate the percentage of full capsids in this study, but the optimal standard serotype will be explored.
Results and Discussion
Protein Analysis Optimization
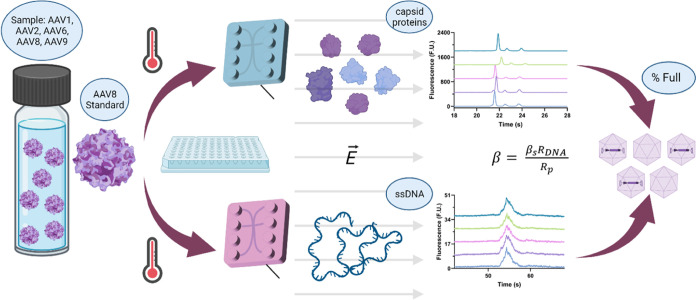
In our mathematical formulation, the protein signal (corrected area) is used to determine Rp. This parameter can be obtained using both protein assays described in the methods: the Protein Express and AAV Pico Protein assays. While there are some differences in the sample treatment and chip loading between these two assays, the critical difference that we believe is responsible for the difference in response of the two is the use of a noncovalent dye (Revvity) in the Protein Express assay and a covalent dye (Revvity) in the AAV Pico Protein assay. Prior to this study, we optimized both assays (not shown in this study) by using an AAV8 sample. Using these protocols, we analyzed AAV1, 2, 6, 8, and 9 samples using the noncovalent (Protein Express) and the covalent (AAV Pico Protein) assays (Figure 2).
Figure 2.
Electrophoretic characterization of the capsid proteins of AAV1, 2, 6, 8, and 9. Electropherograms of the different serotypes using (a) the noncovalent dye with an offset of 130 F.U. and (b) the covalent dye with an offset of 450 F.U. The peak areas for all three capsid proteins are highlighted (c) by serotype and (d) by dye type. Comparing the covalent dye response to the noncovalent, we see a 6-fold increase in peak area. Note that all samples were normalized to 2.5 × 1012 VG/mL at different percentages of full and are expected to have different capsid concentrations. However, while the concentrations vary across serotypes, they were used at the same concentration in both assays, enabling a direct comparison between the two. Predicted VP ratios using (e) the noncovalent and (f) the covalent assay. The VP ratios were estimated by dividing each peak area by the protein MW (Table S1) and normalizing the values.
When the electropherograms yielded by each assay (Figure 2) are compared, we observe over a 6-fold increase in peak areas using the covalent dye from the noncovalent dye, which leads to a greater definition of the viral proteins (VP), in particular of VP1 and VP2. Based on these results, the limit of detection (LOD), as defined by the minimum concentration at which we can detect all observable peaks for the capsid proteins, is 9.9 × 1011 VP/mL for the noncovalent dye and 6.8 × 1010 VP/mL for the covalent dye, a 14.5-fold difference. Despite not assessing the molecular interaction of the dyes directly, we believe that this difference in response stems from the mechanism through which each dye interacts with the capsid proteins. Although the noncovalent dye will bind to the proteins through some unspecific charge attraction, the covalent dye has a more specific target with higher affinity, leading to an increase in binding sites and resulting overall labeling efficiency. Interestingly, although the peak areas vary due to differences in sample concentration, we see the same behavior in both assays where AAV2 and AAV9 appear to have the highest capsid concentration and AAV8 the lowest, suggesting that the two assays are in agreement. This response also suggests that while the labeling efficiency may change, the difference in affinity between the two dyes is unlikely to be serotype-biased.
While the primary motivation behind the analysis of the capsids is to determine Rp, the analysis of the denatured capsids also provides great insight into the VP ratios (Figure 2e,f). While the exact VP ratio may vary, VP stoichiometry is expected to be 10:1:1 for VP3/VP2/VP1.39 However, Snijder et al. demonstrated that depending on the concentration at which the VPs are present, capsid symmetry will not be followed, and the assembly will be stochastic.40 In addition, VP1 contains a domain essential for endosomal escape and, consequently, virus infection.41,42 Therefore, a decreased ratio of VP1 to total capsid proteins can reduce transduction efficiency.43 As such, VP ratios are a critical-to-quality metric, making the ability to monitor them of great therapeutic relevance since changes to AAV capsid protein stoichiometry, in particular VP1 amount, can lead to a loss of potency.41
In this study, the ratios were estimated to be 10.00:0.80:0.68 and 10.00:1.72:1.51 for the noncovalent and covalent assays, respectively (Figure 2e,f). These values were estimated by normalizing the area of each peak, which is relative to the concentration of each capsid protein and to its molecular weight, to enable a comparison of the relative protein molecules in the sample. While there is a difference between the two because of the stochastic nature of the ratios, without an orthogonal method to assess the actual VP ratios of the sample, it is difficult to determine which provides the most accurate ratios. However, because of the repeatability of the results, the estimated ratios have great value for batch-to-batch reproducibility. In addition, a comparison of the peak profiles associated with each VP suggests that the covalent method shows a greater ability to observe changes in peak area while producing the most consistent and narrow peaks (Figure S1). Another VP property that can be monitored using this method is the size. Using the internal LabChip system protein ladders, we were able to estimate the size of the different capsid proteins, with an average % CV of 1.1 and 0.4% for the noncovalent and covalent methods, respectively (Table S1). Interestingly, while the two assays are in close agreement, both seem to overpredict the size of the proteins when compared to the expected sizes of VP3 (59–61 kDa), VP2 (64–67 kDa), and VP1 (79–82 kDa).38 Due to the magnitude of the difference, possible explanations for this difference are probably a combination of post-translational modifications and protein conformations, which may affect their ability to migrate through the gel matrix of the microfluidic chip.2 Notwithstanding, despite the differences in expected and observed VP size, both assays yielded highly repeatable sizing values, with the covalent assay outperforming the noncovalent assay based on a slight difference in coefficient of variation.
Based on the findings presented in this section, the covalent method appears to yield the highest sensitivity, which is essential for detecting subtle yet important changes across protein profiles, and the lowest coefficient of variation, which plays a crucial role in batch-to-batch comparisons. The ability to consistently detect subtle changes is also highly relevant for production optimization, including cell line selection and growth condition optimization. Therefore, the remainder of the study will discuss only the results obtained with the covalent assay.
ssDNA Analysis Optimization
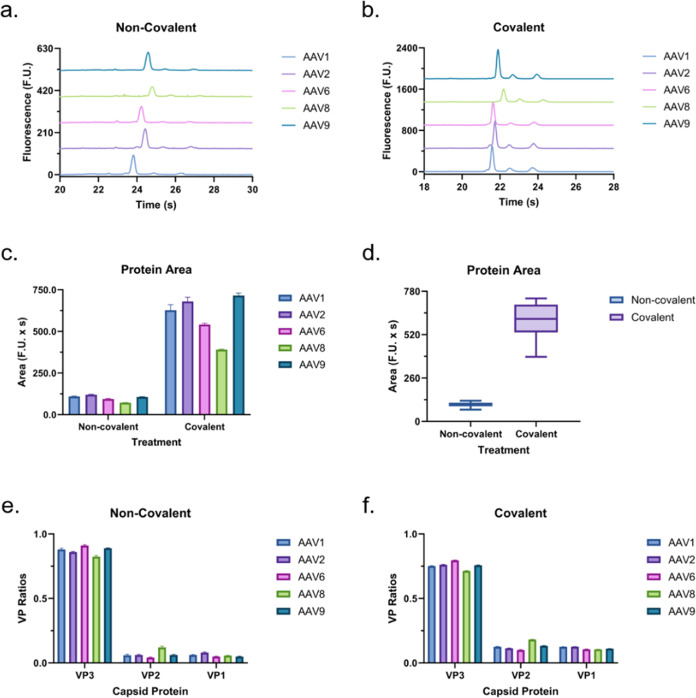
In our previous study, we used proteinase K to digest the capsid for ssDNA release prior to analysis.23 Despite effectively releasing the genomic content of the AAV capsids in a reproducible manner, the treatment requires ∼1.5 h of treatment, which takes a significant toll on the turnaround of the method. Therefore, we optimized an assay that requires only a short (∼10 min) heat treatment without the use of digesting enzymes. In Figure 3, we compare the performance of the two using AAV1, 2, 6, 8, and 9 at a concentration of 2.5 × 1012 VG/mL. Since the genomic concentrations are normalized across all serotypes, we expect all samples to yield equal peak areas here. When we compare the two sample treatments, the heat treatment samples produce slightly higher and sharper peaks, which may be easier to detect at low concentrations. However, the average areas were almost identical. Since the areas produced by each approach are nearly identical in magnitude, so is their LOD of 1.6 × 1011 GC/mL. In contrast, the areas produced by the proteinase K treatment have less variation across the different serotypes. Both sample treatments yielded nearly identical migration times for all serotypes. Overall, the higher variation in peak area when using the heat treatment was outweighed by the benefits of the peak shape, sensitivity, and reduced incubation time. Therefore, the results in the remainder of the study will include only heat-extracted ssDNA data. In addition, while not exploited here, a key benefit of the proposed ssDNA analysis method is that the electrophoretic separation of the genomic content within the microfluidic chip enables genomic-fragment-based analysis of the contents of the capsid so long as the concentrations are within our limit of detection.
Figure 3.
AAV ssDNA electropherogram profiles post-treatment with (a) proteinase K (proK, electropherograms offset by 8 F.U.) and (b) heat (electropherograms offset by 10 F.U.). Comparison of peak (c) area, (d) migration time, and (e) fwhm using each treatment separated by serotype. Average peak (f) area, (g) migration time, and (h) fwhm for each treatment.
Determining Optimal Standard and Assay
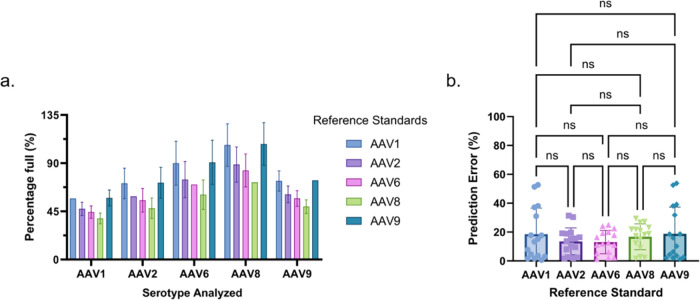
Once we determined the optimal capsid protein assay and ssDNA extraction method, we used eq 6 to assess the performance of five different AAV serotypes (1, 2, 6, 8, and 9) as the reference standard used to estimate the percentage of full capsids in AAV samples of different serotypes. For this, we analyzed the different serotypes simultaneously and used the protein (Figure 2) and ssDNA (Figure 3) data of each to estimate the percentage of full capsids in the other serotypes (Figure 4a). Using the expected percentage of full capsids in each sample, we calculated the average prediction error for each serotype as a standard (Figure 4b). While there was no statistically significant difference across the performance of each serotype based on their prediction deviation, AAV2, AAV6, and AAV8 yielded a narrower distribution. Interestingly, the range of capsid Tm covered by the serotypes used in this section spans from 66.33 to 84.5 °C, whereas AAV2, 6, and 8 have capsid Tm values of 66.33 to 71.6 °C, 77.31 to 78.5 °C, and 70.59 to 73 °C, respectively (Table 1). In contrast, AAV1 and AAV9 have capsid Tm values of 82–84.5 and 76.17–77.0 °C, respectively. The results over this temperature range suggest that while temperature appears to play a critical role in the ability for a serotype to be used as a cross-serotype reference standard, the difference in prediction error between AAV6 and AAV9 despite their similar capsid Tm indicates other biochemical features may play a role. Notwithstanding, given its low prediction error, relevant Tm (close to the middle of the range evaluated), and clinical significance, we decided to use AAV8 as the reference standard.
Figure 4.
Comparison of the predictive performance of each serotype as a reference standard to estimate the percentage of full capsids in the other samples of different serotypes using eq 6. (a) Predictions separated by serotype being analyzed, where each color represents the serotype used as the reference standard for those estimations. Note that serotypes were not used to predict themselves, and the expected value was used instead (i.e., the percentage of full capsids in the sample using an AAV1 reference standard is the expected percentage of full capsids in the sample). (b) Summarized prediction error of each serotype as a reference standard.
Application: Assay Performance
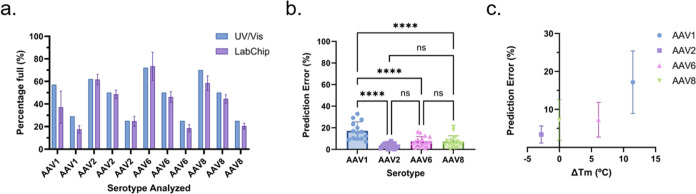
To test the performance of the proposed method, we decided to look at a range of serotypes that covered the operational capsid Tm range of the assay and with varying percentages of full capsids. To this end, we analyzed two or three samples of varying percentages of full capsids of AAV1, 2, 6, and 8, covering a range from 66.33 to 84.5 °C (Table 1). Based on the earlier findings presented in this study, the final assay uses AAV8 as the reference standard, the covalent dye for protein analysis, the heat treatment for the ssDNA analysis, and the mathematical formulation to make the predictions. In addition, to prevent bias when analyzing AAV8 due to the reference standard selection, independently generated AAV8 preparations were used as the reference standard and as the sample being analyzed. Following the proposed method, the average prediction error was ±6.1% with a CV of 15.3% (Figure 5a, Table S2), which is within our ≤20% cutoff value.
Figure 5.
(a) Comparison of the proposed method (LabChip) to UV/vis (Stunner). Comparison of prediction errors relative to each (b) serotype (AAV1, 2, 6, and 8) and (c) ΔTm to AAV8 reference standard. For the ΔTm estimations, an average of the ranges provided in Table 1 was used for each serotype. No error bars are present in the UV/vis data, as this was provided as an average by the provider.
While the overall prediction error and CV were within the desired ranges, when we break the errors down by serotype (Figure 5b), we see that the AAV1 predictions underperform compared with the other three serotypes tested in this section. We see an interesting trend when we take it a step further and look at the capsid ΔTm between the reference standard and the samples. When the ΔTm increases and is positive, the prediction error increases significantly (Figure 5c). On the other hand, while the data set is too limited in that direction to draw a meaningful conclusion, it appears that when the ΔTm is negative, the prediction accuracy is less affected by the difference in ΔTm. We also performed an additional comparison between the LabChip and UV/vis predictions (Figure S2), where most predictions fell within ±10% of each other with the exception of the AAV1 estimations. These findings further support that Tm, which should not affect the UV/vis-based estimation, may be responsible for the larger errors associated with some serotypes using the LabChip technique.
Application: AAV5
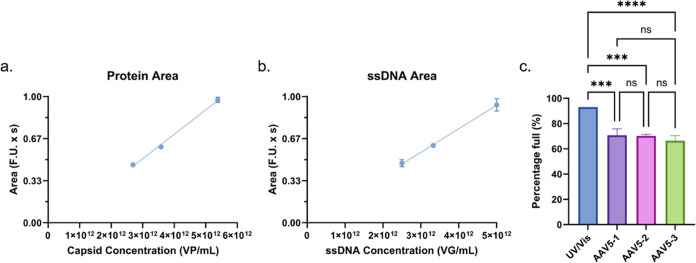
Lastly, while acknowledging that AAV5 (ΔTm = −17.5 °C) is outside the optimal operational range (ΔTm = ± 12 °C) of the proposed assay, given its clinical relevance, we decided to assess the performance and limitations of this method to analyze AAV5. As expected, when the protein (Figure 6a) and ssDNA (Figure 6b) of three AAV5 dilutions were analyzed, they both showed a strong linear relation between the peak area and concentration. Moreover, when the protein and ssDNA data for these samples was used to predict the percentage of full capsids in the samples, as highlighted in Figure 6c, the three samples yielded highly consistent results of 71.0 ± 5.4, 70.3 ± 1.2, and 66.4 ± 3.8% for AAV5–1, AAV5–2, and AAV5–3, respectively. When the predictions for the three samples are combined, the average prediction is 69.1 ± 4.1%. Still, despite the reproducible results, when the predictions are compared to the UV/vis estimation, which was 93%, there is a difference of 23.9%. However, since the results showed to be repeatable and there is a strong linear relation between sample concentration and peak area for AAV5, upon further analysis, a case could be made for using a correction factor in order to increase the accuracy in the estimation of full capsids in AAV5 samples.
Figure 6.
(a) Protein and (b) ssDNA peak area over concentration for three dilutions of AAV5 at 93% full per UV/vis measurement, where the protein response has an R2 of 0.99 and the ssDNA response has an R2 of 0.98 (c) Predicted percentage of full capsids in the three AAV5 dilutions at 93% full (AAV5–1 is 1:4, AAV5–2 is 1:3, and AAV5–3 is 1:4 of an initial stock of 1 × 1013 VG/mL) using an AAV8 standard. No error bars are present in the UV/vis data as this was provided as an average by the provider.
Conclusions
In this study, we improved the sensitivity of the protein analysis 12.5-fold to an LOD of 3.4 × 1011 VP/mL by using a covalent-fluorescent dye labeling strategy. Mechanistically, the noncovalent method should be more general and provided VP ratios closer to the expected 10:1:1 ratio but had a larger LOD when compared to the covalent method. The AAV Pico Protein assay yielded a more sequence-dependent ratio but has higher sensitivity and similar repeatability to the noncovalent method. In our view, the covalent method may be developed along with specifications to monitor AAV capsid production. In contrast, the sensitivity of the ssDNA assay did not change with an LOD of 1.6 × 1011 GC/mL. The sample preparation time decreased 8-fold to 10 min, bypassing the need for enzymes. Once the methods to collect the protein and ssDNA information were determined, different serotypes were evaluated for their potential as reference standards. Ultimately, based on the capsid Tm, percentage full prediction error, and clinical relevance, AAV8 was selected as the default reference standard.
The performance of the methods integrated through the reference standard and mathematical formulation was then tested against samples of AAV1, 2, 6, and 8 at different percentages of full capsids, which yielded an average error of ±6.1% with a CV of 15.3% within a ΔTm range of ±12 °C. The analysis of AAV5 was also attempted, but the difference in capsid Tm led to an average prediction error of ±23.9%, which was above our desired threshold of ±20%. Interestingly, the only instance where the % CV, but not the average error, was above the desired threshold of 20% was with AAV1. Given that AAV1 has the greatest capsid ΔTm relative to AAV8, we explored the relationship between the capsid ΔTm and prediction error. Not surprisingly, the greater the ΔTm, the greater the error. While we have limited data in the negative direction, the data also appear to indicate that negative ΔTm may be less impacted by differences in capsid Tm. However, based on the difference in the prediction accuracy of AAV6 and AAV9, which have very similar capsid Tm, additional serotype-specific properties may also play a role.
Overall, this improved method enables the rapid characterization (<5 min/sample) of the percentage of full capsids in a wide range of serotypes. While traditionally the combination of two independent methods to determine the genome-to-capsid ratio may lead to a compounded error, the mathematical formulation we developed uses the reference standard to combine the assays in a way that mitigates this error. In contrast, analyzing the protein and ssDNA content separately enables us to look closer at VP ratio and ssDNA purity and integrity while only requiring 10 μL of sample.
The cross-serotype functionality and ability to address multiple critical quality attributes (CQAs) simultaneously are of great relevance, as there are at least 331 clinical trials that include AAV, and a great analytical bottleneck in their development is the lack of reliable, high-throughput characterization methods. An important example of this bottleneck is the lack of robust high-throughput analytical tools for assessing the percentage of full capsids in an AAV sample, with AUC, a nonscalable technique, as the current gold standard. Lastly, having an integrated platform that enables the assessment of numerous CQAs, such as total protein concentration, VP ratio, ssDNA concentration and integrity, and percentage of full capsids, can decrease the method transfer burden from analytical development to quality assurance and quality control.
Acknowledgments
This work was funded by Revvity. A.T. is a paid scientific advisor/consultant and lecturer from Revvity. The authors thank Sirion Biotech (Revvity) for the samples and the UV/vis data.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c09006.
Methods for capsid protein and ssDNA characterization; comparison of VP properties across the two methods (Figure S1); sizing accuracy of VPs using the noncovalent and covalent methods (Table S1); percentage full prediction accuracy of AAV8 reference standard (Table S2); and comparison of UV/vis (Stunner) and LabChip Percentage Full Predictions (Figure S2) (PDF)
Author Contributions
§ These authors contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Grieger J. C.; Samulski R. J. Adeno-Associated Virus Vectorology, Manufacturing, and Clinical Applications. Methods Enzymol. 2012, 507, 229–254. 10.1016/B978-0-12-386509-0.00012-0. [DOI] [PubMed] [Google Scholar]
- Srivastava A.; Lusby E. W.; Berns K. I. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J. Virol. 1983, 45, 555–564. 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.; Samulski R. J. Engineering adeno-associated virus vectors for gene therapy. Nat. Rev. Genet. 2020, 21, 255–272. 10.1038/s41576-019-0205-4. [DOI] [PubMed] [Google Scholar]
- Srivastava A. In vivo tissue-tropism of adeno-associated viral vectors. Curr. Opin. Virol. 2016, 21, 75–80. 10.1016/j.coviro.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zincarelli C.; Soltys S.; Rengo G.; Rabinowitz J. E. Analysis of AAV Serotypes 1–9 Mediated Gene Expression and Tropism in Mice After Systemic Injection. Mol. Ther. 2008, 16, 1073–1080. 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- Dobnik D.; Kogovšek P.; Jakomin T.; et al. Accurate Quantification and Characterization of Adeno-Associated Viral Vectors. Front. Microbiol. 2019, 10, 1570. 10.3389/fmicb.2019.01570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock M.; Alvira M. R.; Chen S.-J.; Wilson J. M. Absolute Determination of Single-Stranded and Self-Complementary Adeno-Associated Viral Vector Genome Titers by Droplet Digital PCR. Hum. Gene Ther. Methods 2014, 25, 115–125. 10.1089/hgtb.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta-Hanawa B.; Yamaguchi T.; Uchida E. Two-Dimensional Droplet Digital PCR as a Tool for Titration and Integrity Evaluation of Recombinant Adeno-Associated Viral Vectors. Hum. Gene Ther. Methods 2019, 30, 127–136. 10.1089/hgtb.2019.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuck D.; Kern A.; Kleinschmidt J. A. Development of AAV serotype-specific ELISAs using novel monoclonal antibodies. J. Virol. Methods 2007, 140, 17–24. 10.1016/j.jviromet.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Ayuso E.; Blouin V.; Lock M.; et al. Manufacturing and Characterization of a Recombinant Adeno-Associated Virus Type 8 Reference Standard Material. Hum. Gene Ther. 2014, 25, 977–987. 10.1089/hum.2014.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock M.; McGorray S.; Auricchio A.; et al. Characterization of a Recombinant Adeno-Associated Virus Type 2 Reference Standard Material. Hum. Gene Ther. 2010, 21, 1273–1285. 10.1089/hum.2009.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X.; Chen W. C.; Argento C.; et al. Analytical Strategies for Quantification of Adeno-Associated Virus Empty Capsids to Support Process Development. Hum. Gene Ther. Methods 2019, 30, 144–152. 10.1089/hgtb.2019.088. [DOI] [PubMed] [Google Scholar]
- Kohlbrenner E.; Weber T. Production and Characterization of Vectors Based on the Cardiotropic AAV Serotype 9. Methods Mol. Biol. 2017, 1521, 91–107. 10.1007/978-1-4939-6588-5_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham B.; Nass S.; Kong E.; et al. Analytical Ultracentrifugation as an Approach to Characterize Recombinant Adeno-Associated Viral Vectors. Hum. Gene Ther. Methods 2015, 26, 228–242. 10.1089/hgtb.2015.048. [DOI] [PubMed] [Google Scholar]
- Maruno T.; Usami K.; Ishii K.; Torisu T.; Uchiyama S. Comprehensive Size Distribution and Composition Analysis of Adeno-Associated Virus Vector by Multiwavelength Sedimentation Velocity Analytical Ultracentrifugation. J. Pharm. Sci. 2021, 110, 3375–3384. 10.1016/j.xphs.2021.06.031. [DOI] [PubMed] [Google Scholar]
- Gimpel A. L.; Katsikis G.; Sha S.; et al. Analytical methods for process and product characterization of recombinant adeno-associated virus-based gene therapies. Mol. Ther. Methods Clin. Dev. 2021, 20, 740–754. 10.1016/j.omtm.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson E. E.; Keifer; David Z.; Asokan A.; Jarrold M. F. Resolving Adeno-Associated Viral Particle Diversity With Charge Detection Mass Spectrometry. Anal. Chem. 2016, 88, 6718–6725. 10.1021/acs.analchem.6b00883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh N. L.; Berguig G. Y.; Karim O. A.; et al. Comprehensive characterization and quantification of adeno associated vectors by size exclusion chromatography and multi angle light scattering. Sci. Rep. 2021, 11, 3012 10.1038/s41598-021-82599-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.; et al. Developing an Anion Exchange Chromatography Assay for Determining Empty and Full Capsid Contents in AAV6.2. Mol. Ther.--Methods Clin. Dev. 2019, 15, 257–263. 10.1016/j.omtm.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D.; Hwang P.; Li T.; Piszczek G. Rapid characterization of adeno-associated virus (AAV) gene therapy vectors by mass photometry. Gene Therapy 2022, 29, 691–697. 10.1038/s41434-021-00311-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackmann E. K.; Fulton A. L.; Beebe D. J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189. 10.1038/nature13118. [DOI] [PubMed] [Google Scholar]
- Whitesides G. M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- Coll De Peña A.; Masto L.; Atwood J.; Tripathi A. Electrophoresis-Mediated Characterization of Full and Empty Adeno-Associated Virus Capsids. ACS Omega 2022, 7, 23457–23466. 10.1021/acsomega.2c01813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Wang Y.; Sosic Z.; et al. Identification of adeno-associated virus capsid proteins using ZipChip CE/MS. Anal. Biochem. 2018, 555, 22–25. 10.1016/j.ab.2018.06.006. [DOI] [PubMed] [Google Scholar]
- Katsikis G.; Hwang I. E.; Wang W.; et al. Weighing the DNA Content of Adeno-Associated Virus Vectors with Zeptogram Precision Using Nanomechanical Resonators. Nano Lett. 2022, 22, 1511–1517. 10.1021/acs.nanolett.1c04092. [DOI] [PubMed] [Google Scholar]
- Kotterman M. A.; Schaffer D. V. Engineering adeno-associated viruses for clinical gene therapy. Nat. Rev. Genet. 2014, 15, 445–451. 10.1038/nrg3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa S. S.; Shaimardanova A. A.; Solovyeva V. V.; Rizvanov A. A. Various AAV Serotypes and Their Applications in Gene Therapy: An Overview. Cells 2023, 12, 785. 10.3390/cells12050785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch R. C.; Janicki H.; Völker I.; et al. Displaying High-affinity Ligands on Adeno-associated Viral Vectors Enables Tumor Cell-specific and Safe Gene Transfer. Mol. Ther. 2013, 21, 109–118. 10.1038/mt.2012.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis Jeune V.; Joergensen J. A.; Hajjar R. J.; Weber T. Pre-existing Anti–Adeno-Associated Virus Antibodies as a Challenge in AAV Gene Therapy. Hum. Gene Ther. Methods 2013, 24, 59–67. 10.1089/hgtb.2012.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme C.; Adjali O.; Mingozzi F. Unraveling the Complex Story of Immune Responses to AAV Vectors Trial After Trial. Hum. Gene Ther. 2017, 28, 1061–1074. 10.1089/hum.2017.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett A.; Patel S.; Mietzsch M.; et al. Thermal Stability as a Determinant of AAV Serotype Identity. Mol. Ther. Methods Clin. Dev. 2017, 6, 171–182. 10.1016/j.omtm.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacouret S.; Bouzelha M.; Shelke R.; et al. AAV-ID: A Rapid and Robust Assay for Batch-to-Batch Consistency Evaluation of AAV Preparations. Mol. Ther. 2017, 25, 1375–1386. 10.1016/j.ymthe.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayaprolu V.; Kruse S.; Kant R.; et al. Comparative Analysis of Adeno-Associated Virus Capsid Stability and Dynamics. J. Virol. 2013, 87, 13150–13160. 10.1128/JVI.01415-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A. In vivo tissue-tropism of adeno-associated viral vectors. Curr. Opin. Virol. 2016, 21, 75. 10.1016/j.coviro.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addgene: Adeno-associated virus (AAV) Guide. https://www.addgene.org/guides/aav/.
- ClinicalTrials.gov. https://clinicaltrials.gov/.
- Srivastava A. Rationale and strategies for the development of safe and effective optimized AAV vectors for human gene therapy. Mol. Ther. Nucleic Acids 2023, 32, 949–959. 10.1016/j.omtn.2023.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wörner T. P.; et al. Adeno-associated virus capsid assembly is divergent and stochastic. Nat. Commun. 2021, 12, 1642 10.1101/2020.10.09.332619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson F. B.; Ozer H. L.; Hoggan M. D. Structural Proteins of Adenovirus-Associated Virus Type 3. J. Virol. 1971, 8, 860. 10.1128/jvi.8.6.860-863.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder J.; van de Waterbeemd M.; Damoc E.; et al. Defining the Stoichiometry and Cargo Load of Viral and Bacterial Nanoparticles by Orbitrap Mass Spectrometry. J. Am. Chem. Soc. 2014, 136, 7295. 10.1021/ja502616y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girod A.; Wobus C. E.; Zádori Z.; et al. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J. Gen. Virol. 2002, 83, 973–978. 10.1099/0022-1317-83-5-973. [DOI] [PubMed] [Google Scholar]
- Venkatakrishnan B.; Yarbrough J.; Domsic J.; et al. Structure and dynamics of adeno-associated virus serotype 1 VP1-unique N-terminal domain and its role in capsid trafficking. J. Virol. 2013, 87, 4974–4984. 10.1128/JVI.02524-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama H.; Ishii K.; Maruno T.; Torisu T.; Uchiyama S. Characterization of Adeno-Associated Virus Capsid Proteins with Two Types of VP3-Related Components by Capillary Gel Electrophoresis and Mass Spectrometry. Hum. Gene Ther. 2021, 32, 1403–1416. 10.1089/hum.2021.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.