Abstract
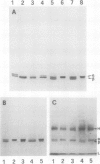
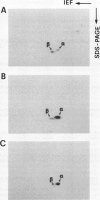
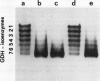
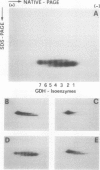
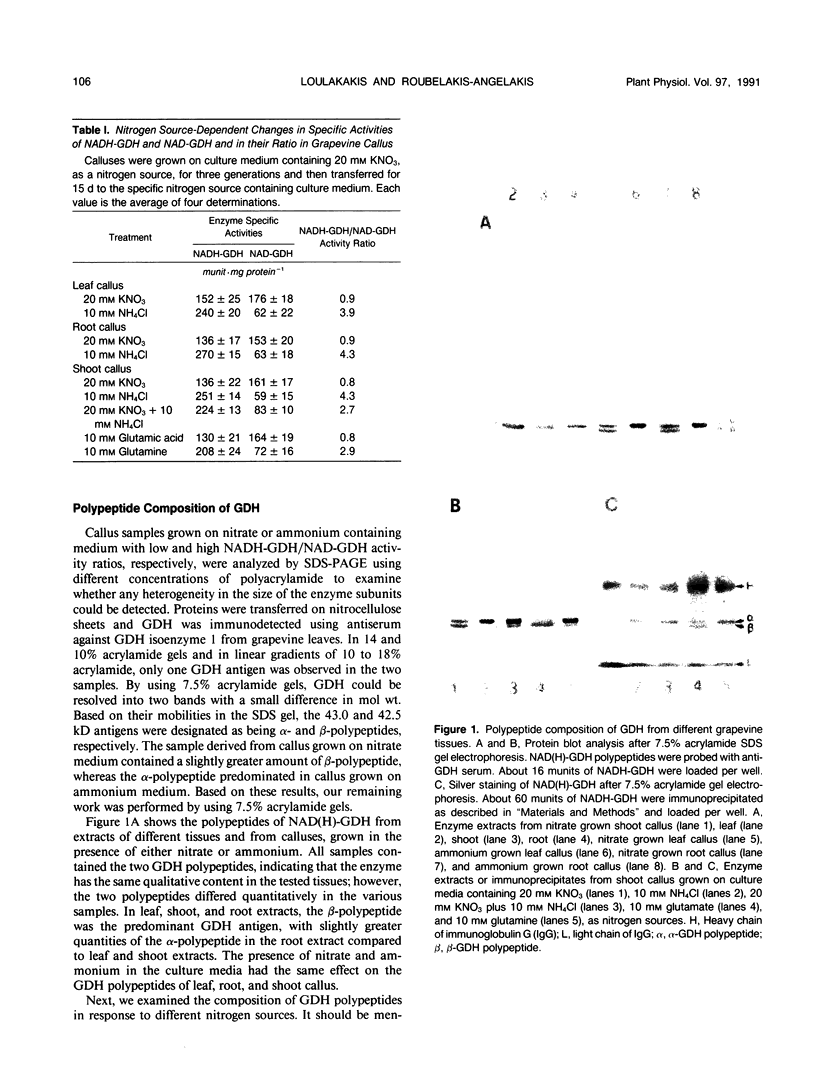
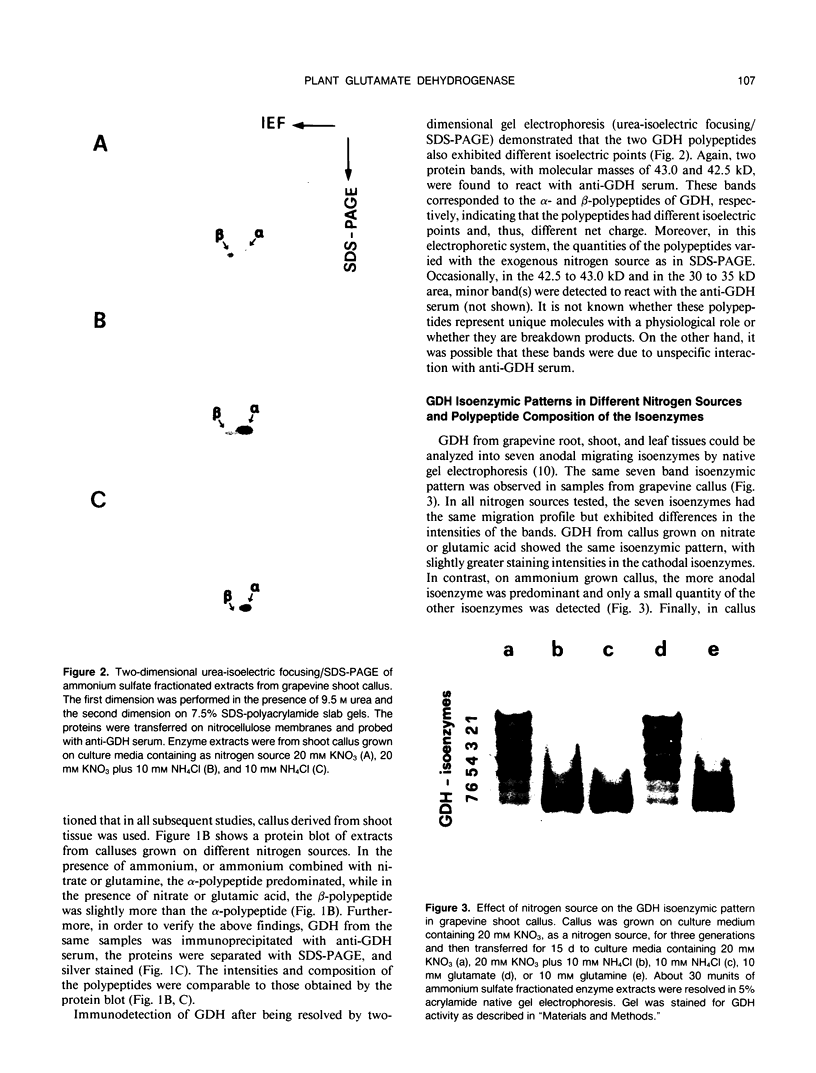
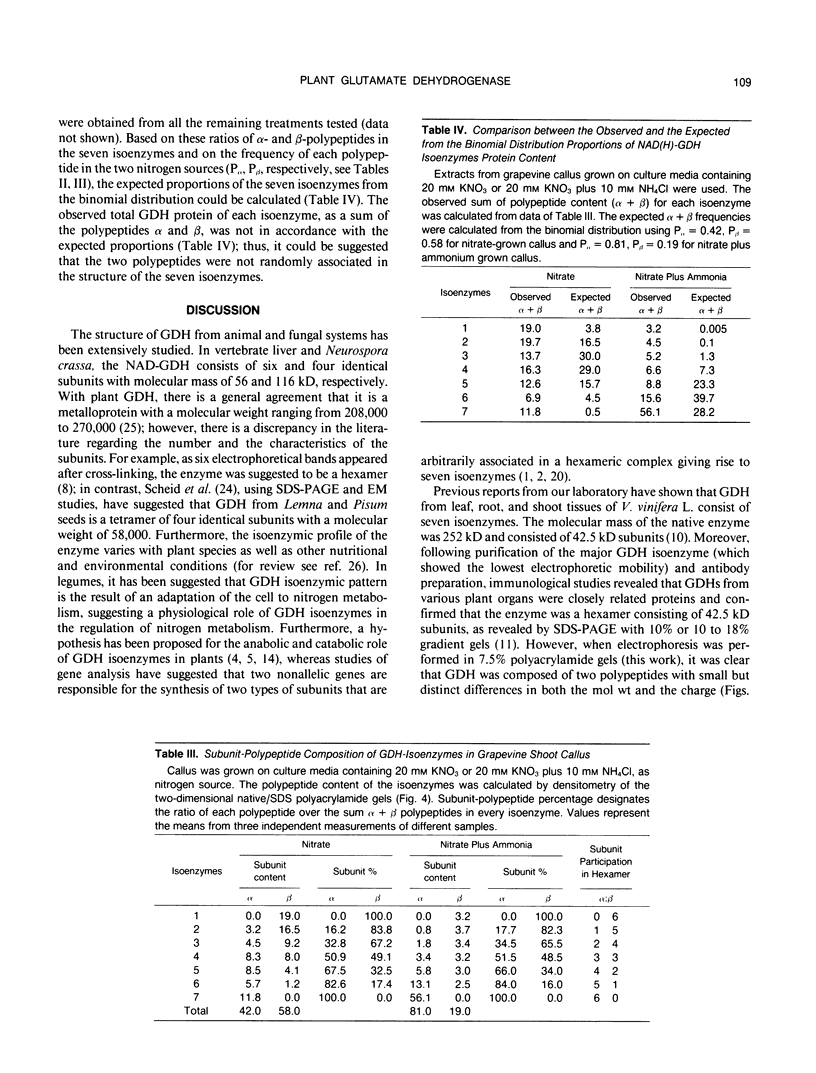
The structure and function of NAD(H)-glutamate dehydrogenase in plants was studied by using grapevine (Vitis vinifera L. cv Sultanina) callus grown under different nitrogen sources. The enzyme consists of two subunit-polypeptides, α and β, with similar antigenic properties but with different molecular mass and charge. The two polypeptides have molecular masses of 43.0 and 42.5 kilodaltons, respectively. The holoenzyme is hexameric and is resolved into seven isoenzymes by native gel electrophoresis. Two-dimensional native/SDS-PAGE revealed that the 1 and 7 isoenzymes are homohexamers and the isoenzymes 2 through 6 are hybrids of the two polypeptides following an ordered ratio. The total quantity of α- and β-polypeptides and the isoenzymic pattern was altered by the exogenous nitrogen source. The sample derived from callus grown on nitrate or glutamic acid contained a slightly greater amount of β-polypeptide and of the more cathodal isoenzymes, whereas α-polypeptide and the more anodal isoenzymes predominated in callus grown in the presence of either ammonium or glutamine. The anabolic reaction was correlated with the α- and the catabolic reaction with the β-polypeptide; this could suggest that each isoenzyme exhibits anabolic and catabolic function of different magnitude. The isoenzymic patterns did not obey the expected binomial distribution proportions.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- DAVIS B. J. DISC ELECTROPHORESIS. II. METHOD AND APPLICATION TO HUMAN SERUM PROTEINS. Ann N Y Acad Sci. 1964 Dec 28;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Loulakakis C. A., Roubelakis-Angelakis K. A. Immunocharacterization of NADH-Glutamate Dehydrogenase from Vitis vinifera L. Plant Physiol. 1990 Sep;94(1):109–113. doi: 10.1104/pp.94.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975 May 25;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Pahlich E., Joy K. W. Glutamate dehydrogenase from pea roots: purification and properties of the enzyme. Can J Biochem. 1971 Jan;49(1):127–138. doi: 10.1139/o71-018. [DOI] [PubMed] [Google Scholar]
- Scheid H. W., Ehmke A., Hartmann T. Plant NAD-dependent glutamate dehydrogenase. Purification, molecular properties and metal ion activation of the enzymes from Lemna minor and Pisum sativum. Z Naturforsch C. 1980 Mar-Apr;35(3-4):213–221. doi: 10.1515/znc-1980-3-408. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray W., Boulikas T., Wray V. P., Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem. 1981 Nov 15;118(1):197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]
- Yamaya T., Oaks A., Matsumoto H. Characteristics of glutamate dehydrogenase in mitochondria prepared from corn shoots. Plant Physiol. 1984 Dec;76(4):1009–1013. doi: 10.1104/pp.76.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]