Abstract
Toxoplasma gondii is a protozoan parasite responsible for widespread infections in humans and animals. Two major asexual forms are produced during the life cycle of this parasite: the rapidly dividing tachyzoite and the more slowly dividing, encysted bradyzoite. To further study the differentiation between these two forms, we have generated a large number of expressed sequence tags (ESTs) from both asexual stages. Previously, we obtained data on ∼7,400 ESTs from tachyzoites (J. Ajioka et al., Genome Res. 8:18–28, 1998). Here, we report the results from analysis of ∼2,500 ESTs from bradyzoites purified from the cysts of infected mice. We also report the results from analysis of 760 ESTs from parasites induced to differentiate from tachyzoites to bradyzoites in vitro. Comparison of the data sets from bradyzoites and tachyzoites reveals many previously uncharacterized sequence clusters which are largely or completely specific to one or other developmental stage. This class includes a bradyzoite-specific form of enolase. Combined with the previously identified bradyzoite-specific form of lactate dehydrogenase, this finding suggests significant differences in flux through the lower end of the glycolytic pathway in this stage. Thus, the generation of this data set provides valuable insights into the metabolism and growth of the parasite in the encysted form and represents a substantial body of information for further study of development in Toxoplasma.
Toxoplasma gondii is a member of the protozoan phylum Apicomplexa, which also includes the causative agents of malaria (Plasmodium spp.), coccidiosis (Eimeria spp.), and cryptosporidiosis (Cryptosporidium spp.). T. gondii is a major pathogen of a broad range of warm-blooded animals, including humans, livestock, and domestic pets (11). The parasite is of clinical importance both for the devastating disease it causes in the developing fetus and as an opportunistic infection in patients immunocompromised through disease or transplantation (19).
The parasite has a complex life cycle that includes sexual and asexual stages. The sexual cycle occurs exclusively in the guts of felines, while asexual growth can occur in almost any tissue of its broad range of hosts. The asexual cycle has two major forms: the rapidly dividing tachyzoite and the more slowly dividing, encysted bradyzoite. Tachyzoites are not normally responsible for host-to-host transmission and instead serve to disseminate infection within a given animal by invading and rapidly multiplying in a wide range of nucleated cells. In apparent response to immune pressure from the host, T. gondii tachyzoites differentiate into bradyzoites, which grow within cyst-like structures in the host tissue. When ingested, bradyzoites are infectious both to cats (resulting in entry into the sexual cycle) and other intermediate hosts, where further asexual growth can occur. Spontaneous reactivation of the disease through rupture of the cysts and dissemination of T. gondii tachyzoites can result in fatal encephalitis in patients with AIDS.
Tachyzoites and bradyzoites differ in a number of surface antigens (6) as well as important metabolic enzymes (10, 33, 34). Because of the difficulty of obtaining large amounts of tissue cysts from infected animals, however, it has been difficult to characterize bradyzoites in detail. Tachyzoites can be induced to differentiate in vitro through a variety of stresses (reviewed in reference 4) into forms which resemble bradyzoites by both morphological and antigenic criteria. Such in vitro bradyzoites have proved invaluable both in enabling the identification of bradyzoite-specific genes (16) and in providing initial insights into the mechanisms that control gene expression in this stage (5). However, the extent to which in vitro bradyzoites resemble parasites found in vivo is unclear.
To understand more about this important stage in the asexual cycle of the parasite, we have generated a large number of bradyzoite expressed sequence tags (ESTs) that complement the data already obtained from the tachyzoite stage (1). We also report here the results of an EST analysis from tachyzoites induced to switch to bradyzoites in vitro following 6 days of growth at high pH (29). As with a similar study of mammalian cells (17) in various states of differentiation, this analysis reveals many genes that appear to be developmentally regulated in addition to a large number of putative housekeeping genes that are expressed constitutively.
MATERIALS AND METHODS
Parasites.
The λgt11 in vivo cyst library (26) was generated from T. gondii ME49, a prototypical type II strain (28). RNA for the library from differentiating parasites was isolated from a cloned derivative of the ME49 strain designated PDS.
RNA preparation from differentiating parasites.
Human foreskin fibroblasts cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% Nu-Serum and 2 mM glutamine and cultured at 37°C in 5% CO2. Monolayers were infected with ∼105 tachyzoites of the PDS strain per cm2. Four hours later, all remaining extracellular parasites were removed by repeated rinsing of the monolayer with prewarmed DMEM. The culture medium was replaced with RPMI 1640 medium, pH 8.1, containing 5% fetal bovine serum, 50 mM HEPES, and 20 mg of gentamicin per liter, and the cells were incubated in a humid air atmosphere at 37°C. The pH of the culture medium equilibrated in the range of 8.1 to 8.2 and was monitored daily. The parasites were allowed to differentiate and encyst under these conditions and were harvested on day 6 after infection. The medium was changed every two days during this time.
The resulting in vitro cysts were harvested by scraping the monolayer with a rubber policeman. The cysts and host cells were pelleted by centrifugation for 12 min at 400 × g at 20°C, the supernatant was removed, and the pellets were resuspended in three aliquots of 15 ml (each) of phosphate-buffered saline. Each aliquot was subjected to three pulses of 2 s in a Waring blender (50-ml metal cup) to disrupt host cells, mixed with 2 volumes of DMEM, and pelleted as described above. The cysts were washed twice more in 40 ml of DMEM to remove host cell debris, resuspended in 6 ml of RPMI 1640, and prewarmed to 37°C. To release the bradyzoites from the cysts, an equal volume of “digestive fluid” (1% NaCl, 0.52% HCl, 0.1 mg of pepsin per ml) was added to the suspension and the mix was incubated at 37°C for 1 min. The solution was then neutralized by slowly adding 1 volume of 1% Na2CO3 and 3 volumes of RPMI 1640. The released bradyzoites were again pelleted as described above and resuspended in a total volume of 0.3 ml of phosphate-buffered saline. Total RNA was extracted from the parasite suspension with Ultraspec Total RNA isolation reagent (Biotecx Lab, Inc.) by following the manufacturer’s recommendations.
Library construction. (i) ME49 in vivo bradyzoite cDNA library.
Construction of the ME49 in vivo bradyzoite cDNA library in λgt11 is described in detail in reference 26. Briefly, tissue cysts of ME49 parasites were isolated from the brains of CBA/Ca mice 6 weeks after oral infection (14) and digested briefly with pepsin-HCl to release bradyzoites. Total RNA was isolated from approximately 8 × 107 bradyzoites by the guanidine isothiocyanate method (7). Single-stranded cDNA was made from poly(A)+ RNA with an NotI-oligo(dT) adapter primer [5′-CAATTCGCGGGCCGC(T)15-3′], and its second strand was synthesized with DNA polymerase I and RNase H. Flushed cDNA was ligated to Universal PCR adapters (Clontech) containing internal EcoRI sites. PCR amplification was carried out for 35 cycles, and products were digested with EcoRI and NotI. Fragments were ligated directionally into the corresponding sites of the λgt11 Sfi-Not vector.
The primary phage library was used to infect Escherichia coli LE392 and make high-titer plate lysates for phage DNA isolation by standard methods (2). Inserts were excised by EcoRI and NotI digestion, separated on an agarose gel, and selected for sizes between 0.7 and 2.0 kbp. The isolated fragments were ligated into the corresponding sites in pBluescript SKII− (Stratagene) and transformed into E. coli DH10. The size distribution of the inserts was determined by PCR amplification in 20 individual clones with the T3 and T7 primers. This subcloned library was plated to a density of about 200 colonies per plate, and 4,224 individual colonies were picked by hand and transferred to 11 384-well dishes containing Luria-Bertani medium–100 μg of ampicillin per ml–8% glycerol. Inoculated dishes were sealed and incubated for 14 h at 37°C. Locations of wells without growth were recorded, and the original plates were frozen at −80°C for shipping following replication into fresh plates with a 384-pin block.
(ii) ME49 in vitro bradyzoite library.
cDNA was synthesized from poly(A)+ RNA by using the cDNA Synthesis Kit (Stratagene) according to the manufacturer’s instructions. After second-strand synthesis and ligation of EcoRI adapters, double-stranded cDNA was digested with XhoI and ligated into λUniZAP XR. Individual clones for sequencing were selected from the primary library obtained after packaging.
Cycle sequencing, processing, and database submission. (i) ME49 in vivo bradyzoite cDNA library.
Individual clones were transferred to 96-well plates containing Luria-Bertani medium and grown overnight. Identification numbers consisting of TgESTzz followed by a plate number, row letter, and column number were assigned to each clone. Bacteria from each well were diluted in water, and PCR amplification of insert fragments was performed with T3 and T7 primers in MJ Research PTC 200 thermal cyclers. Amplification products were diluted in water and used as templates for 5′ sequencing with DYEnamic ET T7 dye primers.
Cycle sequencing, gel image analysis, and DNA sequence extraction were performed as described previously (1). Processed sequence data were annotated with similarity information, sequence quality information, and library information and then submitted to dBEST/GenBank.
dBEST entries for this project are clearly annotated to indicate the library source from which ESTs were derived. Data from a pilot scale study (153 ESTs) of the bradyzoite library prior to size selection (GenBank accession no. AA274250 through AA274402) was also submitted but was not included in the analysis described here. ESTs derived from the in vivo library after size selection are designated “size-selected” in the library title field to indicate this fact.
A small number of human and mouse cDNAs derived from the host cells from which Toxoplasma was isolated were revealed by sequencing of these libraries. dBEST entries have been annotated to indicate such contaminants. A note warning of the possibility that ESTs from these libraries may correspond to previously unidentified host sequences has also been included with the dBEST entries for these clones. Furthermore, since the in vivo bradyzoite library was obtained by PCR amplification, it is possible that genomic contaminants may also be represented.
(ii) ME49 in vitro bradyzoite library.
Analysis of this library was performed as previously described (1).
Cluster analysis and comparison to the tachyzoite EST database.
The in vivo bradyzoite size-selected EST data set (2,353 ESTs) was downloaded from dBEST and processed with READSEQ using BCM Search Launcher. The data file in FASTA format was assembled into 610 contigs containing pointers to 2,258 ESTs with TIGR_Assembler and N2TOOL (from the ICAASS suite, J. Parsons, European Bioinformatics Institute) after exclusion of 17 sequences less than 60 nucleotides long. Contigs were then reBLASTED against dBEST (BLASTn) and GenBanknr (BLASTx with RepeatMasker) to correlate contig identities, EST numbers from the tachyzoite and bradyzoite data sets, and putative homologies to each contig. A complete listing of the assembled bradyzoite contigs and matches to known proteins identified by BLASTx searches can be found at the Boothroyd laboratory home page (6a).
Nucleotide sequence accession numbers.
Sequences for the in vivo bradyzoite EST project have been submitted to GenBank with accession no. AA519069 to AA520977 and AA531619 to AA532062.
RESULTS
Library generation.
We constructed a cDNA library from mRNA isolated from the ME49 strain of parasites grown in vitro in human foreskin fibroblasts and subjected to high pH (∼8.1) for 6 days (4, 29). Parasites subjected to such treatment are induced to express bradyzoite-specific genes such as BAG1 (3), SAG4 (also called P18) (30, 31), and BSR4 (also called P36) (16, 31). Switching was monitored by immunofluorescence with monoclonal antibody T8 4A12 (31) to detect expression of P36. The results showed that >95% of the parasites were successfully induced to express this marker (as against <5% in control cultures grown under normal tachyzoite conditions). This analysis does not reveal the extent of induction (i.e., how far along the parasites are on the pathway of tachyzoite-to-bradyzoite differentiation). In vitro bradyzoites are clearly immature, since they fail to express the tissue cyst antigen P21 at time points at least up to 72 h under “switch” conditions. In some strains this antigen can be detected at later time points (30), but its expression was not monitored in this study.
From the in vitro-bradyzoite cDNA library, 1,436 clones were subjected to sequence analysis, of which 761 yielded usable traces. To determine if this library would be informative, we compared the frequencies of known tachyzoite-stage-specific genes in this and the tachyzoite EST data set (1). We also examined the representations of three genes previously established to be induced during in vitro switching (29, 30, 33). The results indicated that there were still substantial representations of tachyzoite-specific genes: SAG1 and SAG2 transcripts were present at 0.9 and 0.4%, respectively, versus 1.9 and 0.8% in the ME49 tachyzoite EST data set. The drop of about twofold is consistent with the differentiating state of these parasites. It was disappointing, however, to find only a single EST (TgESTzz09e11:SAG4) for the known bradyzoite-specific genes. The abundance of SAG4 and LDH2 was expected to be low, and a frequency of 1 in 761 for a SAG4 transcript was not surprising. BAG1, however, is a relatively abundant transcript in mature bradyzoites which is strongly induced early in switching (29). While its frequency in a comparatively small number of ESTs cannot be predicted, the absence of this transcript and the background of tachyzoite sequences suggested that additional sequencing would not reveal a high proportion of ESTs in which we were interested, i.e., genes that would be upregulated in immature bradyzoites, which might then serve as markers for the differentiation process. An additional problem that we encountered during construction of the library was that there were clear differences in the susceptibilities of tachyzoites and encysted bradyzoites to techniques designed to solubilize them and isolate total RNA (21). As such, the resulting library may not have accurately represented the population of switching parasites. Despite this limitation, the data set from the in vitro library may still contain pointers to interesting genes. Indeed, our analysis of the in vivo data set (see below) revealed a number of singletons in the in vitro ESTs that are apparently restricted to the bradyzoite form and that may indeed represent transcripts from genes induced early after induction. Preliminary analysis, clustering, and BLASTx homology information for this library can be found in the Toxoplasma database of clustered ESTs (32).
Because of these limitations, we focused our effort on ESTs from a library from mature bradyzoites isolated from the brains of infected mice (33). Based on an initial analysis of this library reported in the original publication and preliminary sequencing in the current study of a set of about 150 clones, it was clear that there was a strong bias in favor of truncated sequences clustered at the 3′ ends of the respective genes. This bias is likely to have arisen from a number of technical problems encountered during the original construction of the library, which include (i) the difficulty of purifying cysts from infected animals and treating them to remove host cell contamination, (ii) the harsh treatment necessary to remove the bradyzoites from the cysts (acid-pepsin digestion), and (iii) preferential amplification of small fragments during PCR amplification of the cDNA prior to cloning. To overcome the small size and 3′ bias, the library was amplified and inserts were excised by EcoRI and NotI digestion and size fractionated. Inserts in the 0.7- to 2.0-kb range were isolated from the gel and used to prepare a new directional library in pBluescript SK−. This size range was chosen because it ensured that most ESTs would include coding sequence. An initial sample of 20 clones from this library was characterized for insert size and then sequenced. As expected, the insert sizes ranged from 600 to 1,400 bp, with an average of ∼850 bp. By examining the five ESTs corresponding to known genes in this set, we observed that all traces overlapped the coding region, confirming that size selection had successfully eliminated the most extreme 5′- truncated clones. This result was further confirmed upon analysis of specific transcripts from the full-scale project (see below). Furthermore, in this set of 20 ESTs, 2 ESTs corresponded to known bradyzoite-specific genes (MAG1 and BAG1), suggesting that a major investment in analysis of this library would be fruitful. Based on this, a further 3,747 ESTs were sequenced from this library, with 2,353 (62.8%) providing high-quality sequence data. These data were assembled into 613 separate contigs, of which 235 are singletons. Of the 613 separate contigs, 205 (33%) gave matches to known proteins with a BLASTx high score of >70 (this is a measure of the quality of the alignment; >70 indicates likely significance).
Table 1 shows a comparison of the frequencies of ESTs of the most abundant tachyzoite- and bradyzoite-specific genes identified previously (although note that “specific” refers to expression of the protein product and that the basis of this specificity is frequently not known). These data show that most of the expected stage-specific genes are appropriately represented in the ME49 tachyzoite and bradyzoite libraries (e.g., ESTs for tachyzoite-specific genes are more abundant in the tachyzoite data set than in the bradyzoite data set). Although we identified two ESTs (TgESTzz27h08 and TgESTzz64g06) corresponding to SAG1 (whose gene product [SAG1 or P30] is not normally detectable in bradyzoites [12]), these two ESTs include sequences upstream of the major transcription initiation site identified in tachyzoites. Thus, they do not necessarily indicate that bradyzoites express low levels of mature SAG1 transcripts. The origin and significance of these transcripts, which presumably represent initiation or readthrough from an upstream site, are unclear. The only gene mapped to this region is SRS1 (13), whose polyadenylation site lies ∼1.5 kb upstream of the 5′-most SAG1 transcription start site. The two bradyzoite ESTs extend only ∼200 (TgESTzz27h08) and ∼100 (TgESTzz64g06) bp upstream of the SAG1 transcription start site.
TABLE 1.
Summary of EST frequencies of abundant Toxoplasma genes for ME49 tachyzoite and bradyzoite data sets
| Pattern of expressiona | Geneb | ME49 tachyzoite cDNA library (n = 1,806)
|
ME49 bradyzoite cDNA library (n = 2,353)
|
||
|---|---|---|---|---|---|
| No. of ESTs | % of total no. of cDNAs | No. of ESTs | % of total no. of cDNAs | ||
| Tachyzoite specific | SAG1 | 39 | 2.15 | 2c | 0.08 |
| SAG2 | 24 | 1.32 | 0 | 0 | |
| Constitutive | SAG3 | 3 | 0.17 | 1 | 0.04 |
| GRA1 | 38 | 2.10 | 14 | 0.59 | |
| GRA2 | 28 | 1.55 | 43 | 1.83 | |
| GRA3 | 5 | 0.28 | 3 | 0.13 | |
| GRA4 | 6 | 0.33 | 6 | 0.25 | |
| GRA5 | 27 | 1.49 | 44 | 1.87 | |
| Bradyzoite specific | SAG4 | 0 | 0 | 23 | 1.02 |
| BSR4/P36 | 0 | 0 | 0 | 0 | |
| BAG1 | 0 | 0 | 100 | 4.42 | |
| MAG1 | 1 | 0.05 | 18 | 0.76 | |
| LDH2 | 0 | 0 | 26 | 1.11 | |
| Unknown | SRS 1 to 4d | 17 | 0.94 | 0 | 0 |
Patterns of expression based on protein detection and/or reverse transcription-PCR and Northern blot analysis prior to this study.
Genes are arranged into groups according to their patterns of expression based on protein detection and/or other criteria.
Do not represent mature SAG1 transcripts (see the text).
Sum of ESTs for all members of the family of SAG1-related sequences (SRS).
Interestingly, we were unable to identify ESTs corresponding to the BSR4/P36 antigen or the other SAG1 related sequence (SRS) genes (20) in the in vivo data set. The absence of sequences corresponding to BSR4 is surprising, since this is a major surface antigen in bradyzoites. Previous data indicate that while SRS3/P35 is restricted to tachyzoites, SAG3 was previously reported to be expressed in both tachyzoites and bradyzoites (31). One EST corresponding to SAG3 occurred in the bradyzoite set (0.042%), compared to three ESTs occurring in tachyzoites (0.17%). These small numbers preclude any clear conclusion about the relative abundance of the SAG3 transcript. BLAST analysis of the expression of the bradyzoite-specific surface antigen SAG4 (P18 [22]) revealed both this gene and a closely related homolog (TgESTzz67e09; P = 2.6 × 10−33) for which 21 ESTs were assembled into an apparently complete cDNA. The open reading frame (ORF) it encodes has 50% identity to SAG4, is present at approximately equivalent levels (21 versus 23 ESTs for SAG4), and is also bradyzoite specific.
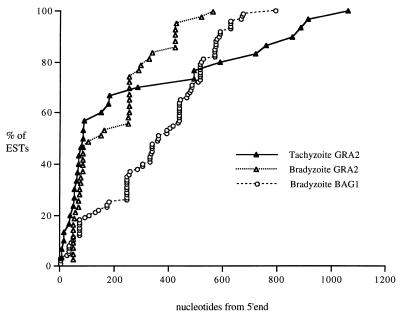
The fact that the libraries being compared here were constructed at different times and in different laboratories (and one was the result of amplification and subcloning) means that direct comparisons of frequencies should be viewed with caution. Overall, however, the changes in EST frequency for the known Toxoplasma genes detailed in Table 1 reflect the known differences between the two stages (15, 31). To further examine overlaps between the two data sets and the reliability of putative stage-specific designations, we performed BLASTn analyses for two genes, GRA2 and BAG1, to map their relative start positions on the full-length cDNA. These data are plotted in Fig. 1. ESTs for GRA2 were highly represented in both tachyzoite and bradyzoite libraries, and ESTs from both data sets span almost the entire cDNA, with ∼50% of sequences in both data sets having 5′ ends close to the putative transcription start site (∼1,050 bp; GenBank L01753 [27]). BAG1 expression is restricted to the bradyzoite data set, and ESTs to this transcript also span the full-length cDNA, with clustering at the presumed 5′ end. For these genes at least, overlaps between ESTs allow sampling of the entire transcript and direct comparisons of frequencies. However, it should be noted that digestion of cDNAs during construction or subcloning of the in vivo bradyzoite library will have shortened cDNAs that contain internal EcoRI or NotI sites and thus may render comparison of other cDNAs more difficult.
FIG. 1.
Pattern of start positions of ESTs on known transcripts for GRA2 and BAG1 genes. Start positions were calculated from BLASTn of dBEST with the assembled cDNAs for each gene. For GRA2, EST start sites were plotted relative to the 5′ end of exon 1 (27), and for BAG1, EST start sites were plotted from the 5′ end of the assembled BAG1 contig. The 5′ end of the BAG1 contig maps 27 nucleotides 5′ to the 5′ end of the mRNA reported by Parmley et al. (25).
Table 2 shows a partial listing from the in vivo bradyzoite data set of high-scoring BLASTx matches to known proteins in GenBank. Based on EST frequencies, there may be stage-specific expression of many of these genes, although clearly the small sample size makes such conclusions tentative, especially in cases where only a few ESTs are found (e.g., the NTPase homolog). Interpretation of expression patterns from EST frequency is further complicated by the observation (based on reverse transcription-PCR and Northern blot analysis) that some transcripts are equally abundant in both stages even when the products they encode are stage specific. Regulation for these genes may be translational or posttranslational (e.g., LDH1 [34], MAG1 [24, 30], and BSR4 [16]). Nevertheless, a number of interesting genes listed in Table 2 appear to be restricted to the bradyzoite form based on their EST frequencies. Among these, the homologs of DNase IV (apurinic endonuclease; TgESTzz34g03; P = 1.2 × 10−42) and deoxyribose phosphate aldolase (TgESTzz32g04; P = 5.7 × 10−7) may be indicative of reliance on specific pathways in DNA repair and nucleotide metabolism that offer therapeutic potential. Likewise, a number of homologs of enzymes in the metabolism of oxygen radicals are also present. These differences require experimental confirmation but support the intuitive notion that life within a cyst is different from that in a vacuole and more stressful with respect to exposure to reactive metabolites.
TABLE 2.
Partial listing of top matches to known proteins in GenBank identified by BLASTx homology searches
| Contig | EST | Gene name | BLASTx P valuea | BLASTx high scorea | No. of bradyzoite ESTs | No. of tachyzoite ESTs |
|---|---|---|---|---|---|---|
| Genes for metabolic enzymes | ||||||
| bs4_98 | TgESTzz32g04 | Deoxyribose phosphate aldolase | 5.7 × 10−7 | 115 | 22 | 0 |
| bs4_320 | TgESTzz34g03 | Apurinic endonuclease | 1.2 × 10−42 | 124 | 14 | 0 |
| bs4_10 | TgESTzz54b03 | Ketoacyl reductase | 2.1 × 10−6 | 81 | 12 | 0 |
| bs4_48 | TgESTzz33a12 | Enolase | 4.1 × 10−81 | 702 | 13 | 0 |
| bs4_619 | TgESTzz66d07 | NTPase homolog | 6.1 × 10−22 | 211 | 1 | 0 |
| bs4_26/507 | TgESTzz38f05 | Oxoglutarate-malate translocator | 4.3 × 10−16 | 116 | 10 | 0 |
| bs4_760 | TgESTzz59c08 | Dihydroorotate dehydrogenase | 9.8 × 10−45 | 204 | 1 | 0 |
| bs4_826 | TgESTzz35c04 | Pyrroline-5-carboxylate reductase | 1.4 × 10−20 | 178 | 1 | 0 |
| bs4_359 | TgESTzz26e05 | Methionine aminopeptidase | 2.1 × 10−65 | 394 | 4 | 0 |
| bs4_246 | TgESTzz36a03 | Molybdenum cofactor | 4.9 × 10−16 | 176 | 4 | 0 |
| Biosynthetic precursor E | ||||||
| bs4_435 | TgESTzz64b03 | Hsc-70-interacting protein of SCN6 | 3.5 × 10−8 | 125 | 7 | 0 |
| bs4_398 | TgESTzz68f03 | Heat shock protein | 1.5 × 10−38 | 290 | 3 | 0 |
| bs4_57 | TgESTzz35f09 | Thiolesterase | 3.0 × 10−18 | 135 | 3 | 0 |
| bs4_286 | TgESTzz37c01 | Thioredoxin | 9.6 × 10−19 | 154 | 3 | 0 |
| bs4_201 | TgESTzz54c12 | Calcium-independent phospholipase A2 | 5.6 × 10−37 | 185 | 3 | 1 |
| bs4_547 | TgESTzz68h05 | Glutathione peroxidase | 5.0 × 10−13 | 118 | 3 | 0 |
| bs4_197 | TgESTzz35a02 | Enoyl coenzyme A hydratase | 6.4 × 10−13 | 111 | 2 | 0 |
| bs4_885 | TgESTzz34c10 | Cyclic AMP protein kinase regulatory chain | 1.2 × 10−9 | 112 | 1 | 0 |
| bs4_182 | TgESTzz46e12 | SNF2L transcription activator | 1.7 × 10−30 | 127 | 7 | 0 |
| bs4_09 | TgESTzz34g08 | Protein phosphatase 2C | 6.3 × 10−9 | 93 | 5 | 0 |
| bs4_624 | TgESTzz33b10 | Karyopherin alpha-2 subunit | 7.8 × 10−74 | 253 | 2 | 0 |
| bs4_802 | TgESTzz36d09 | Cyclophilin | 1.2 × 10−47 | 349 | 1 | 0 |
| bs4_713 | TgESTzz68f05 | Cul-3 | 5.0 × 10−16 | 143 | 1 | 0 |
| bs4_791 | TgESTzz38e01 | Golgi adapter Ap-1 (47 kDa) | 5.7 × 10−52 | 260 | 1 | 0 |
| bs4_445 | TgESTzz68f10 | Coatamer beta′ subunit | 1.1 × 10−14 | 113 | 2 | 0 |
| Parasite-specific genes | ||||||
| bs4_31 | TgESTzz67e09 | SAG4 homolog | 2.6 × 10−33 | 126 | 21 | 0 |
| bs4_309 | TgESTzz69c05 | ROP8 homolog | 0.0085 | 90 | 9 | 2 |
| bs4_537 | TgESTzz48e02 | ROP8 homolog | 1.1 × 10−16 | 179 | 2 | 0 |
| bs4_305 | TgESTzz34c07 | ROP2 homolog | 3.1 × 10−12 | 97 | 6 | 1 |
| bs4_68 | TgESTzz35e11 | P22 (SAG2) homolog | 0.71 | 71 | 12 | 0 |
| bs4_88 | TgESTzz53c11 | Sporozoite antigen 19 kDa | 3.4 × 10−47 | 222 | 24 | 8 |
The BLASTx P value indicates the probability that the match could have occurred by chance. The BLASTx high score is a measure of the best, ungapped alignment (>70 is often significant). Neither the P value nor the high score alone can be relied upon to always pick up significant matches, and so both are presented here.
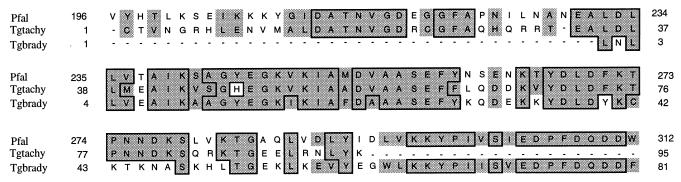
The existence of an apparently bradyzoite-specific contig encoding enolase (TgESTzz33a12; P = 4.1 × 10−81) prompted us to examine the regions of overlap of this contig with the enolase-coding region and with contigs assembled from the tachyzoite data set (Fig. 2). While contigs from both forms overlap in the central portion of the ORF with high homology to P. falciparum enolase (tachyzoite, P = 1.4 × 10−27; bradyzoite, P = 4.1 × 10−81), the two Toxoplasma contigs are only 64% conserved at the nucleotide level and clearly represent different forms of the same enzymatic activity. The existence of bradyzoite-specific forms of lactate dehydrogenase (34) and enolase suggests that flux into or through the lower arm of the glycolytic pathway is different between the two stages. Previous studies have suggested that while both forms appear to maintain a high glycolytic flux, bradyzoites lack a functional tricaboxylic acid cycle (9). This would place greater emphasis on conversion of pyruvate to lactate for NAD+ regeneration in this stage. The two asexual life cycle stages of Toxoplasma may also rely upon different energy sources whose products enter the glycolytic pathway at different points. These differences also reflect different anabolic requirements, such as the need for bradyzoites to synthesize sugar-nucleotide precursors (e.g., UDP-N-acetylglucosamine) as cyst wall components (23). The metabolic consequences of these differences are unclear but intriguing.
FIG. 2.
Comparison of regions of overlap of tachyzoite and bradyzoite forms of enolase revealed by contig assembly of ESTs. Deduced amino acid sequences are compared to the corresponding region from P. falciparum enolase. Boxed and shaded residues indicate identity and conservative changes relative to the P. falciparum sequence, respectively. Dashed regions indicate regions for which no EST data are available.
DISCUSSION
We have generated a total of 3,267 ESTs from in vivo and in vitro bradyzoites of T. gondii. The 2,350 ESTs derived from the in vivo cyst cDNA library fall into more than 600 clusters with homology to some 200 known genes, including several that appear specific to this stage. Prior to this study, only three genes expressed specifically in bradyzoites had been identified and cloned. The present study has therefore greatly extended our knowledge of genes expressed during this stage. In combination with the 7,400 ESTs from Toxoplasma tachyzoites, these new sequences provide valuable information for the further study of the biology of this important parasite.
The value of the EST approach in studying gene expression in T. gondii is borne out by the data in Tables 1 and 2. In a number of cases, we have been able to (i) assemble multiple ESTs into large contigs encompassing all or significant portions of the ORF of a gene and (ii) make crude estimates of the relative levels of expression of a transcript for a given gene based on EST frequencies. In a few cases, this information allows us to make deductions about the likely importance of this gene and/or its probable function, as a starting point for further investigation.
One limitation of the approach is exemplified by our attempts to generate information from the in vitro library of differentiating parasites. In this case, problems stem from (i) the fact that the in vitro bradyzoites are early in the process of switching and thus a high background of tachyzoite sequences persists and (ii) the fact that parasite preparations contain forms with different susceptibilities to chaotropic agents. Overall, these factors yielded a cDNA library with a high background of tachyzoite genes. Problems which stem from the background of tachyzoite-specific and constitutively expressed, highly abundant genes might be overcome in the future by using subtracted and normalized libraries or by alternative manual approaches such as differential display (18). These methods should be combined with optimized protocols for extracting mRNAs from encysting parasites. Nevertheless, the small number of sequences obtained from this library may still provide clues about genes involved in the switching process.
Currently, the Toxoplasma EST database is limited to genes expressed in the asexual stages of the parasite life cycle. The solution to this limitation would be to generate cDNA libraries from the sexual stages and sporozoites and/or to do genomic sequencing. With respect to the second of these, Toxoplasma has a haploid genome size of ∼8 × 107 bp (8) and introns are moderately common. As such, the likely return on a small-scale genome sequencing effort would be relatively low compared to that seen here. It should be noted that useful information about the genes of Toxoplasma is also likely to be forthcoming from the Plasmodium falciparum genome project (20). Indeed, comparison of the two data sets is likely to be highly complementary and informative, especially where the fast evolutionary clock makes identification of Plasmodium genes difficult without the bridge provided by information from Toxoplasma.
In conclusion, the EST approach is an extremely cost-effective way to generate a very large amount of information on gene expression in a developmentally regulated system. Both developmental and constitutive classes of ESTs may reveal metabolic pathways critical to the parasite that serve as targets for therapeutic intervention. In the particular case being studied here, the project has produced a great many gene sequences from a stage of the parasite that is difficult to study and understanding of which is absolutely critical to control infection, especially in immunocompromised patients, where reactivation of chronic infection is a major cause of serious or even fatal disease.
ACKNOWLEDGMENTS
We thank our colleagues in our respective labs, Anthony Mossop for help with ICAASS and Doug Vollrath and David Roos (University of Pennsylvania) for helpful comments and for provision of key materials. The inception of this project is in part due to discussions held at a National Cooperative Drug Discovery Group meeting in 1995; the strong support and encouragement of Alex Fairfield, Keith Joiner, Barbara Laughon, and David Roos are gratefully acknowledged, as is the Washington University Genome Center EST Team.
This work was supported by grants from the NIH (AI30230 and AI41014). Construction of the in vivo cyst library was funded by the U.S. Department of Agriculture (grant 91-37204-6878). A.H. was supported by fellowships from the Swiss National Science Foundation and the Roche Research Foundation.
I. D. Manger and A. Hehl contributed equally to this work.
REFERENCES
- 1.Ajioka J, Boothroyd J C, Brunk B P, Hehl A, Hillier L, Manger I D, Overton G C, Marra M, Roos D, Wan K L, Waterston R, Sibley L D. Sequencing of ESTs from the protozoan parasite Toxoplasma gondii: efficient identification of genes and identification of phylogenetically restricted sequences of the Apicomplexa. Genome Res. 1998;8:18–28. doi: 10.1101/gr.8.1.18. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. 1 to 3. New York, N.Y: John Wiley and Sons, Inc.; 1995. [Google Scholar]
- 3.Bohne W, Gross U, Ferguson D J, Heesemann J. Cloning and characterization of a bradyzoite-specifically expressed gene. Mol Microbiol. 1995;16:1221–1230. doi: 10.1111/j.1365-2958.1995.tb02344.x. [DOI] [PubMed] [Google Scholar]
- 4.Bohne W, Parmley S F, Yang S, Gross U. Bradyzoite-specific genes. Curr Top Microbiol Immunol. 1996;219:81–91. doi: 10.1007/978-3-642-51014-4_9. [DOI] [PubMed] [Google Scholar]
- 5.Bohne W, Wirsing A, Gross U. Bradyzoite-specific gene expression in Toxoplasma gondii requires minimal genomic elements. Mol Biochem Parasitol. 1997;85:89–98. doi: 10.1016/s0166-6851(96)02814-9. [DOI] [PubMed] [Google Scholar]
- 6.Boothroyd J C, Hehl A B, Knoll L J, Manger I D. The surface of Toxoplasma: more and less. Int J Parasitol. 1998;28:3–9. doi: 10.1016/s0020-7519(97)00182-3. [DOI] [PubMed] [Google Scholar]
- 6a.Boothroyd laboratory home page.http://cmgm.stanford.edu/micro/boothroyd/toxo1.html.
- 7.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 8.Cornelissen A W, Overdulve J P, van der Ploeg M. Determination of nuclear DNA of five eucoccidian parasites, Isospora (Toxoplasma) gondii, Sarcocystis cruzi, Eimeria tenella, E. acervulina and Plasmodium berghei, with special reference to gamontogenesis and meiosis in I. (T.) gondii. Parasitology. 1984;88:531–553. doi: 10.1017/s0031182000054792. [DOI] [PubMed] [Google Scholar]
- 9.Denton H, Brown S M, Roberts C W, Alexander J, McDonald V, Thong K W, Coombs G H. Comparison of the phosphofructokinase and pyruvate kinase activities of Cryptosporidium parvum, Eimeria tenella and Toxoplasma gondii. Mol Biochem Parasitol. 1996;76:23–29. doi: 10.1016/0166-6851(95)02527-8. [DOI] [PubMed] [Google Scholar]
- 10.Denton H, Roberts C W, Alexander J, Thong K W, Coombs G H. Enzymes of energy metabolism in the bradyzoites and tachyzoites of Toxoplasma gondii. FEMS Microbiol Lett. 1996;137:103–108. doi: 10.1111/j.1574-6968.1996.tb08090.x. [DOI] [PubMed] [Google Scholar]
- 11.Dubey J P. Toxoplasma, Neospora, Sarcosystis, and other tissue cyst-forming coccidia of humans and animals. In: Kreier J P, editor. Parasitic protozoa. Vol. 6. New York, N.Y: Academic Press; 1993. pp. 1–158. [Google Scholar]
- 12.Gross U, Bohne W, Luder C G, Lugert R, Seeber F, Dittrich C, Pohl F, Ferguson D J. Regulation of developmental differentiation in the protozoan parasite Toxoplasma gondii. J Eukaryot Microbiol. 1996;43:114S–116S. doi: 10.1111/j.1550-7408.1996.tb05033.x. [DOI] [PubMed] [Google Scholar]
- 13.Hehl A B, Krieger T, Boothroyd J C. Identification and characterization of SRS1, a Toxoplasma gondii surface antigen upstream of and related to SAG1. Mol Biochem Parasitol. 1997;89:271–282. doi: 10.1016/s0166-6851(97)00126-6. [DOI] [PubMed] [Google Scholar]
- 14.Huskinson-Mark J, Araujo F G, Remington J S. Evaluation of the effect of drugs on the cyst form of Toxoplasma gondii. J Infect Dis. 1991;164:170–171. doi: 10.1093/infdis/164.1.170. [DOI] [PubMed] [Google Scholar]
- 15.Kasper L H. Identification of stage-specific antigens of Toxoplasma gondii. Infect Immun. 1989;57:668–672. doi: 10.1128/iai.57.3.668-672.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knoll L J, Boothroyd J C. Isolation of developmentally regulated genes from Toxoplasma gondii by a gene trap with the positive and negative selectable marker hypoxanthine-xanthine-guanine phosphoribosyltransferase. Mol Cell Biol. 1998;18:807–814. doi: 10.1128/mcb.18.2.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee N H, Weinstock K G, Kirkness E F, Earle-Hughes J A, Fuldner R A, Marmaros S, Glodek A, Gocayne J D, Adams M D, Kerlavage A R, Fraser C M, Venter J C. Comparative expressed-sequence-tag analysis of differential gene expression profiles in pc-12 cells before and after nerve growth factor treatment. Proc Natl Acad Sci USA. 1995;92:8303–8307. doi: 10.1073/pnas.92.18.8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang P, Averboukh L, Pardee A B. Methods of differential display. In: Adolph K W, editor. Methods in molecular genetics. Vol. 5. 1994. pp. 3–16. Gene and chromosome analysis, part C. Academic Press, Inc., San Diego, Calif. [Google Scholar]
- 19.Luft B J, Remington J S. Toxoplasmic encephalitis in AIDS. Clin Infect Dis. 1992;15:211–222. doi: 10.1093/clinids/15.2.211. [DOI] [PubMed] [Google Scholar]
- 20.Malarial Genome Project home page.http://www.sanger.ac.uk/Projects/P_falciparum/.
- 20a.Manger, I. D., A. B. Hehl, and J. C. Boothroyd. The surface of Toxoplasma tachyzoites is dominated by a family of GPI-anchored antigens related to SAG1. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 21.Manger, I. D., A. B. Hehl, and J. C. Boothroyd. Unpublished data.
- 22.Odberg-Ferragut C, Soete M, Engels A, Samyn B, Loyens A, Van Beeumen J, Camus D, Dubremetz J F. Molecular cloning of the Toxoplasma gondii sag4 gene encoding an 18 kDa bradyzoite specific surface protein. Mol Biochem Parasitol. 1996;82:237–244. doi: 10.1016/0166-6851(96)02740-5. [DOI] [PubMed] [Google Scholar]
- 23.Ortega, E., and J. C. Boothroyd. Unpublished data.
- 24.Parmley, S. F. Unpublished results.
- 25.Parmley S F, Weiss L M, Yang S. Cloning of a bradyzoite-specific gene of Toxoplasma gondii encoding a cytoplasmic antigen. Mol Biochem Parasitol. 1995;73:253–257. doi: 10.1016/0166-6851(95)00100-f. [DOI] [PubMed] [Google Scholar]
- 26.Parmley S F, Yang S, Harth G, Sibley L D, Sucharczuk A, Remington J S. Molecular characterization of a 65-kilodalton Toxoplasma gondii antigen. Mol Biochem Parasitol. 1994;66:283–296. doi: 10.1016/0166-6851(94)90155-4. [DOI] [PubMed] [Google Scholar]
- 27.Prince J B, Araujo F G, Remington J S, Burg J L, Boothroyd J C, Sharma S D. Cloning of cDNAs encoding a 28 kilodalton antigen of Toxoplasma gondii. Mol Biochem Parasitol. 1989;34:3–13. doi: 10.1016/0166-6851(89)90014-5. [DOI] [PubMed] [Google Scholar]
- 28.Sibley L D, Howe D K. Genetic basis of pathogenicity in toxoplasmosis. Curr Top Microbiol Immunol. 1996;219:3–15. doi: 10.1007/978-3-642-51014-4_1. [DOI] [PubMed] [Google Scholar]
- 29.Soete M, Camus D, Dubremetz J F. Experimental induction of bradyzoite-specific antigen expression and cyst formation by the RH strain of Toxoplasma gondii in vitro. Exp Parasitol. 1994;78:361–370. doi: 10.1006/expr.1994.1039. [DOI] [PubMed] [Google Scholar]
- 30.Soete M, Fortier B, Camus D, Dubremetz J F. Toxoplasma gondii: kinetics of bradyzoite-tachyzoite interconversion in vitro. Exp Parasitol. 1993;76:259–264. doi: 10.1006/expr.1993.1031. [DOI] [PubMed] [Google Scholar]
- 31.Tomavo S, Fortier B, Soete M, Ansel C, Camus D, Dubremetz J F. Characterization of bradyzoite-specific antigens of Toxoplasma gondii. Infect Immun. 1991;59:3750–3753. doi: 10.1128/iai.59.10.3750-3753.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toxoplasma Database of Clustered Expressed Sequence Tags.http://daphne.humgen.upenn.edu:1024/toxodb/ver_1/toxodb.html.
- 33.Yang S, Parmley S F. A bradyzoite stage-specifically expressed gene of Toxoplasma gondii. Mol Biochem Parasitol. 1995;73:291–294. doi: 10.1016/0166-6851(95)00124-j. [DOI] [PubMed] [Google Scholar]
- 34.Yang S, Parmley S F. Toxoplasma gondii expresses two distinct lactate dehydrogenase homologous genes during its life cycle in intermediate hosts. Gene. 1997;184:1–12. doi: 10.1016/s0378-1119(96)00566-5. [DOI] [PubMed] [Google Scholar]