Abstract
The imatinib-sensitive fusion gene FIP1L1::PDGFRA is the most frequent molecular abnormality identified in patients with eosinophilic myeloid neoplasms. Rapid recognition of this mutation is essential given the poor prognosis of PDGFRA-associated myeloid neoplasms prior to the availability of imatinib therapy. We report a case of a patient in whom delayed diagnosis resulted in cardiac transplantation for eosinophilic endomyocardial fibrosis. The delay in diagnosis was due, in part, to a false-negative result in fluorescence in situ hybridization (FISH) testing for FIP1L1::PDGFRA. To explore this further, we examined our cohort of patients presenting with confirmed or suspected eosinophilic myeloid neoplasms and found 8 additional patients with negative FISH results despite a positive reverse-transcriptase polymerase chain reaction test for FIP1L1::PDGFRA. More importantly, false-negative FISH results delayed the median time to imatinib treatment by 257 days. These data emphasize the importance of empiric imatinib therapy in patients with clinical features suggestive of PDGFRA-associated disease.
Keywords: Clinical studies, Eosinophilia, Myeloproliferative disorders
Introduction
Hypereosinophilic syndromes (HES) are a group of disorders characterized by peripheral eosinophilia >1.5 × 109/L and clinical manifestations attributable to the eosinophilia [1]. Although the underlying causes of HES are heterogeneous, participants with suspected or confirmed primary eosinophilic myeloid neoplasms (HESN) account for 10–20% of patients presenting with HES in most series [2]. Historically, these patients were the most difficult to treat with mortality rates exceeding 30% within 5 years of presentation [3]. In this regard, identification of the IP1L1::PDGFRA fusion gene in 2003 [4] represented a turning point with near complete hematologic and molecular responses to the tyrosine kinase inhibitor, imatinib, in patients harboring this mutation. PDGFRA fusion genes can be detected by reverse-transcriptase polymerase chain reaction (RT-PCR), fluorescence in situ hybridization (FISH), or sequence analysis. Each method has relative advantages and disadvantages with respect to sensitivity, specificity, and the ability to detect alternative fusion partners.
After documenting the presence of the FIP1L1:: PDGFRA fusion by RT-PCR in several patients referred for steroid-resistant HES and a negative test by FISH [5], we conducted a retrospective chart review to determine the frequency of this occurrence. Clinical and laboratory data were assessed from 606 patients enrolled on a natural history study of eosinophilia (NCT00001406) between April 4, 1994, and December 31, 2021 (online suppl. Fig. 1; for all online suppl. material, see https://doi.org/10.1159/000528046). All patients underwent comprehensive evaluation, including testing for FIP1L1::PDGFRA by RT-PCR [3]. Sixty-five patients with confirmed or suspected eosinophilic myeloid neoplasms (HESN) were identified using previously described criteria [6], of which 37 (57%) were positive for FIP1L1::PDGFRA by RT-PCR and/or FISH testing, 19 had other molecular abnormalities, and 9 had clinical and laboratory features of HESN with no mutation identified (online suppl. Fig. 2).
Demographic, clinical, and laboratory characteristics of the 65 confirmed or presumed HESN patients grouped by mutational status are provided in Table 1. Of the parameters examined, male predominance, serum tryptase, and B12 levels were significantly different between the groups. Of note, only one FIP1L1::PDGFRA-positive patient had normal tryptase and B12 levels. As expected, all 33 patients who tested positive for FIP1L1::PDGFRA and were treated with imatinib had a rapid and complete clinical and hematologic response (p < 0.001, compared to the other two groups combined, Fisher’s exact test). Of 4/10 PDGFRA-negative patients with other molecular abnormalities who underwent an imatinib trial and responded, three had translocations involving PDGFRB. The fourth, who has a V617F JAK2 mutation, had only a transient (<6 months) response. Five (71%) of the 7 mutation-negative presumed HESN patients who received imatinib also responded completely. However, 4 of the 5 responders were treated with imatinib prior to testing for FIP1L1::PDGFRA and/or translocations involving PDGFRB.
Table 1.
Demographic and clinical characteristics of patients with HESN
| FIP1L1::PDGFRA (n = 37) | Other mutation (n = 19) | No mutation (n = 9) | |
|---|---|---|---|
|
| |||
| Demographics | |||
| Median age (range) | 45 (17–77) | 54 (27–81) | 38 (17–74) |
| Male sex (%)* | 36 (97) | 8 (42) | 6 (67) |
| Race (%) | |||
| African American | 11 (30) | 2 (11) | 1 (11) |
| Asian | 3 (8) | 2 (11) | 0 (0) |
| Multiple races | 2 (<1) | 0 (0) | 8 (89) |
| White | 1 (<1) | 15 (79) | 0 (0) |
| Hispanic | 23 (62) | 0 (0) | 0 (0) |
| Laboratory parameters | |||
| Geo mean peak AEC, cells × 109/L (range) | 12.59 (3.36–97.00) | 7.32 (1.56–33.00) | 12.72 (3.94–113.75) |
| Geo mean peak serum B12, pg/mL (range)** | 2,823 (467–27529) | 1,413 (388–5,157) | 932 (289–3,109) |
| Serum B12 ≥2,000 pg/mL (%)* | 26/30 (87) | 9/19 (47) | 3/7 (43) |
| Geo mean peak serum tryptase, ng/mL (range)** | 23.9 (4.3–58.0) | 17.2 (3.2–629.0) | 7.2 (1.0–15.7) |
| Serum tryptase ≥12 ng/mL (%)* | 18/28 (64) | 5/18 (28) | 1/7 (14) |
| Organ involvement | (n = 35)*** | (n = 19) | (n = 9) |
| Dermatologic (%) | 19 (54) | 10 (53) | 3 (33) |
| Gastrointestinal (%) | 11 (31) | 7 (37) | 1 (11) |
| Pulmonary (%) | 24 (69) | 9 (47) | 5 (56) |
| Cardiac (%) | 13 (37) | 4 (21) | 3 (33) |
| Neurologic (%) | 8 (23) | 5 (26) | 2 (22) |
| Thromboembolic (%) | 5 (14) | 3 (16) | 3 (33) |
| Splenomegaly (%) | 19 (54) | 11 (58) | 3 (33) |
| Imatinib responsiveness* (yes/no/not tried) | 33/0/4 | 4/6/9 | 5/2/2 |
p < 0.001, Fisher’s exact test, PDGFRA-positive group compared to the two other groups combined.
p = 0.01, Kruskal-Wallis test.
Data were not available for 2 participants.
Both RT-PCR and FISH testing for FIP1L1::PDGFRA were performed in 39 of the 65 patients with presumed or confirmed HESN (online suppl. Table 1). Twelve were positive for FIP1L1::PDGFRA in both tests, and 8 were positive only by RT-PCR (p = 0.008; two-sided McNemar test). The remaining 19 patients were negative in both tests. An illustrative case follows.
Case Report
A 66-year-old male with a history of congestive heart failure and refractory arrhythmias without evidence of coronary artery disease, multiple thromboembolic episodes despite anticoagulation, splenectomy, thyroidectomy, and peripheral eosinophilia underwent orthotopic heart transplantation. On gross examination, his cardiac explant showed biventricular dilation and hypertrophy. Microscopically, severe atherosclerosis, not appreciated on prior catheterization, was noted in portions of the right coronary and left anterior descending arteries, with “patchy subendocardial fibrosis in the lateral and posterior left ventricle” and “sporadic hypereosinophilia” in both ventricles, consistent with eosinophilic endomyocardial fibrosis. Bone marrow biopsy was performed 2 years posttransplant to assess persistent eosinophilia and was notable for moderate cellularity with 30% eosinophils and Pelger-Huet cells (subsequent exome sequencing revealed no evidence of a pathogenic variant in LBR). At 8 years posttransplant, his AEC had risen to 20.45 × 109/L prompting a second bone marrow biopsy, which was normocellular with marked eosinophilia and spindle-shaped mast cells. Cytogenetics were normal, FISH testing for FIP1L1::PDGFRA (CHIC2) and translocations involving PDGFRB and FGFR were negative, as was PCR testing for D816V KIT and V617f JAK2. He was treated with prednisone 40 mg daily without response. Hydroxyurea (up to 2.5 g daily) was added but discontinued due to neutropenia and lack of efficacy.
He was referred to the National Institutes of Health for further evaluation. At the time, he was relatively asymptomatic on no specific therapy for HES but mycophenolate 1.5 g orally twice daily and tacrolimus 3 mg orally twice daily for immunosuppression posttransplant. Laboratory testing showed leukocytosis (12.83 × 109/L) with eosinophilia and basophilia (5.95 × 109/L and 0.26 × 109/L, respectively), anemia (Hgb 11.4 g/dL), thrombocytosis (platelets 565 × 109/L), elevated serum tryptase level (40.4 ng/ mL; normal <11.5 ng/mL), and normal serum B12 level. Blood eosinophils showed atypical nuclear lobulation and uneven granulation, and rare immature forms were noted. Bone marrow biopsy was hypercellular (70%) with moderate fibrosis (2–3+/4), marked eosinophilia with left shift, and increased scattered spindle-shaped CD2negCD25pos mast cells without aggregates. The aspirate differential was notable for 24% eosinophils and 1% CD34+ blasts. Taken together, these findings were suggestive of a chronic eosinophilic myeloid neoplasm (HESN). Testing for FIP1L1::PDGFRA was positive in the bone marrow and blood by RT-PCR. He was treated with prednisone 60 mg daily for 3 days (for prevention of imatinib-associated myocardial necrosis) followed by imatinib 400 mg daily with rapid and sustained response. Repeat bone m arrow performed 2 months after the initiation of imatinib was normocellular for age (30–40%) without appreciable eosinophilia, mastocytosis, or reticulin fibrosis. His disease manifestations were well controlled, and he remained in complete hematologic and molecular remission for 23 months on imatinib therapy until his sudden death of unknown cause. No autopsy was performed.
Discussion
Although definitive proof is lacking, it is likely that the cardiac findings that led to transplant in this patient were the result of delayed diagnosis of FIP1L1::PDGFRA-associated HESN. To investigate the role of false-negative FISH testing in diagnostic delay, the time from the first test for FIP1L1::PDGFRA to the initiation of imatinib therapy was compared between FIP1L1::PDGFRA-positive patients with an initial negative FISH result and those whose initial test (FISH or RT-PCR) was positive. As shown in the lower panel of Figure 1, a negative initial FISH test delayed the median time to treatment by 257 days (p < 0.001; Mantel-Cox test). This difference was not due to delays in considering the diagnosis, since the geometric mean time from first AEC >1.5 × 109/L to performance of the first test was similar between patients irrespective of the results of the initial testing (102 days vs. 34 days, p = 0.2, Mann-Whitney test; Fig. 1, upper panel).
Fig. 1.
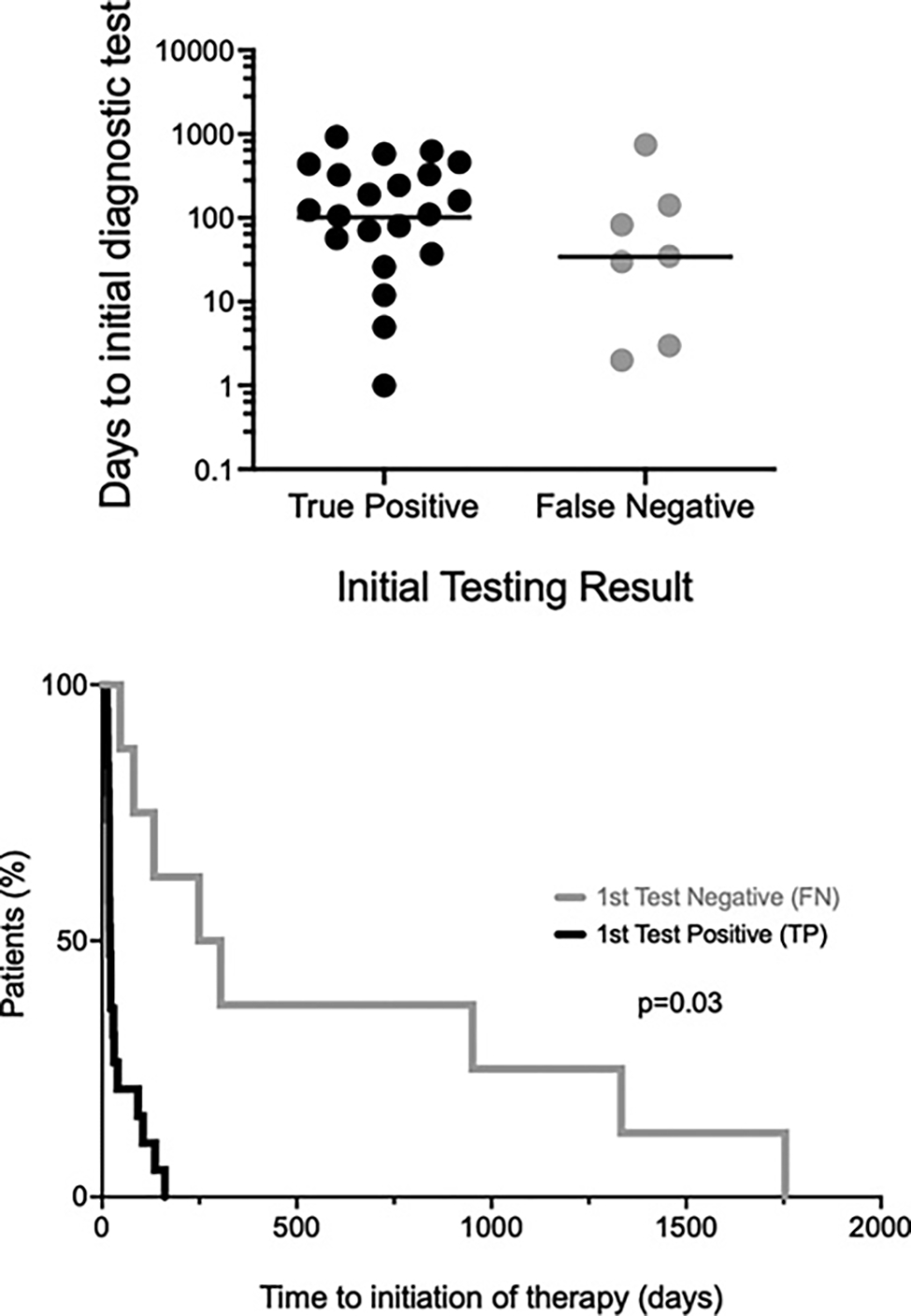
False-negative testing for FIP1L1::PDGFRA by fluorescence in situ hybridization leads to diagnostic delay. The time in days to first test (FISH or PCR) for FIP1L1::PDGFRA following documentation of AEC >1.5 × 109/L are shown in the upper panel for participants whose initial test result was positive (n = 21; black) compared to those with an initial false-negative result (n = 7; gray). Ten patients were excluded from the analysis because testing was not available at the time of their initial AEC >1.5 × 109/L. The horizontal lines represent geometric mean values. The time from first test to initiation of imatinib is shown for the same two groups in the bottom panel, expressed as the % of patients initiating imatinib over time (p = 0.03, Mantel-Cox test). Eleven patients, all of whom had initial positive tests, were excluded because they were not treated with imatinib or were treated prior to the discovery of FIP1L1::PDGFRA.
Neither clinical nor demographic features were different between the patients who tested positive or false-negative by FISH (Table 2). Of note, 9/12 (75%) of the FISH-positive patients had testing performed on a bone marrow sample compared to 4/8 (50%) of those who had a false-negative test. Although this difference was not statistically significant (and the two FISH-positive patients who underwent testing in both bone marrow and peripheral blood had positive tests in both samples), a higher percentage of cells expressing FIP1L1::PDGFRA in the bone marrow compared to peripheral blood has been suggested as a potential explanation for the relative insensitivity of FISH testing in peripheral blood [7] and could have contributed to the false negative results in some of the patients in the current series. To explore this further, we interrogated whole-genome sequences from FISH-positive and negative HES cases for the variant allele fraction (VAF) of the PDGFRA-FIP1L1 fusion. Assuming one mutated allele in an otherwise diploid background, the VAF serves as a measure of tumor content in the sample. Whole-genome sequence was available for 15 of the 20 PDGFRA-FIP1L1-positive patients, including 11 of the patients who underwent both FISH and PCR testing, and, as expected [3, 4, 7], revealed variable breakpoints inside the FIP1L1 locus. Breakpoint data were available for 7 patients who underwent both FISH and PCR testing and showed no obvious differences between the two groups (Table 2). FISH-positive cases (n = 6) had significantly higher fusion VAFs (median 0.36) compared to FISH-negative cases (n = 5, median 0, Mann-Whitney U test p = 0.03) (Fig. 2). Although the sequencing depth limits the detection sensitivity of the fusion [8], this result suggests that samples with relatively low tumor cell content are more likely to lead to false-negative FISH results.
Table 2.
Clinical and molecular findings in patients with positive versus false-negative FISH testing for FIP1L1::PDGFRA
| FISH+PCR+ | FISH-PCR+ | |
|---|---|---|
| FIP1L1::PDGFRA | FIP1L1::PDGFRA | |
| (n = 12) | (n = 8) | |
|
| ||
| Demographics | ||
| Median age (range) | 49 (17–60) | 41 (32–66) |
| Sex (M/F) | 11/1 | 8/0 |
| Laboratory parameters | ||
| Geo mean peak AEC, cells × 109/L (range) | 17.57 (2.26–97.0) | 9.26 (3.49–20.9) |
| Source of FISH sample | ||
| Bone marrow only | 7 | 4 |
| Blood only | 3 | 4 |
| Both | 2 | 0 |
| Fusion breakpoint (Hg19)* | chr4:54289693 | chr4:54270316 |
| chr4:54261987 | chr4:54284574 | |
| chr4:54271718 | ||
| chr4:54302694 | ||
| chr4:54302694 | ||
| Variant allele frequency** | 0.36 | 0 |
Fusion breakpoint analysis was available for 7 subjects.
p = 0.03.
Fig. 2.
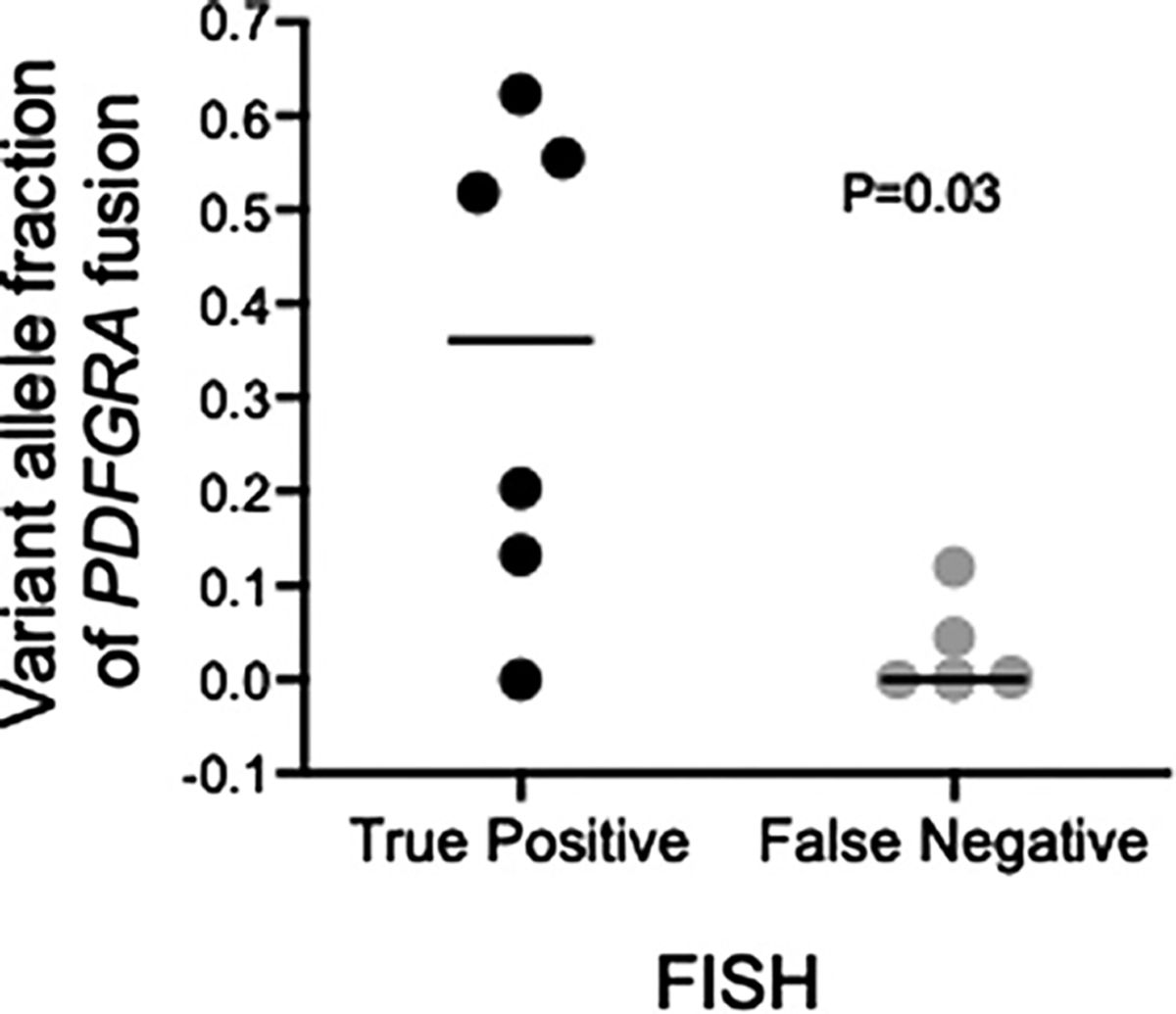
FIP1L1::PDGFRA fusion variant allele fraction (VAF; number of reads supporting the breakpoint divided by total read coverage at the locus) from whole-genome sequencing for FISH-positive (black) and FISH-negative (gray) cases. The VAF is calculated as the fraction of reads supporting the variant divided by the total reads at the breakpoint. Given the mean sequencing coverage, this approach had a >90% power to detect a VAF of 0.04–0.08 in the samples without fusion detected by WGS [8]. Each dot represents one sample. Horizontal bars indicate the median values for each group. p value calculated with the Mann-Whitney U test.
Imatinib response rates in published series of presumed PDGFRA-negative patients with HES range from 9 to 60% [6, 9–12], suggesting that mutations in imatinib-sensitive genes are common in this patient population. Although PDGFRB fusion genes and PDGFRA fusion genes involving partners other than FIP1L1 may account for a small proportion of these cases [13–15], our data and the data from one prior study comparing the results of RT-PCR and FISH testing [16] suggest that false-negative FISH testing for FIP1L1::PDGFRA is likely to be a much more common explanation. Moreover, whereas the prior study did not provide data on clinical outcomes, our study clearly demonstrates that a significant delay in diagnosis and initiation of imatinib therapy in the setting of a false-negative FISH test can, as in the case described, result in life-threatening eosinophilic complications. The fact that some patients with HES and features suggestive of a myeloid neoplasm have no mutation detected by either FISH or RT-PCR but respond dramatically and completely to imatinib further highlights the importance of empiric imatinib therapy, especially in male patients with elevated serum tryptase and/or B12 levels.
Supplementary Material
Funding Sources
This work was supported in part by the Intramural Research Program of NIAID. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261201500003I or 75N91019D00024. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Footnotes
Conflict of Interest Statement
None of the authors report conflicts of interest.
Statement of Ethics
This study protocol was reviewed and approved by NIH Institutional Review Board, protocol number 94-I-0079. Written informed consent was obtained from all participants. Written informed consent from the patient described in the case report for publication of the details of his medical case was obtained prior to his death.
Data Availability Statement
Some of the data that support the findings of this study are not publicly available due to their containing information that could compromise the privacy of research participants but are available from the corresponding author upon reasonable request.
References
- 1.Valent P, Klion AD, Roufosse F, Simon D, Metzgeroth G, Leiferman KM, et al. Proposed refined diagnostic criteria and classification of eosinophil disorders and related syndromes. Allergy. 2023;78(1):47–59. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gotlib J, Cools J. Five years since the discovery of FIP1L1-PDGFRA: what we have learned about the fusion and other molecularly defined eosinophilias. Leukemia. 2008;22(11):1999–2010. [DOI] [PubMed] [Google Scholar]
- 3.Klion AD, Noel P, Akin C, Law M, Gilliland DG, Cools J, et al. Elevated serum tryptase levels identify a subset of patients with a myeloproliferative variant of idiopathic hypereosinophilic syndrome associated with tissue fibrosis, poor prognosis, and imatinib responsiveness. Blood. 2003;101(12):4660–6. [DOI] [PubMed] [Google Scholar]
- 4.Cools J, DeAngelo DJ, Gotlib J, Stover EH, Legare RD, Cortes J, et al. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N Engl J Med. 2003;348(13):1201–14. [DOI] [PubMed] [Google Scholar]
- 5.Constantine GM, Ware J, Brown T, Thumm L, Kamal N, Kumar S, et al. Platelet-derived growth factor receptor-alpha-positive myeloid neoplasm presenting as eosinophilic gastrointestinal disease. J Allergy Clin Immunol Pract. 2020;8(6):2089–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khoury P, Desmond R, Pabon A, Holland-Thomas N, Ware JM, Arthur DC, et al. Clinical features predict responsiveness to imatinib in platelet derived growth factor receptor alpha-negative hypereosinophilic syndrome. Allergy. 2016;71(6):803–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Score J, Walz C, Jovanovic JV, Jones AV, Waghorn K, Hidalgo-Curtis C, et al. Detection and molecular monitoring of FIP1L1-PDGFRA-positive disease by analysis of patient-specific genomic DNA fusion junctions. Leukemia. 2009;23(2):332–9. [DOI] [PubMed] [Google Scholar]
- 8.Rheinbay E, Meifang Q, Bouyssou JM, Oler AJ, Thumm L, Makiya M, et al. Genomics of PDGFR-rearranged hypereosinophilic syndrome. bioRxiv. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Metzgeroth G, Walz C, Erben P, Popp H, Schmitt-Graeff A, Haferlach C, et al. Safety and efficacy of imatinib in chronic eosinophilic leukaemia and hypereosinophilic syndrome: a phase-II study. Br J Haematol. 2008; 143(5):707–15. [DOI] [PubMed] [Google Scholar]
- 10.Ogbogu PU, Bochner BS, Butterfield JH, Gleich GJ, Huss-Marp J, Kahn JE, et al. Hypereosinophilic syndrome: a multicenter, retrospective analysis of clinical characteristics and response to therapy. J Allergy Clin Immunol. 2009;124(6):1319–25.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jain N, Cortes J, Quintás-Cardama A, Manshouri T, Luthra R, Garcia-Manero G, et al. Imatinib has limited therapeutic activity for hypereosinophilic syndrome patients with unknown or negative PDGFRalpha mutation status. Leuk Res. 2009;33(6):837–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baccarani M, Cilloni D, Rondoni M, Ottaviani E, Messa F, Merante S, et al. The efficacy of imatinib mesylate in patients with FIP1L1-PDGFRalpha-positive hypereosinophilic syndrome. Results of a multicenter prospective study. Haematologica. 2007;92(9):1173–9. [DOI] [PubMed] [Google Scholar]
- 13.Fang H, Tang G, Loghavi S, Greipp P, Wang W, Verstovsek S, et al. Systematic use of fluorescence in situ hybridization (FISH) and clinicopathological features in the screening of PDGFRB rearrangements of patients with myeloid/lymphoid neoplasms. Histopathology. 2020;76(7):1042–54. [DOI] [PubMed] [Google Scholar]
- 14.Jan M, Grinshpun DE, Villalba JA, Dal Cin P, Sykes DB, Iafrate AJ, et al. A cryptic imatinib-sensitive G3BP1-PDGFRB rearrangement in a myeloid neoplasm with eosinophilia. Blood Adv. 2020;4(3):445–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reiter A, Gotlib J. Myeloid neoplasms with eosinophilia. Blood. 2017;129(6):704–14. [DOI] [PubMed] [Google Scholar]
- 16.Olsson-Arvidsson L, Norberg A, Sjögren H, Johansson B. Frequent false-negative FIP1L1-PDGFRA FISH analyses of bone marrow samples from clonal eosinophilia at diagnosis. Br J Haematol. 2020;188(5):e76–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Some of the data that support the findings of this study are not publicly available due to their containing information that could compromise the privacy of research participants but are available from the corresponding author upon reasonable request.


