Abstract
Objective:
Colitis-associated cancer (CAC) treatment lacks effective small-molecule drugs and efficient targeted delivery systems. Here, we loaded M13 (an anti-cancer drug candidate) to colon-targeting ginger-derived nanoliposomes (NL) and investigated if orally administered M13-NL could enhance the anticancer effects of M13 in CAC mouse models.
Methods:
The biopharmaceutical properties of M13 were assessed by physicochemical characterizations. The in vitro immunotoxicity of M13 was assessed against PBMCs using FACS and the mutagenic potential of M13 was evaluated by the Ames assay. The in vitro efficacy of M13 was tested in 2D- and 3D-cultured cancerous intestinal cells. AOM/DSS-induced CAC mice were used to evaluate the therapeutic effects of free M13 or M13-NL on CAC in vivo.
Results:
M13 has beneficial physiochemical properties, including high stability, and no apparent immunotoxicity or mutagenic potential in vitro. M13 is effective against the growth of 2D- and 3D-cultured cancerous intestinal cells in vitro. The in vivo safety and efficacy of M13 were significantly improved by using NL for drug delivery (p < 0.001). Oral administration of M13-NL exhibited excellent therapeutic effects in AOM/DSS-induced CAC mice.
Conclusion:
M13-NL is a promising oral drug formulation for CAC treatment.
Keywords: biopharmaceutical properties, colon-targeted drug delivery, glutathione conjugation, self-assembled Nanoliposomes
1. Introduction
Colitis-associated cancer (CAC) is a type of colorectal cancer induced by inflammatory bowel disease (IBD), which is a chronic disease characterized by disruption of the intestinal epithelial barrier and influx of immune cells [1,2]. Two clinical forms of IBD are ulcerative colitis and Crohn’s disease). The risk for colorectal cancer is at least 2-fold higher in ulcerative colitis patients compared with the normal population [3,4]. Current clinically available therapies for CAC often require intravenous injections of anti-inflammatory and/or anticancer drugs at high dosages, resulting in systemic drug biodistribution [5]. Long-term treatment of CAC, which needs repeated intravenous injections, affects patient compliance and leads to potential systemic toxicity, such as allergic reactions and nausea [6]. Therefore, there is a considerable need for a safe and effective therapeutic strategy to achieve long-term targeted delivery of drugs to the diseased intestinal tissues of CAC.
Compared to parenteral routes (intravenous, subcutaneous, and intramuscular injections), oral administration of therapeutic drugs to treat CAC has the potential of localizing the drugs to the diseased gastrointestinal tract without affecting other healthy organs [7]. The oral approach not only improves safety and efficacy but also has the advantage of treatment convenience, which increases patient compliance [8]. Oral delivery is therefore ideal for managing chronic intestinal diseases, including IBD and CAC. The challenge for oral drugs is that most chemotherapeutic drugs suffer from poor water solubility and high metabolic instability, and an efficient targeted drug delivery system is lacking. Two major approaches to overcoming these challenges of oral drug delivery include modifying the drug structure to improve its properties and loading the drug into a targeted delivery system. However, modifying the structure may induce unpredictable toxicity and a delivery system should be designed with consideration of its biocompatibility.
Our laboratory and other groups have shown that the natural product, 6-shogaol, is a potent anti-inflammatory and anticancer lead compound, but that its biopharmaceutical properties are poor (high metabolic instability and limited water solubility). To improve these properties of 6-shogaol, we synthesized the tripeptide-conjugated 6-shogaol, M13 (5-glutathione-6-shogaol). M13 is a major phase II metabolite of 6-shogaol in vivo; it is a detoxified product that is generated via hepatic metabolism [9,10]. Given that phase II hepatic metabolism converts lipophilic 6-shogaol into amphiphilic M13, our first hypothesis was that M13 might offer better biopharmaceutical properties and biosafety than its lead compound, 6-shogaol.
To implement the oral drug treatment of M13, we employed non-toxic plant-derived lipid nanoparticles as a delivery system. Plant-derived nanoparticles from broccoli, grapefruit, and ginger have the advantages of offering biocompatibility (low immunogenicity) and easy large-scale production [11]. Orally administered ginger-derived nanoparticles (GDNPs) were previously found to target the colon and reduce acute and chronic intestinal inflammation in mice [12]. Here, we extracted total lipids from GDNPs and reassembled them into nanoliposomes (NLs) [13]. Such NLs have been shown to target the colon with encapsulated cargos (such as doxorubicin, 6-shogaol, and siRNA-CD98) upon oral administration, improving the efficacy of the loaded therapeutics [13–15]. Our second hypothesis was that loading M13 into NL (M13-NL) would further improve the beneficial properties of M13 by enhancing its bioavailability and avoiding side effects, and that M13-NL will be a potent and safe formulation to treat CAC.
To test these hypotheses, we investigated the physicochemical properties of M13 and tested its anticancer activities in 2D- and 3D-cultured cancerous intestinal cells. Further, we evaluated the safety and efficacy of orally administered M13-NL on the azoxymethane/dextran sulfate sodium (AOM/DSS)-induced CAC mouse model.
2. Materials and methods
2.1. Chemical synthesis and physicochemical characterization of M13
Chemical synthesis of M13 was carried out as previously described by Zhu et al with some improvements (Supplementary Figure S 1) [9]. Briefly, NaHCO3 (catalyst, 0.036 mmol) was added to a mixture of 6-shogaol (0.36 mmol) and reduced L-glutathione (2.18 mmol) in methanol/water (30 mL, 1:1, v/v). The mixture was stirred at room temperature for 12 h. The final residue was purified by column chromatography (C18 Cartridges) with 70% acetonitrile in water, and the M13 (white powder) was obtained after concentration and freeze-drying. The purity of M13 was verified by HPLC method as described previously [13,16]. A C18 column (2.1×50 mm, 5 μm, 80 A) was employed for the separation, and the UV detection wavelength was set at 282 nm. The structure of M13 was verified using a Q-TOF mass spectrometer (ESI-Q-TOF/MS).
The kinetic solubility of M13 in pure water was determined as described in previous literature by Saal and Petereit [17]. A weighed of M13 (3 mg, 6 mg, and 12 mg, respectively) was dissolved in 100 mL pure water, and then 200 μL mixture solutions were pipetted into one well of a 96-well plate. Following shaking of the suspension on an orbital shaker at 600 rpm for 24 h at room temperature, it was separated by centrifugation (1000 g for 3 min). Immediately after the filtration step, 50 μL of the filtrate was transferred into 50 μL of a 1:1 (v:v) mixture of DMSO with phosphate buffer pH 7.4. The concentration of M13 was determined by HPLC with UV detection and calibrated by a 3-standard curve (1, 20, 200 μM) using an external standard from the same batch of M13.
The thermodynamic (or equilibrium) solubility of M13 in pure water was determined as described below. About 2.5 mg of M13 was added into the lower chamber of miniuniprep vials, and 450 μL of pure water was added into each chamber. After water addition, filter pistons of miniuniprep vials were placed and compressed to the position of the liquid level to allow for the contact of water and compound with the filter during incubation. The thermodynamic solubility samples were vortexed for 2 min. Incubation was carried out at room temperature for 24 h with shaking at 600 rpm. Miniunipreps were compressed to prepare the filtrates for injection into the HPLC system. All vials were inspected for visible undissolved material before filtering and for leakage after filtering. Dilute supernatant with water by a factor of 100 folds to make diluents. Three UV standard solutions were injected into HPLC from low to high concentration (M13; 1, 20, 200 μM), followed by testing of the diluents and supernatant. Testing M13 samples were injected in duplicate.
The values of ionization/protonation constants (pKa) of M13 were determined by UV metric method (with co-solvent) and pH metric method (with co-solvent) and performed on a pKa log P/D tester. For the UV metric method, 5 μL M13 solution (10 mmol/L in DMSO) and 25 μL UV buffer were added into a sample vial, 1.5 mL of 80% (v/v) MeOH (dissolve about 5.59 g KCl in 100 mL deionized water and fill up to 500 mL with analytical grade MeOH) was added into the sample vial automatically. The sample solution was pre-acidified with 0.5 M HCl by the instrument automatically, then titrated three times from low to high pH (2 to 12) with 0.5 M KOH; data was refined to get the aqueous pKa value. For the pH metric method (with co-solvent), about 1 mg of M13 powder was weighed and added into a sample vial, then 1.5 mL 80% (v/v) MeOH was added into the vial automatically. The sample solution was pre-acidified with 0.5 M HCl by the instrument, then titrated three times from low to high pH (2 to 12) with 0.5 M KOH; the data was refined to get the aqueous pKa value.
The 1-octanol-phosphate buffer (pH 7.4) distribution coefficient (Log D7.4) of M13 was determined by a shake-flask method at room temperature. Briefly, the 500 mL of 1-octanol and 500 mL of phosphate buffer (pH 7.4) were saturated mutually and stood for 24 hours to separate, the bottom layer was labeled as 1-octanol saturated buffer; the upper layer was labeled as buffer-saturated 1-octanol. About 9.65 mg of Labetalol powder was placed into a 50 mL centrifuge tube, and 9.65 mL DMSO was transferred into the above tube to prepare the internal standard (IS). Then 402 μL of IS was dissolved in 50 mL of 1-octanol saturated buffer and incubated with 949 mL of 50% methanol-water solution to prepare the 1-octanol IS solution (used as the blank); 421 μL of IS was dissolved in 5.263 mL of buffer-saturated 1-octanol and incubated with 994 mL of 50% methanol-water solution to prepare the IS buffer (pH 7.4) solution. The M13 solution (10 mM in DMSO; 2 μL/well) was transferred to the 96-well polypropylene cluster tubes. Buffer-saturated 1-octanol (150 μL/well) and 1-octanol saturated buffer (150 μL/well) were added to each well. Each of the tubes was vigorously mixed on its sides for 1 min and then shaken for 1.1 h at a speed of 600 rpm at room temperature and centrifuged at 3500 g for 10 min. We diluted the buffer layer sample by a factor of 20 fold and the 1-octanol layer sample by a factor of 200 fold with IS solution. The M13 content in both the layers was measured by liquid chromatography-tandem mass spectrometry (LC-MS/MS) in multiple reaction monitoring (MRM) mode and Log D7.4 was calculated using the formula:
2.2. Ames test (Salmonella typhimurium reverse mutation assay) of M13
The Ames test was conducted using a modification of Organization for Economic Cooperation and Development (OECD) Guideline 471 [18]. Two Salmonella typhimurium histidine auxotrophs (TA98 and TA100) were prepared by Creative Proteomics, Inc. Tester strain TA98 is reverted from histidine dependence (auxotrophy) to histidine independence (prototrophy) by frameshift mutagens. Tester strain TA100 is reverted by mutagens that cause both frameshift and base pair substitution mutations. The experiments were carried out using the S. typhimurium TA100 and TA98 in the presence and absence of a post-mitochondrial supernatant (S9 fraction) prepared from the phenobarbital-induced Sprague Dawley rat liver. Prior to each test, about 50 μL of frozen bacterial stock suspension was inoculated into 10 mL of Nutrient Broth No. 2 and grown overnight at 37 °C in an incubator shaker.
About 100 μL of bacteria were mixed with 100 μl of M13 solution (in PBS) with different contents (156, 312, 625, 1250, 2500, and 5000 μg/plate) and 0.5 ml S9 mix or 0.5 ml PBS were pre-incubated in sealed 0.75-ml tubes at 37 °C for 20 min, then were mixed with 2 mL top agar complemented with a histidine solution and poured directly onto minimal glucose agar plates. The plates were solidified at room temperature before incubation at 37 °C for 72 h. The bacteria without any treatment was used as a negative control. The concurrent strain-specific positive controls employed for the assays without metabolic activation (-S9) were 2-nitrofluorene (2NF, 1 μg/plate) for TA98, and sodium azide (NaN3, 1 μg/plate) for TA100. The concurrent positive controls used for the assays with metabolic activation (+S9) were 2-aminoanthracene (2AA, 1 μg/plate) for TA98 and 2AA (2 μg/plate) for TA100. About 100 μL of PBS served as the vehicle control (i.e. PBS group) for all tests with or without S9 metabolic activation. The colony numbers on the plates of each group were determined by manual counting. The mean number of revertants per plate, the standard deviation, and the mutation factor (i.e. mean numbers of revertants on the test item plate/mean number of revertants on the vehicle control plate) values were calculated for each group. All tests were conducted using triplicate plates for each concentration in each experiment. There were two independent experiments for the tests of negative control, positive control, and PBS groups.
2.3. In vitro evaluation of safety and toxicity (InVEST) assay of M13
InVEST assay was performed to investigate the effects of M13 against 25 targets of 7 target families that may cause adverse drug reactions in humans including G-protein-coupled receptors (GPCRs), ion channels, phosphodiesterase, protease, and nuclear receptors (Table 1). M13 was tested at the final assay concentration of 20 μM in duplicate. The assay formats include radio ligand filter binding assays for GPCRs and ion channels, enzymatic assays for phosphodiesterase and protease, and fluorescence polarization binding/cell-based activity assays for nuclear receptor AHR. Reference compounds were tested in parallel with each target in full concentration-response (8 or 10 points category response curves) with variable starting concentration and the increment between concentrations. The results were expressed as % activity as compared with the appropriate baseline activity. A value of 100 indicated no effect of M13 on the target.
Table 1.
InVEST test of M13 against 25 targets from five target classes
| Target classes | Targets | Assay format | % Activity1) | Reference compound | IC50 (M) of reference compound | |
|---|---|---|---|---|---|---|
|
| ||||||
| Data 1 | Data 2 | |||||
| GPCR2) | A1 adenosine | Radioligand filter binding | 85 | 87 | CPDPX | 2.61E-09 |
| GPCR | A2A adenosine | Radioligand filter binding | 99 | 100 | CGS 21680 | 5.69E-08 |
| GPCR | alpha1A adrenergic | Radioligand filter binding | 93 | 93 | prazocin | 9.27E-09 |
| GPCR | alpha2A adrenergic | Radioligand filter binding | 92 | 100 | RX 821002 | 1.15E-08 |
| GPCR | beta1 adrenergic | Radioligand filter binding | 120 | 116 | alprenolol | 1.02E-08 |
| GPCR | CCK | Radioligand filter binding | 94 | 92 | lorglumide | 3.78E-09 |
| GPCR | D1 dopamine | Radioligand filter binding | 100 | 116 | SCH 23390 | 9.01E-10 |
| GPCR | D2S dopamine | Radioligand filter binding | 106 | 89 | 7-OH DPAT | 1.97E-08 |
| GPCR | D3 dopamine | Radioligand filter binding | 96 | 100 | 7-OH DPAT | 1.36E-09 |
| GPCR | histamine H1 | Radioligand filter binding | 102 | 99 | pyrilamine | 3.34E-09 |
| GPCR | Muscarinic M1 | Radioligand filter binding | 111 | 113 | pirenzepine | 4.94E-09 |
| GPCR | Muscarinic M2 | Radioligand filter binding | 93 | 115 | AF-DX 384 | 1.79E-08 |
| GPCR | Muscarinic M3 | Radioligand filter binding | 100 | 103 | 4-DAMP | 1.11E-08 |
| GPCR | Opioid delta | Radioligand filter binding | 98 | 104 | DADLE | 6.85E-09 |
| GPCR | Opioid mu | Radioligand filter binding | 98 | 96 | DAMGO | 5.91E-09 |
| GPCR | Opioid kappa | Radioligand filter binding | 95 | 93 | U-69,593 | 8.88E-09 |
| GPCR | 5-HT1A | Radioligand filter binding | 102 | 100 | 8-OH DPAT | 3.71E-09 |
| GPCR | 5-HT1B | Radioligand filter binding | 100 | 102 | GR125743 | 4.85E-09 |
| GPCR | 5-HT2A | Radioligand filter binding | 99 | 98 | ketanserin | 4.66E-09 |
| GPCR | 5-HT2B | Radioligand filter binding | 109 | 113 | rauwolscine | 7.08E-09 |
| Ion channel | 5-HT3 | Radioligand filter binding | 98 | 100 | GR 65630 | 1.09E-09 |
| Phosphodiesterases | PDE3A | Enzymatic activity | 93 | 94 | IBMX | 2.21E-05 |
| Phosphodiesterases | PDE4A | Enzymatic activity | 93 | 94 | IBMX | 3.00E-05 |
| Protease | Thrombin alpha | Enzymatic activity | 96 | 97 | gabexate mesylate | 2.26E-06 |
| Nuclear receptor | AHR-activation | Fluorescence polarization binding/cell-based activity | 100 | 101 | FICZ | 1.05E-08 |
| Nuclear receptor | AHR-inhibition | Fluorescence polarization binding/cell-based activity | 98 | 101 | GNF-351 + 10 nM FICZ | 2.90E-07 |
Results are expressed as % activity as compared with appropriate baseline activity. A value of 100 indicates no effect of test compound. Data 1 and 2 represent 2 repetitions.
GPCR, G-protein-coupled receptor.
2.4. Evaluation of immunotoxicity of M13 on human CD4+ and CD8+ T cell proliferation
The frozen human peripheral blood mononuclear cells (PBMCs) were thawed and resuspended in T-cell expansion media at a cell concentration of 107/mL, transferred to a T75 flask and rested overnight at 37 °C, 5% CO2. The next day, cells will be labeled with proliferation dye, resuspended in T-cell expansion media, and plated in 96-well plates at a density of 1×104 cells/well and incubated overnight. M13 powders were dissolved in DMSO and then diluted 1000 times with culture media to prepare a final test concentration of 20 μM. Cells were then incubated with M13 solution at 37 °C, 5% CO2 for 4 days. Cells with the blank culture medium were used as a standard control. Cells with an equivalent amount of solvent (i.e. culture media with 0.1% DMSO) were used as solvent control (SC). Cells with a CD3/CD28 T cell activator were used as a positive control (25 μL/mL). Interleukin-2 (IL-2), as a stimulator to activate the immune response of PBMCs, was added or not added to each group. Triplicate samples will be initiated for each testing condition. At the end of the 4-day incubation, cells were harvested and stained with antibodies against CD4 [allophycocyanin (APC)] and CD8 (APC/Cy7) and with the viability dye 7-Amino-Actinomycin D (7-AAD). All samples were analyzed on a flow cytometer and the percentage of viable CD4+ and CD8+ T cell subsets and the proliferative response of CD4+ and CD8+ cells were counted.
2.5. Assembly of M13-loaded nanoliposomes (M13-NL)
Ginger-derived natural nanoparticles (GDNPs) were obtained from fresh ginger (Zingiber officinale Rosc.) using the ultracentrifugation method reported in our previous papers [13]. Total lipids extraction was extracted from GDNPs using a modified liquid-liquid extraction method reported by Sung et al [19]. Assembly of M13-NL was carried out using a method reported previously [16]. In general, 1 mL M13 (5.0 mg/mL in ethanol) was mixed with 1 mL total lipids (5.0 mg/mL in dichloromethane) and dried under reduced pressure to obtain a thin lipid-M13 complex film. Then the dried film was suspended in 5 mL PBS buffer. After a bath sonication for 5 min, an equal volume of PBS buffer was added and sonicated for another 5 min. Finally, the suspension was passed through a liposome extruder 20~25 times (at 55 °C) with a 200 nm polycarbonate membrane. Empty NL (as control) was made by the identical protocol without the addition of M13 as described above.
2.6. Cell cultures
PBMCs were maintained in T-cell expansion medium. HIEC-6 cells were maintained in OptiMEM 1 reduced serum medium supplemented with 20 mM HEPES, 10 mM GlutaMAX, 10 ng/mL epidermal growth factor, 4% (v/v) fetal bovine serum (FBS), and 1% penicillin-streptomycin (PS). HCT-116 cells were maintained in McCoy's 5a medium supplemented with 10% (v/v) FBS and 1% PS. Caco-2/BBe cells were grown in DMEM with 10% (v/v) FBS and 1% PS (v/v). Human colon cancer stem cells (HC-CSC) were maintained in complete growth media with serum and antibiotics.
2.7. Cytotoxicity assay in 2D cell cultures
The MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) tetrazolium assay was performed on four different intestinal cell cultures (HIEC-6, HCT-116, Caco-2/BBe, and HC-CSC) in the logarithmic phase to determine the cytotoxicity of free M13 and M13-NL on cancer cells (HCT-116, Caco-2/BBe, and HC-CSC) and normal cell line (HIEC-6). Briefly, cells were seeded in 96-well plates at a density of 1×104 cells/well and incubated overnight. Cells were then incubated with different amounts of free M13, NL, or M13-NL (0, 1, 2, 5, 10, 20, 30, and 40 μg/mL) for 24 h. After M13-, NL- or M13-NL-containing medium was removed, cells were thoroughly rinsed once with PBS. Cells were then incubated with MTT (0.5 mg/mL final concentration in the medium) at 37 °C for 4 h. Thereafter, the medium was discarded and 100 μL dimethyl sulfoxide was added into each well prior to spectrophotometric measurements at 570 nm using a microplate reader. The cell survival curves of 2D cells responding to different concentrations of M13-treated are drawn according to the results of the MTT assay and the standard deviations between 2D cells from the same dosage group (n = 3) are depicted. The extrapolated IC50 values were calculated with GraphPad Prism 8.0 applying a non-linear regression sigmoidal dose-response (variable slope) curve fitting model based on the results of the MTT assay.
2.8. Three-dimensional (3D) printing and culturing of Caco-2/BBe cells
The hydrogel (bio-ink) in a cartridge was allowed to warm up to room temperature before being used. About 1.1 × 107 Caco-2/BBe cells were isolated from the culture flask and suspended in 100 μL media. A total of 1 mL bio-ink was mixed (10:1) with 1.1 × 107 cells/100 μL media (i.e. concentration of approximately 1 × 107 cells/mL) and loaded into a new cartridge for droplet printing by using the BioX 3D printer and according to the instrument manufacturer's protocols. In brief, the 1.1 mL bio-ink/cells mixture was placed in the print head and capped with a 22G (inner diameter: 0.41 mm) printing nozzle. Each bio-ink/cells droplet was printed in a 96-well plate using the Droplet print function at room temperature. A pressure of 25 kPa and a print speed of 5 mm/s was used. After printing, we added 100 μL of crosslinking agent to each well and removed the entire volume after 45 s. Then we washed each well with 200 μL of DMEM to completely remove the crosslinking solution. Then 200 μL of fresh DMEM with 10% (v/v) FBS and 1% PS (v/v) was added to each well and refreshed every 3 to 4 days.
2.9. Cytotoxicity assay in 3D cultured Caco-2/BBe cells
The 3D droplet-based Caco-2/BBe cells were treated with different concentrations of M13 (0, 5, 10, 20, 40, 80, 100, 150, and 200 μM) for 11 days in the 96-well plate, with initial dosing at day 0 (Start day of 3D cultivation) and re-dosing at day 4 and 7. The morphology of the 3D Caco-2/BBe cells was evaluated on days 0, 4, 7, and 11 by bright field images acquired with a confocal fluorescence microscopy. The diameter (denoted by “d”) of each spheroid was measured by the same microscopy during the experiment (n = 4). Spheroid volume (denoted by “V”) was calculated using the formula:
A Live-Dead cell assay kit was used to assess cell viability by measuring and comparing green [live cells stained with acridine orange (AO)] and red [dead cells stained with propidium iodide (PI)] fluorescence signal intensity of 3D Caco-2/BBe cells at day 11. After incubating for 11 days, the DMEM was removed and the 3D Caco-2/BBe cells were washed with warm PBS 3 times. Then 100 μL of fresh DMEM and 2 μL of assay reagent were added to each well and the cells were incubated at 37 °C for another 10 min. Images of 3D Caco-2/BBe cells were acquired using a confocal fluorescence microscopy with quantitative analysis using ImageJ software. The cells without staining were used as a blank control; the cells without M13 treatment were used as a negative control; the cells without M13 treatment and fixed with 70% ethanol before staining was used as a positive control. The survival rate of 3D Caco-2/BBe cells was assessed by calculating the mean fluorescence intensity of green fluorescence in cells of each group treated with different concentrations of M13 relative to the negative control group (n = 4).
The intracellular ATP content of 3D Caco-2/BBe cells on day 11 was measured using a 3D cell viability assay according to the manufacturer’s instructions. By setting the average ATP content of the negative control to 100%, the cell viabilities of M13-treated 3D spheres were expressed as the percentages of the negative control, and the standard deviations between 3D spheres from the same dosage group (n = 4) were depicted. The extrapolated EC50, 95% confidence interval, and the Hill slope values were calculated applying a non-linear regression sigmoidal dose-response (variable slope) curve fitting model based on ATP content.
2.10. In vitro cancer-related protein expressions
Caco-2/BBe cells were cultured to approximately 80% confluence in T25 flasks and incubated with free M13 (5 μg/mL), NL (5 μg/mL), or M13-NL (10 μg/mL, i.e. contain 5 μg/mL M13 and 5 μg/mL NL) for 24 h. Cells with a blank culture medium were used as a negative control. After free M13-, NL-, or M13-NL-containing medium was removed, cells were thoroughly rinsed twice with 10 mL of PBS. Then, 1 mL of lysis buffer containing 10 μg/mL aprotinin, 10 μg/mL leupeptin, and 10 μg/mL pepstatin were added into each tube and rock the lysates gently at 4°C for 30 min. Then, the cell lysates were centrifuged at 14,000 g for 5 min at 4°C. The supernatant was collected in new microtubes.
Cancer-related protein expressions were investigated using a Proteome Profiler Human XL oncology array kit according to the instructions of the supplier. In brief, cell lysates (200 μg) were incubated with each array overnight at 4°C on a rocking platform shaker. After removing the cell lysates and washing the membranes 3 times with wash buffer, the arrays with a detection antibody cocktail were incubated for 1 h at room temperature. Then, 2 mL of streptavidin–HRP solution was added to each membrane for a 30 min incubation, followed by three washes. The labeled protein spots were visualized using Chemi Reagent Mix for a 1 min incubation and then detected by using the ChemiDoc MP imaging system and Image Lab software (version 4.1). The intensities of the resulting spots were calculated with Image Studio Lite software (Version 5.2) and correspond to the mean ± standard deviation of four independent experiments.
2.11. In vitro expression and phosphorylation profiling of cell cycle proteins
Caco-2/BBe cells were cultured and incubated with free M13, NL, or M13-NL for 24 h in the same way as described above. Cells with a blank culture medium were also used as a negative control. After removing the medium, cells were thoroughly rinsed triple with 10 mL of PBS. The preparation of cell lysates was performed using an antibody array assay kit according to the instructions of the supplier. The protein concentration of cell lysates was determined with the Protein Assay Kit performed according to the manufacturer’s protocol.
The cell cycle control phospho-antibody array kit containing 238 highly specific antibodies was used to detect the cell cycle control proteins in the cell lysates of each group. Slide scanning was performed using a GenePix 4000B microarray scanner. Then the intensities of the resulting spots from three independent experiments were calculated and analyzed using the Image Studio™ Lite software. Data were normalized by dividing the average intensity signal of replicate spots by the mean intensity of all antibodies on the array.
2.12. The Single-dose maximum tolerated dose (MTD) oral toxicity study of M13 in vivo
Female CD-1 mice were housed in a clean facility maintained at 22 ± 2 °C and 30–70% relative humidity with a 12-h light/dark cycle. Diet and water were supplied ad libitum. Mice (8-week-old) were randomly allocated to two groups (n=9 per group). The mice of one group were orally administered with a single dose of M13 in PBS solution with 1000 mg/kg at the start point (day 0) and subjected to regular water and diet for 7 days. The mice of the other group were orally administered with PBS and used as a control. All mice are monitored for mortality, clinical signs, body weights, and food consumption during these days. The blood of mice was collected before euthanasia on day 7 and followed for hematological and biochemical analysis. The major organs (small intestine, large intestine, liver, kidney, heart, spleen, and lung) of mice were immediately collected for histological analysis. Mice experiments were performed following ARRIVE guidelines 2.0 and with approval from the Georgia State University Institutional Animal Care and Use Committee (IACUC, Protocol # A20039).
2.13. Hematological and biochemical analysis of the blood
Before euthanasia, the mice were under anesthesia with 3% isoflurane, and about 0.3~0.4 mL of fresh blood was immediately collected via the retro orbital sinus by blood-collecting tubes. Then hematological analyses were performed using an automatic hematology analyzer and biochemical analyses were performed using a biochemical analyzer with a comprehensive diagnostic profile reagent rotor.
2.14. Hematoxylin and eosin (H&E) staining
To conduct histological analysis, the major organs of mice were fixed in 10% neutral-buffered formalin for 2 days followed by embedding in paraffin to make sections. Sections of 4~5 μm thickness were obtained from different organ samples of each group by microtome slicing. After deparaffinization, the sections were subjected to staining with H&E and 4’,6-diamidino-2-phenylindole (DAPI). Images were acquired using confocal fluorescence microscopy.
2.15. In vivo M13 quantification by LC-MS/MS
Female C57BL/6J mouse was fed with 0.2 mL of M13 (0.5 mg/mL in PBS) or M13-NL (constructed by 0.5 mg/mL of M13 and 0.5 mg/mL of lipids in PBS). After two hours, two groups (M13 and M13-NL groups, n=5 per group) of mice were euthanized and the middle parts of the colons were collected and weighed. Then, 10 times (1 mL solution for 100 μg of the colon) of icy cold 70% Acetonitrile (which contains 0.2% formic acid) was added to each sample, and 10 autoclaved glass beads (diameter = 3 mm) were added to each sample tube. All the samples were submitted to a tube tissue homogenizer for 15 min homogenization. The tissue suspension was then passed through a 0.22 nylon spin filter and transferred into a 1.5 mL HPLC injection vial. The filtrated solution was submitted to the LC-MS/MS analysis, the LC-MS interface was set at positive ionization mode (ESI+) and M13 was detected with multiple reaction monitoring (MRM) with ion transition: 584.3→137.0, separation column (2.1 × 50 mm C18), mobile phase: (A) 95% H2O/5% Methanol (add 0.1% Formic acid), (B) Methanol.
2.16. Anti-cancer activities of M13 and M13-NL in AOM/DSS-induced CAC mice
Female C57BL/6J mice were used to establish an orthotopic CAC mouse model. Mice were housed in a clean facility maintained at 22 ± 2 °C and 30–70% relative humidity with a 12-h light/dark cycle. Diet and water were supplied ad libitum. To induce colon carcinogenesis using AOM and DSS, mice (8-week-old) were intraperitoneally injected with 10 mg/kg AOM at the start point (day 0) and subjected to regular water and diet for 7 days. Thereafter, mice received 2.0% DSS in their drinking water every day for 7 days, followed by regular drinking water for the next 14 days to allow for recovery. Then DSS administration was repeated one time for another 7 days. After 8 days adaptation period, mice were randomly allocated to four groups (n=7 per group) with different oral administration treatments every two days: (1) AOM/DSS control group (only AOM/DSS treatment and orally administered PBS), (2) free M13 group [AOM/DSS treatment and orally administered free M13 (5 mg/kg in PBS)], (3) NL group [AOM/DSS treatment and orally administered NL (5 mg/kg in PBS)], and (4) M13-NL group [AOM/DSS treatment and orally administered M13-NL (5 mg/kg M13 loaded into 5 mg/kg NL)]. Mice without any treatment and only oral PBS were used as a wild-type (WT) control group (n=5). The body weights of mice were measured every 3 or 4 days during the study. After 9 doses of treatment, the blood of the mice was collected. Then the mice were euthanized, and the lengths of the colons were measured. The colons were cut open longitudinally and the colon tumor number and size were quantified. Finally, colons and other organs (liver, spleen, kidney, and small intestine) were immediately collected. After the spleen weights were measured, all the organs were rinsed extensively in PBS (pH 7.4) to remove residual blood and internal contents. Experiments were performed following ARRIVE guidelines 2.0 and with approval from the Georgia State University Institutional Animal Care and Use Committee (IACUC, Protocol # A20039).
2.17. Immunofluorescence (IF) and immunohistochemistry (IHC) of tissues
For the AOM/DSS-induced CAC mouse model, the 4 μm sections of colon samples were deparaffinated and used for IF or IHC staining. To evaluate the effect of free M13 or M13-NL treatment on colon tumor growth inhibition, TdT-mediated dUTP notch end labeling (TUNEL) assay was performed on the colon simples manually using fluorometric TUNEL system according to the manufacturer’s standard protocols. The DNA fragmentation was labeled and visualized with fluorescein-12-dUT (FITC). The sample blocks were counterstained with DAPI and imaged by a confocal microscopy with quantitative analysis using ImageJ software. TUNEL-positive cells overlapping with DAPI nuclear staining were counted per crypt. In addition, IHC staining (Ki-67 assay) was performed on the colon samples using an automated immunostainer and a rabbit anti-Ki-67 antibody (as the primary antibody) according to the manufacturer’s standard protocols. Ki-67-positive cells were counted per crypt.
To detect the expression levels of related proteins in colon tumors of different treatment groups, we carried out the IHC staining analysis for survivin, EGFR, BCL-x, Dkk-1, EpCAM, CDC25A, and RAD52 proteins, respectively, by using the sections of colon samples. For the survivin, EGFR, BCL-x, and TROP-1 proteins, all antibodies were HRP (horse radish peroxidase)-conjugated; For the Dkk-1 protein, biotin-conjugated Dkk-1 antibody and HRP-conjugated anti-biotin antibody were used as the primary and secondary antibody, respectively; For the CDC25A protein, Anti-CDC25A mouse monoclonal antibody and HRP-conjugated goat anti-mouse polymer antibody were used as the primary and secondary antibody, respectively; For the RAD52 protein, Anti-RAD52 rabbit polyclonal antibody and HRP-conjugated goat anti-rabbit polymer antibody were used as the primary and secondary antibody, respectively. IHC signal detection was performed using DAB HRP Substrate Kit, according to the manufacturer's instructions. Then slides were stained with hematoxylin counterstain and sealed through dehydration. The antibodies/antibodies detection kits used in this study are listed in Supplementary Table S1. Confocal fluorescence microscopy and ImageJ software Imaging were used for quantitative analysis, respectively. For survivin, CDC25A, and RAD52 proteins, IHC staining-positive cells (i.e. nuclear staining) were counted per crypt; For EGFR, BCL-x, Dkk-1, and EpCAM proteins, IHC staining-positive area of intestinal epithelial cells (i.e. cytoplasmic staining) were counted per crypt.
2.18. Intestinal microbiota analysis
For the AOM/DSS-induced CAC mouse model, feces were collected from mice before sacrifice (on day 60) in the therapeutic experiments as described above. Fecal samples (2 feces/mouse) were placed in individual tubes containing DNA stabilization buffer and shipped for DNA extraction, library preparation, and sequencing by One Codex. Shallow shotgun whole-genome sequencing (WGS) for microbiota analysis was performed by Transnetyx with classification performed by One Codex (https://app.onecodex.com/). Data analysis (the number and relative abundance of taxa, α-diversity, β-diversity) was performed on the platform of One Codex by Compare Analyses server (https://app.onecodex.com/custom-plots). The number and relative abundance of taxa at some rank [e.g., species or operational taxonomic units (OTUs)] were analyzed and compared by the bar graph, heatmap, pie graph, and Venn diagram. α-diversity of the samples was measured by observed species based on OTUs, Shannon index, and Simpson index. β-diversity of the samples was measured by principal coordinates analysis (PCoA) derived from Weighted UniFrac and Bray-Curtis distances among different groups.
2.19. Statistics
Data represent mean values ± standard deviation (SD) from at least 3 independent experiments. One-way ANOVA was used to determine statistical significance. All statistical analyses were performed with GraphPad Prism 8.0. P < 0.05 was considered to be statistically significant.
3. Results
3.1. M13 has excellent biopharmaceutical properties and biosafety
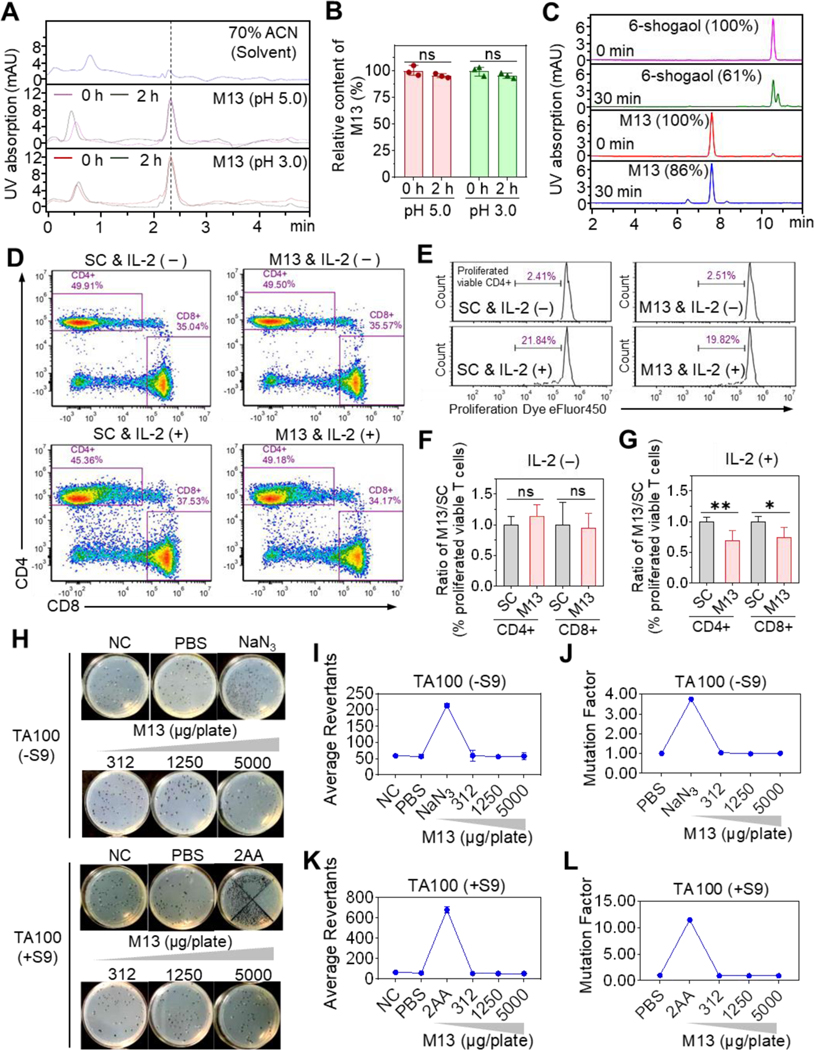
We characterized the biopharmaceutical properties of the synthesized M13 (5-glutathione-6-shogaol). HPLC analysis showed that the relative contents of M13 were consistent before and after incubation at 37 °C for 2 h in acidic PBS (pH = 5.0 or 3.0) (Figures 1A and 1B), indicating that M13 is chemically stable at acidic pHs. After incubation for 30 min with mouse intestinal microsomes, the M13 sample retained 86% of the input, but 6-shogaol retained only 61% of the input (Figure 1C), indicating that M13 exhibits lower metabolic liability than 6-shogaol. M13 also showed high aqueous solubility, as determined by its kinetic solubility (>200 μM) and thermodynamic (or equilibrium) solubility (1390.03 μM), and the partition of M13 between the organic solvent (octanol) and aqueous buffer (characterized with a LogD7.4 of −1.75). The ionization/protonation constants (pKa) were M13 pKa1 = 3.24, M13 pKa2 = 9.12, and M13 pKa3 = 10.42, as determined by UV metric at pH 2–12. This indicates that M13 is only partially ionized at physiological pH (6 to 7), suggesting that it has a high permeability across cell membranes [20,21].
Figure 1. M13 presents excellent biopharmaceutical properties.
(A) HPLC profile of M13 in acidic PBS. (B) Relative M13 content before and after incubation (n = 3). (C) Relative content of M13 or 6-shogaol (determined by HPLC) before and after incubation with mouse intestinal microsomes (n = 3). (D) FACS of cultured PBMCs with gating on viable CD4+ and CD8+ T cells in the solvent control (SC) and M13-treated groups with (+) or without (-) IL-2 addition. (E) Percentage of viable proliferating CD4+ T cells. (F-G) M13-treated-to-SC group ratio of the percentage of proliferating viable T cells with or without IL-2 addition (n=3). (H-L) Representative plate photos (H), colony average revertant numbers (I and K), and mutational factors (J and L) of the Ames test using S. typhimurium strain TA100 with (+S9) or without (-S9) metabolic activation (n = 3). Negative control (NC), no treatment; PBS, vehicle control; NaN3, (1 μg/plate), positive control; 2AA, (2-aminoanthracene, 2 μg/plate), positive control. (ns, not significant; *p < 0.05, **p < 0.01).
To evaluate the immunogenic effects of M13 on human T lymphocytes, we cultured PBMCs in blank medium (standard control), medium containing CD3/CD28 activator (positive control), medium containing solvent (solvent control, SC), and medium containing M13 solution (20 μM). All groups were cultured in the presence or absence of IL-2. After a 4-day incubation, PBMCs were labeled with immunofluorescent dyes and analyzed by flow cytometer. Our results showed that, in resting-stage T lymphocytes (without IL-2 supplementation), the percentages of CD4+ and CD8+ T-cell populations and the T-cell proliferation status were similar in the M13-treated, SC, and standard control groups (Figures 1D–1F, Supplementary Figures S2A and S3). This indicates that M13 treatment is safe for resting lymphocytes. Interestingly, in activated-stage T lymphocytes (with IL-2 supplementation), the M13-treated group exhibited reduced proliferation of activated CD4+ and CD8+ lymphocytes compared with the SC and standard control groups (Figures 1D, 1E, and 1G, Supplementary Figures S2B and S3). This suggests that M13 might suppress CD4+ and CD8+ cells in IBD patients with activated lymphocytes in a fashion similar to the action of anti-TNF-a drugs (e.g., infliximab)[22].
Next, we assessed the mutagenic potential of M13 by using the Salmonella typhimurium reverse mutation assay (the Ames test). Different concentrations of M13 (156 ~ 5000 μg/plate), solvent (PBS), positive controls (NaN3, 2AA, or 2NF), and negative (untreated) controls were tested on two S. typhimurium tester strains (TA98 and TA100) with (+S9) or without (-S9) metabolic activation. We found that M13 did not increase the number of revertant colonies at any tested concentration, and the mutation factors were less than 1.6 for all concentrations of M13 in strains TA98 or TA100 (Figures 1H–1L and Supplementary Figure S4). These results indicate that M13 is not mutagenic.
We also performed a safety screening of M13 using an in vitro evaluation of safety and toxicity (InVEST). We tested the effects of M13 against 25 selected targets, including factors known to be involved in toxicity issues in humans. The results showed that M13 did not inhibit G-protein-coupled receptor (GPCR) targets, an ion channel (5-HT3), a phosphodiesterase, or the protease, thrombin alpha, and did not significantly modulate the activity of the nuclear receptor, AHR (Table 1). Collective, our results indicate that M13 exhibits parameters consistent with excellent safety in vitro.
To further determine the potential acute toxicity of M13 in vivo, we performed a single-dose maximum tolerated dose (MTD) study of M13 in CD-1 mice. Eight-week-old female mice were orally administered with a single dose of M13 (1000 mg/kg) or PBS (as control) at day 0 and subjected to regular water and diet for 7 days (Supplementary Figure S5A). During the experiment, we did not observe any death or difference in the physiological indexes of mice between the M13 and control groups. There was no significant difference in the body weight change, spleen-to-body weight ratio, or blood physiological and biochemical indexes between the two groups of mice on day 7 (Supplementary Figure S5B-S5F), and no pathological sign was found in the anatomical organs. These results demonstrate that oral administration of 1000 mg/kg M13 is safe for CD-1 mice.
3.2. M13 inhibits the growth of 2D- and 3D-cultured cancerous intestinal cells
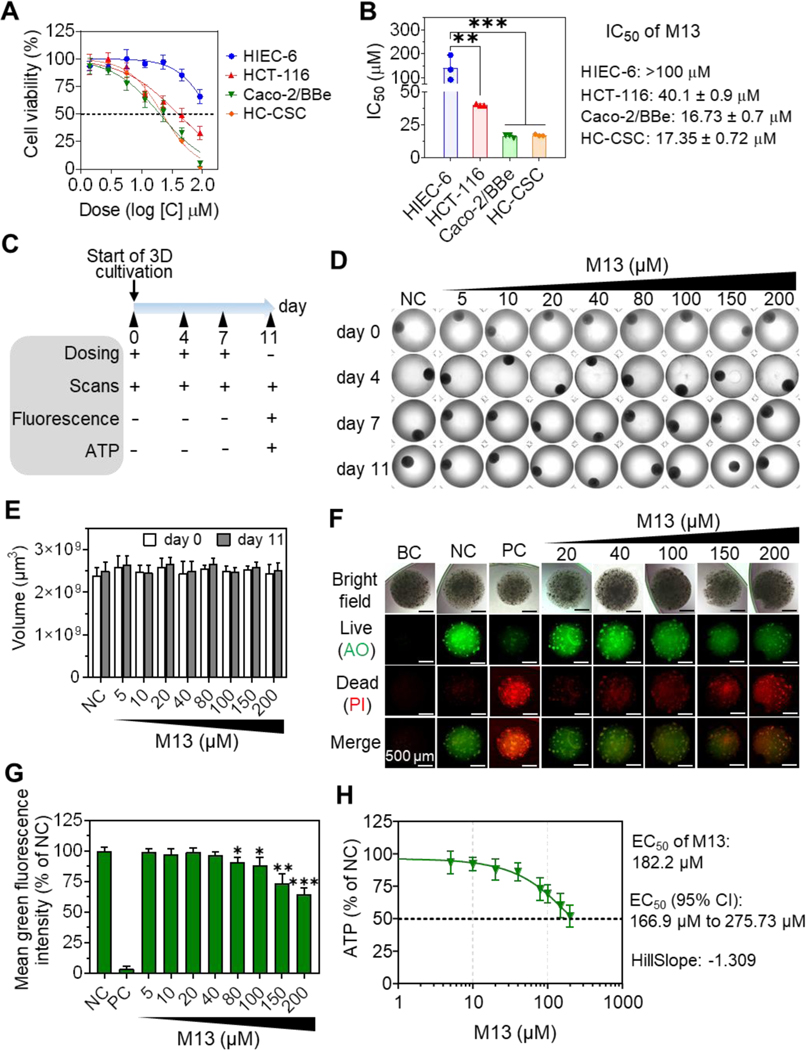
We next used the MTT assay to study the effects of M13 on the viability of four 2D-cultured human intestinal cell lines (HIEC-6, HCT-116, Caco-2/BBe, and HC-CSC). We found that M13 at concentrations ranging from 0.1 to 100 μM dose-dependently decreased the viabilities of all four intestinal cell lines (Figure 2A). Importantly, M13 showed much stronger cytotoxicity toward the cancerous intestinal cell lines (HCT-116, Caco-2/BBe, and HC-CSC) compared to the normal intestinal cell line (HIEC-6) (Figure 2A). As shown in Figure 2B, the IC50 value of M13 against HIEC-6 cells (> 100 μM) was significantly higher (by 2.5-, 6-, and 5.8-fold, respectively) than that of the cancerous HCT-116 (40.1 μM, p < 0.01), Caco-2/BBe (16.73 μM, p < 0.001), and HC-CSC (17.35 μM, p < 0.001) cells. These results demonstrated that M13 preferentially decreases the viability of cancerous intestinal cells over normal intestinal cells.
Figure 2. In vitro anti-cancer effects of M13 in 2D and 3D cultured intestinal cells.
(A) Cell viability curves of cells treated with M13 for 24 h (n = 3). (B) IC50 values (24 h incubation, n = 3). (C) Experimental outline of dosing on droplet-based 3D Caco-2/BBe cells. (+: treatment, -: no treatment) (D) Representative morphologies (Bright-field images) (n = 3). Negative control (NC): no treatment. Well diameter: 6.4 mm. (E) Comparison of the volume changes of 3D Caco-2/BBe cells (n = 4). (F) Representative live/dead cell viability images of 3D cells (acquired on day 11) (n = 4). Blank control (BC), without staining; Positive control (PC), without M13 treatment; AO, acridine orange; PI, propidium iodide. (G) The survival rates of 3D Caco-2/BBe cells (n = 4). (H) Dose-response curve of 3D Caco-2/BBe cells based on ATP contents (extrapolated, n = 4). Error bars represent one standard deviation between two groups or compared with NC (*p < 0.05, **p < 0.01, ***p < 0.001)
To assess the long-term cytotoxicity of M13 against cancer cells, we established a 3D droplet-based Caco-2/BBe cell spheroid model through 3D bioprinting. The diameters of the 3D cell spheres were about 1.65~1.75 mm. The 3D-cultured cells were incubated with different concentrations of M13 for 11 days, and droplet size (calculated using scanned images) and viability (live/dead cell staining and intracellular ATP) were analyzed (Figure 2C).
We observed that as the culture time increased, the color of the 3D cell spheres gradually deepened (Figure 2D and Supplementary Figure S6), but their sizes did not change significantly (Figure 2E), regardless of the group. Live/dead cell staining showed that the number of dead cells (red fluorescence under propidium iodide staining) in the 3D cell spheres was positively correlated with the M13 concentration, and the number of living cells (green fluorescence under acridine orange staining) decreased as the M13 concentration increased (Figure 2F). The cell viability of M13-treated spheres was normalized as a percentage of the negative control (NC, 100% viable, without M13 treatment). This calculation revealed that the green fluorescence intensity of 3D cell spheres treated with M13 at a concentration of ≥ 80 μM was significantly lower (p < 0.05) than that of the NC group (Figure 2G). We further quantified the ATP content in 3D-cultured Caco-2/BBe cells and found that the half-maximal effective concentration (EC50) of M13 was 182.2 μM with a 95% confidence interval (95% CI) (Figure 2H). Together, these results show that Caco-2/BBe cells cultured in 3D spheres are more resistant to M13 than the same cells grown in 2D monolayers, but M13 still clearly exerts a long-term inhibitory effect against the 3D-cultured cancerous intestinal cells.
3.3. M13-NL down-regulates cancer-related proteins and cell cycle proteins in vitro
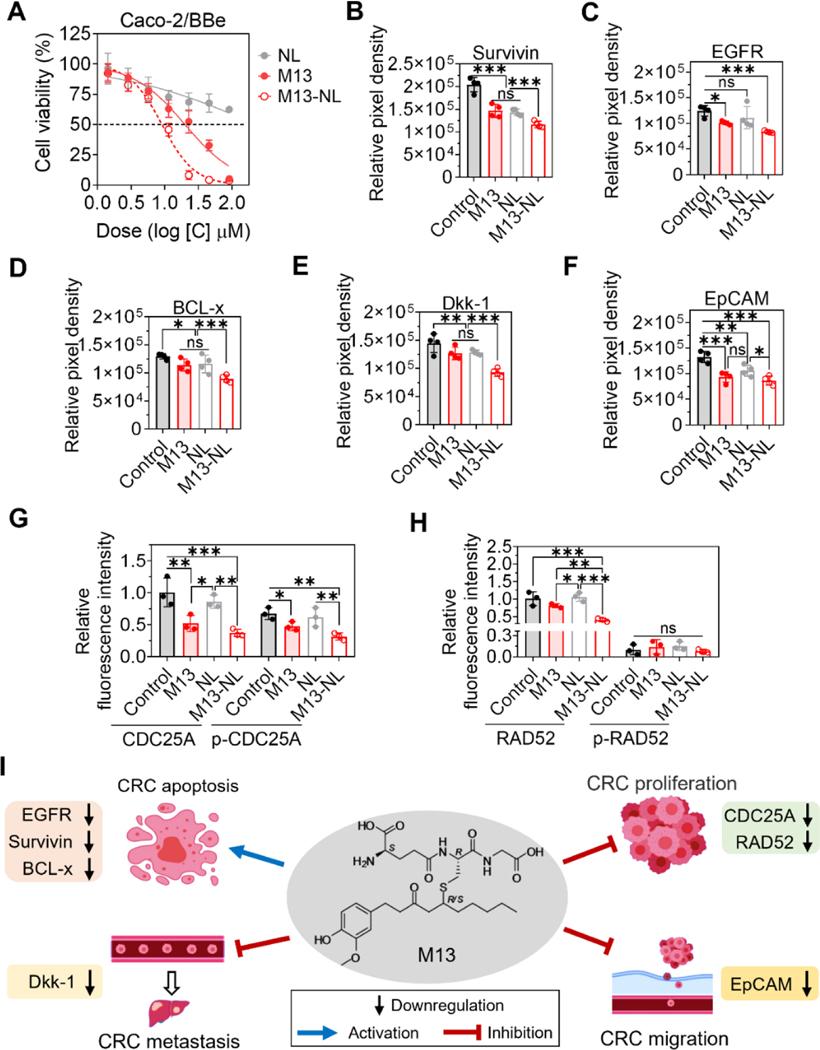
We compared the anticancer activities of free M13, NL, and M13-NL against Caco-2/BBe cells. After a 24-h incubation, M13-NL had stronger cell cytotoxicity than free M13 (Figure 3A), likely reflecting the enhanced uptake of M13-NL by Caco-2/BBe cells. The IC50 value of M13-NL against Caco-2/BBe cells was 8.64 ± 1.02 μM, which was 2-fold lower than that of free M13. Although NL exhibited slight cytotoxicity to Caco-2/BBe cells with increasing concentrations, as expected, the NL group exhibited higher cell viability than the M13 and M13-NL groups (Figure 3A). Overall, NL demonstrated excellent biocompatibility. The above results suggest that NL is a non-toxic drug carrier that can significantly reduce the required drug dose of M13 (p < 0.05), and M13-NL is more potent than free M13 in inhibiting the viability of cancerous intestinal cells in vitro.
Figure 3. Comparisons of anti-cancer effects between M13 and M13-NL and changes of protein expressions in cultured cancerous intestinal cells.
(A) Cell viability of Caco-2/BBe cells treated with free M13, NL, or M13-NL for 24 h (n = 3). (B-H) Point hybridization analysis of the survivin (B), EGFR (C), BCL-x (D), Dkk-1 (E), EpCAM (F), CDC25A and p-CDC25A (G), and RAD52 and p-RAD52 proteins (H) expression in the cell lysates of Caco-2/BBe cells without treatment (control) or treated with free M13 (5 μg/mL), NL (5 μg/mL), or M13-NL (5 μg/mL M13 loaded into NL) (n= 3 or 4). (ns, not significant; *p < 0.05, **p < 0.01, ***p < 0.001). (I) Proposed mechanism of M13 treatment on colorectal cancer cells in vitro.
Next, we sought to identify the mechanism by which M13 exerts its anticancer efficacy. Cell lysates of Caco-2/BBe cells incubated for 24 h with free M13, NL, or M13-NL were hybridized on cancer-related protein array membranes (Proteome Profiler™ Human XL Oncology Array), and cell cycle control/phosphorylation profiling protein antibody microarray slides (Cell Cycle Control Phospho Antibody Array). Representative images of cancer-related protein and cell cycle protein signals are shown in Supplementary Figures S7 and S8, respectively. The results from the human oncology array showed that 22 cancer-related proteins were significantly down-regulated in the free M13- or M13-NL-treated groups (p < 0.001) compared with the control group (without treatment) (Table 2). M13-NL treatment induced much greater down-regulation of these cancer-related proteins than free M13 treatment (p < 0.001).
Table 2.
Statistical analysis of differentially expressed cancer-related proteins in the cell lysates of Caco-2/BBe Cells between free M13 and control, M13-NL and control, and M13-NL and free M13 treatment groups.
| Differentially expressed proteins | Free M13 vs. Control1) and statistical significance2) | M13-NL vs. Control and statistical significance | M13-NL vs. Free M13 and statistical significance |
|---|---|---|---|
| Survivin | Down-regulation, *** | Down-regulation, *** | Down-regulation, *** |
| EGFR | Down-regulation, * | Down-regulation, *** | ns |
| BCL-x | Down-regulation, * | Down-regulation, *** | Down-regulation, *** |
| Dkk-1 | Down-regulation, ** | Down-regulation, *** | Down-regulation, *** |
| TROP1/EpCAM | Down-regulation, *** | Down-regulation, *** | ns |
| CapG | Down-regulation, * | Down-regulation, ** | Down-regulation, * |
| Cathepsin B | Down-regulation, * | Down-regulation, * | ns |
| Cathepsin D | Down-regulation, * | Down-regulation, ** | Down-regulation, * |
| Cathepsin S | Down-regulation, ** | Down-regulation, ** | ns |
| CEACAM-5 | Down-regulation, * | Down-regulation, ** | Down-regulation, * |
| Enolase 2 | Down-regulation, * | Down-regulation, *** | Down-regulation, ** |
| ErbB2/Her2 | Down-regulation, ** | Down-regulation, ** | ns |
| ErbB3/Her3 | Down-regulation, *** | Down-regulation, *** | ns |
| Galectin-3 | Down-regulation, * | Down-regulation, *** | Down-regulation, * |
| HNF-3β | Down-regulation, ** | Down-regulation, *** | Down-regulation, * |
| HO-1/HMOX1 | Down-regulation, *** | Down-regulation, *** | Down-regulation, * |
| CXCL8/IL-8 | Down-regulation, * | Down-regulation, ** | Down-regulation, * |
| CCL20/MIP-3α | Down-regulation, ** | Down-regulation, *** | Down-regulation, ** |
| P27/Kip1 | Down-regulation, ** | Down-regulation, *** | Down-regulation, ** |
| PDGF-AA | Down-regulation, * | Down-regulation, *** | Down-regulation, *** |
| Progranulin | Down-regulation, * | Down-regulation, ** | Down-regulation, * |
| Prostasin/Prss8 | Down-regulation, * | Down-regulation, *** | Down-regulation, ** |
up- or down-regulation
ns, not significant
p < 0.05
p < 0.01
p < 0.001 (n = 4)
Among the 22 cancer-related proteins, survivin, EGFR, BCL-x, Dkk-1, and EpCAM were down-regulated more than 2-fold in the M13-NL-treated cells (Figures 3B–3F). It is noteworthy that the expression of survivin, BCL-x, Dkk-1, and EpCAM was down-regulated in the NL-treated group compared to the control group, indicating that NL exhibits a certain degree of activity in down-regulating the expression of cancer-related proteins in Caco-2/BBe cells. We also identified some differences in the expression levels of 238 cell cycle control/phosphorylation proteins in the free M13- and/or M13-NL-treated groups compared with the control and/or NL groups: CDC25A and phosphorylated CDC25A (p-CDC25A) were down-regulated in the free M13- and M13-NL-treated groups compared with the control and NL groups (p < 0.01) (Figure 3G). Moreover, RAD52 was down-regulated in the M13-NL-treated group compared with the other groups (p < 0.01), but there was no significant between-group change in phosphorylated RAD52 (p-RAD52) (Figure 3H). Since these proteins are involved in various signaling pathways and physiological reactions, we speculate that M13 may significantly down-regulate the expression levels of various cancer-related proteins and cell cycle control/phosphorylation proteins to inhibit cell proliferation (CDC25A and RAD52), migration (EpCAM), and metastasis (Dkk-1), and promote cell apoptosis (survivin, EGFR, and BCL-x) in colorectal cancer cells (Figure 3I) [23–26]32, [27–29].
3.4. Oral administration of M13-NL yields potent anti-cancer effects in AOM/DSS-induced CAC mouse model
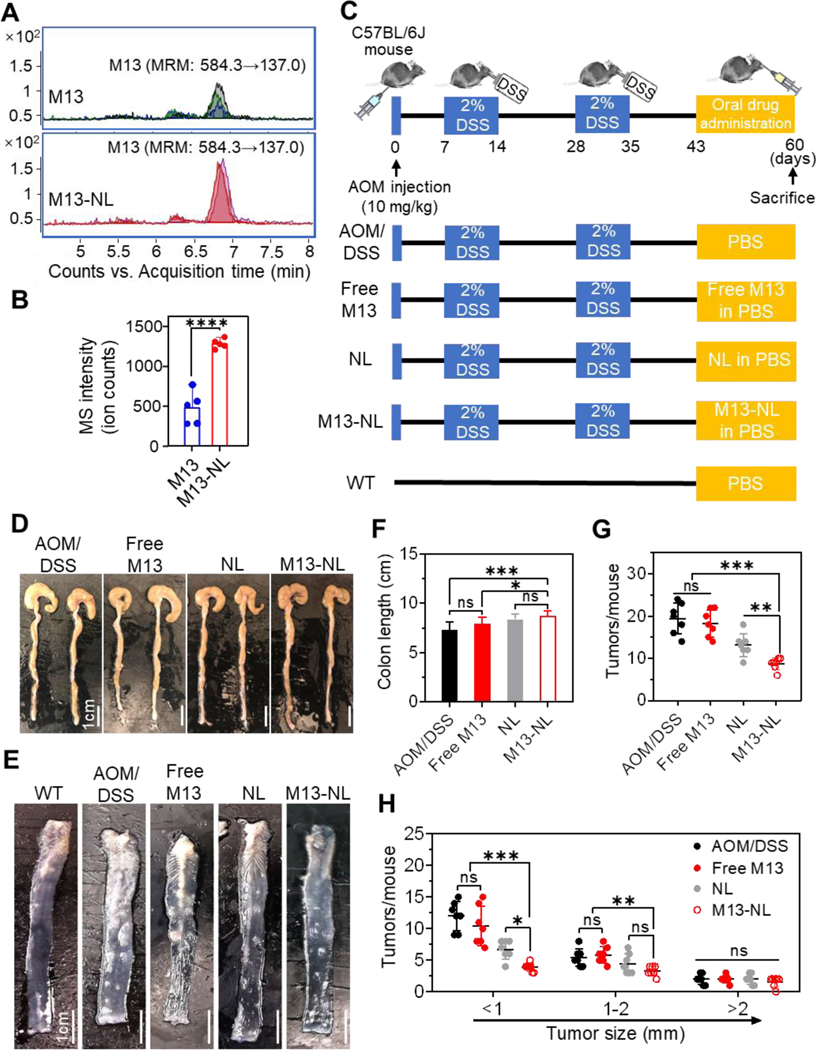
To determine whether NL encapsulation could enhance the bioavailability of M13 in vivo, we orally gavaged C57BL/6J mice (female, n=5 per group) with free M13 or M13-NL (5 mg/kg BW) and used LC-MS/MS to evaluate M13 concentrations in colon tissues collected at 2 h post-administration (Figure 4A). When we compared mass spectrometry signal intensities (ion counts) in the colon tissues of mice, we detected significantly higher contents of M13 after oral administration of M13-NL than free M13 (p < 0.0001) (Figure 4B). These data suggest that M13-NL significantly enhances the in vivo bioavailability of M13 in colon tissues upon oral administration.
Figure 4. Efficacies of M13 or M13-NL treatment on AOM/DSS-induced colon tumors in mice.
(A) Multiple reaction monitoring (MRM) mass spectra of M13 in the colons of mice after orally administered free M13 or M13-NL. (B) Relative signal intensity (ion counts) of M13 from (A) (n = 5). (C) Experimental timeline for AOM/DSS-induced tumorigenesis in the mouse model. Mice were orally administered free M13 (5 mg/kg), NL (5 mg/kg), or M13-NL (5 mg/kg M13 loaded into 5 mg/kg NL) every other day after day 42. No treatment mice were used as the wild-type (WT) control. (D-E) Representative images of the colons (D) and anatomical colon samples (E) (n = 3). (F-H) Comparison of the colon lengths (F), the total number of tumors (G), and the number of the tumor size distribution (H) (n = 7). (ns, not significant; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
Next, an AOM/DSS-induced CAC mouse model (C57BL/6J) was used to test the anticancer effects of free M13, NL, and M13-NL. As shown in Figure 4C, AOM/DSS-induced CAC mice (female, n=7 per group) were orally administered PBS (AOM/DSS group), free M13 (5 mg/kg), NL (5 mg/kg), or M13-NL (5 mg/kg of M13 loaded into 5 mg/kg of NL) via gavage every 2 days. Healthy mice were used as a control group (WT group). After sacrificing the mice at day 60, we found that DSS induced severe inflammation in the colon, resulting in a shortened colon length of mice in the AOM/DSS group (Figure 4D). In comparison, the colon length was significantly increased in the M13-NL-treated group (p < 0.001), but not in the free M13-treated group (Figure 4F), compared to the AOM/DSS group. When the colons of each group were dissected longitudinally (Figure 4E), we found that M13-NL treatment significantly reduced the number of mid-sized tumors (diameter 1–2 mm) compared with the free M13 or AOM/DSS groups (p < 0.01), and significantly reduced the number of small-sized tumors (diameter < 1 mm) compared with the NL group (p < 0.001) (Figures 4G and 4H). In contrast, free M13 treatment failed to reduce the number of tumors compared with the AOM/DSS group. These results indicate that oral M13-NL is better than free M13 and has the strongest anti-tumor activity among the tested groups in the AOM/DSS-induced CAC mouse model.
3.5. Oral administration of M13-NL accelerates the recovery of colon tumor tissues and down-regulates cancer-related proteins and cell cycle proteins in the AOM/DSS-induced CAC mice
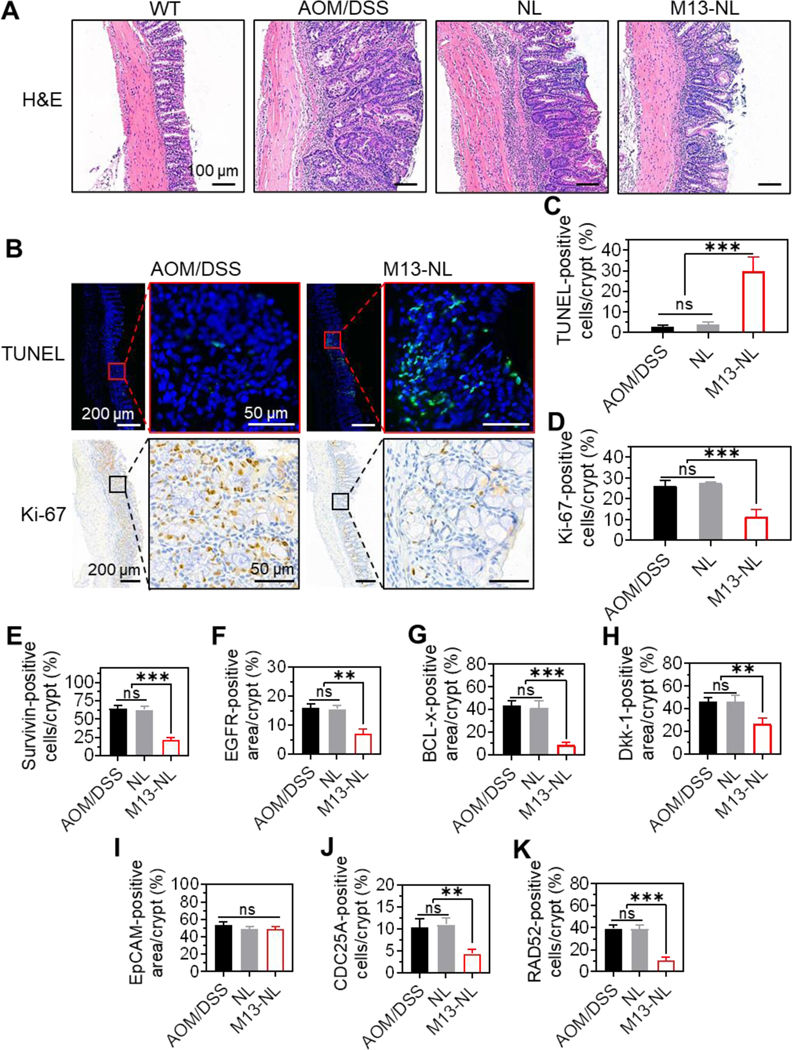
We further performed a histological assessment of colon tissues from AOM/DSS-induced CAC mice. H&E staining clearly showed hyperplasia, dysplasia, loss of cells in epithelial orientation, and expansion of cells into the lamina propria, which are typical signs of adenoma in AOM/DSS mice. In contrast, the tissues from WT mice revealed normal histological structures (Figure 5A). Colon tissues of mice in the NL and AOM/DSS groups showed signs of advanced adenomas, while colon tissues of mice in the M13-NL group showed some signs of early adenoma and appeared less severely affected than those of the AOM/DSS group. Immunofluorescence (IF) staining of apoptotic cells (TUNEL) and immunohistochemical (IHC) staining of Ki-67 were used to investigate the pro-apoptotic and anti-proliferative effects, respectively, of M13-NL in vivo. Tumor sections from the AOM/DSS group showed less green fluorescence from TUNEL-positive cells, whereas those from the M13-NL-treated group showed significantly stronger green fluorescence signals than the AOM/DSS group (p < 0.001) (Figures 5B and 5C).
Figure 5. Histological analysis of M13-NL against colon tumors in AOM/DSS-induced CAC mice.
(A) Representative colon sections from WT, AOM/DSS, NL, and M13-NL groups stained with H&E (n = 3). (B) Representative colon sections from AOM/DSS and M13-NL groups stained with TUNEL (green) and Ki-67 (brown) (n = 3). (C-K) TUNEL- (C), Ki-67 (D), Survivin- (E), EGFR- (F), BCL-x- (G), Dkk-1- (H), EpCAM- (I), CDC25A- (J), and RAD52-positive (K) cells or area were counted and averaged per crypt (n = 5). (ns, not significant; *p < 0.05, **p < 0.01, ***p < 0.001).
IHC staining showed that the Ki-67-positive cells were significantly reduced in the M13-NL-treated group compared with the AOM/DSS group (p < 0.001) (Figures 5B and 5D). Colon tissues from the WT and NL groups were also analyzed by TUNEL and Ki-67 staining (Supplementary Figure S9), and our quantitative analysis showed that there was no significant difference in the percentages of TUNEL- or Ki-67-positive cells per crypt between the AOM/DSS and NL groups (Figures 5C and 5D).
IHC staining showed that M13-NL treatment significantly reduced the expression levels of survivin (p < 0.001), EGFR (p < 0.01), BCL-x (p < 0.001), Dkk-1 (p < 0.01), CDC25A (p < 0.01), and RAD52 (p < 0.001), but not EpCAM, in CAC colon tissues compared with those found in the AOM/DSS and NL groups (Figures 5E–5K and Supplementary Figure S10). Collectively, these data demonstrate that oral administration of M13-NL yields potent anticancer effects by down-regulating various cancer-related proteins and key cell cycle proteins in the colons of CAC mice.
Moreover, changes in body weight and the spleen-to-body weight ratio, H&E staining of major organs, and hematological and biochemical indicators of blood were measured as part of an initial safety evaluation. We found no significant change in body weight or spleen-to-body weight ratio (Supplementary Figure 11), no major organ lesion (Supplementary Figure 12), and no significant change in hematological or biochemical parameters of the blood (Supplementary Figure 13) in mice treated with M13-NL or empty NL. Our results demonstrate that oral treatment with M13-NL will target M13 to the colon without inducing systemic side effects in mice.
3.6. Oral administration of M13-NL improves intestinal microbiome homeostasis in AOM/DSS-induced CAC mice
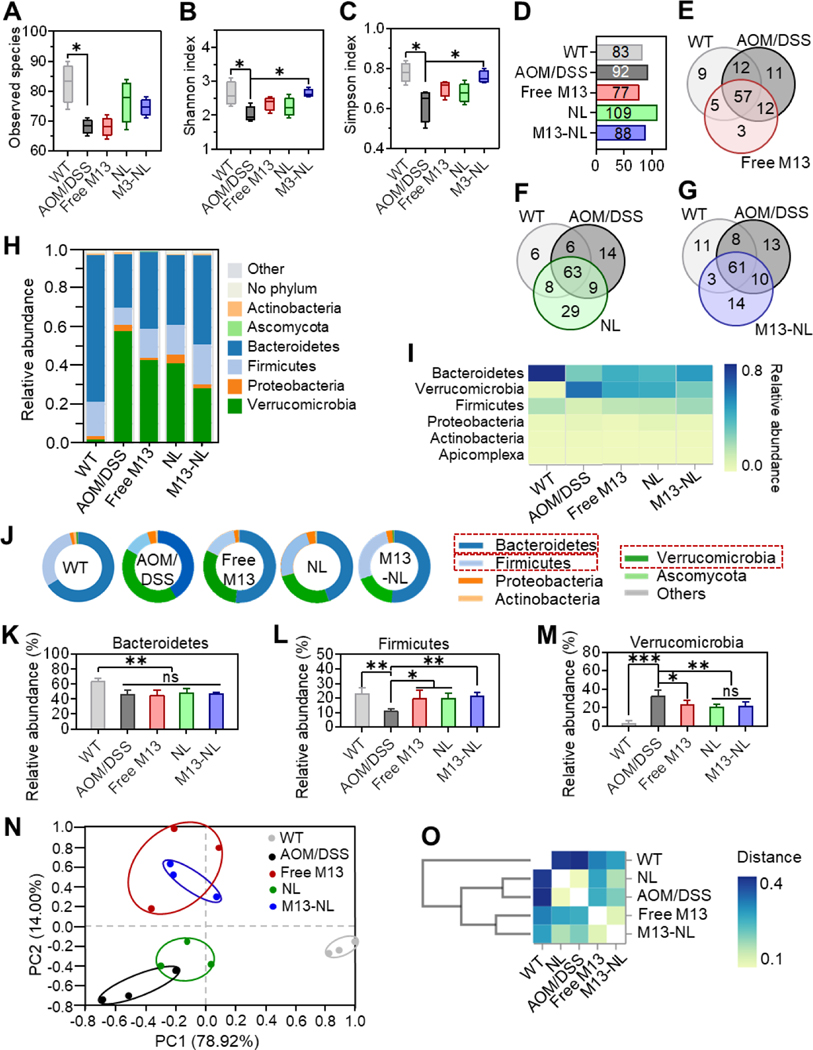
To investigate the impact of free M13, NL, and M13-NL on the intestinal microbiota composition of AOM/DSS-induced CAC mice, we performed whole-genome shotgun sequencing (WGS) of DNA extracted from feces collected from the different treatment and WT groups. As shown in Figures 6A-6C, the observed species, Shannon index, and Simpson index of intestinal microbiota were significantly higher in the healthy mice (WT group) than in the AOM/DSS group (p < 0.05). M13-NL treatment significantly improved the species diversity and community abundance (Shannon index and Simpson index) compared with those of the AOM/DSS group (p < 0.05) (Figures 6B and 6C). Venn diagram analysis of the species composition revealed that the richness of the intestinal microbiome differed between the free M13, NL, and M13-NL treatment groups versus the WT or AOM/DSS groups (Figures 6D-6G). Only three unique species were observed after free M13 treatment; this was far fewer than the 29 and 14 unique species obtained after NL and M13-NL treatment, respectively (Figures 6E-6G). This result is consistent with the low bioavailability of orally administrated free M13 in the colon. At the phylum level (Figures 6H-6J), Bacteroidetes and Firmicutes were the most prevalent taxa in the WT group, followed by Proteobacteria and Verrucomicrobia; in contrast, Verrucomicrobia was the most prevalent taxa in the AOM/DSS group, followed by Bacteroidetes and Firmicutes. We found that the intestinal microbiota compositions of mice from the free M13 and NL groups resembled that of the AOM/DSS group, whereas that from the M13-NL group differed from that of the AOM/DSS group. Among the experimental groups, the relative abundances of microbiota at the class and order levels were similar to those obtained at the phylum level (Supplementary Figures S14 and S15). Compared with the AOM/DSS group, the percentage of Bacteroidetes was not significantly different in the three treatment groups, whereas that of Firmicutes was significantly up-regulated (p < 0.05) and that of Verrucomicrobia was significantly down-regulated (p < 0.05) in the three treatment groups (Figures 6K-6M). These changes in the fecal microbial content of the free M13, NL, and M13-NL-treated groups were generally consistent with their therapeutic outcomes against CAC. β-Diversity was measured via principal coordinates analysis (PCoA) derived from Weighted UniFrac and Bray-Curtis distances to visualize the dissimilarity distances and variation among the different groups at the phylum level. The PCoA results suggested that the gut microbial compositions in the free M13 (red dots) and M13-NL (blue dots) groups were close to each other and closer to those of the WT group (gray dots) than those of the untreated-AOM/DSS group (black dots), while the microbiota in the NL group (green dots) was close to that of the AOM/DSS group (black dots) (Figure 6N). The PCoA results were further confirmed by cluster analyses (Bray-Curtis distances) between each group (Figure 6O), which suggested that treatment with M13 or M13-NL was likely to shift the gut microbial compositions away from those in the untreated AOM/DSS group and potentially towards the healthy gut microbial compositions found in WT mice.
Figure 6. Remodeling effects of experimental groups on gut microbiota in AOM/DSS-induced CAC mice.
(A-C) Observed species (A), Shannon (B), and Simpson indices (C) were calculated based on operational taxonomic units (OTUs) level of intestinal flora (n = 3). (D) The total number of species at the OTUs level (> 0.01%). (E-G) Venn diagrams of common and unique microbial species of mice among WT, AOM/DSS, and Free M13 groups (E), WT, AOM/DSS, and NL groups (F), and WT, AOM/DSS, and M13-NL groups (G), respectively. (H) Relative abundance at the phylum level. (I) Heatmap of the relative abundance at the phylum level. (J) The microbial composition at the phylum level. (K-M) Comparison of the relative abundances of Bacteroidetes (H), Firmicutes (I), and Verrucomicrobia (J) at the phylum level (n = 3). (N) Principal coordinates analysis (PCoA) at the phylum level (n = 3). (O) Cluster analysis (distance plot) of the microbiome at the phylum level. (ns, not significant; *p < 0.05, **p < 0.01, ***p < 0.001).
4. Discussion
The failure to develop a drug candidate stems from its poor biopharmaceutical properties, such as low aqueous solubility and/or high chemical instability, metabolic instability, mutagenic potential, and immunogenicity. Although the lead compound 6-shogaol presents potent anti-inflammatory and anticancer activities, it has failed to progress to clinical trials due to its poor biopharmaceutical properties, such as high hydrophobicity, poor absorption, high metabolic instability, and concomitant rapid elimination [10,30]. The tripeptide-conjugated 6-shogaol, M13, is a phase II metabolite of 6-shogaol, which we hypothesized would have better aqueous solubility, chemical stability, and metabolic stability than the lead compound. This study demonstrated that M13 has improved physiochemical properties, including high aqueous solubility and chemical stability, and is not easily ionized at physiological pH. M13 is stable in a mimicked gastric environment (pH=3 or 5) upon incubation at 37 °C for 2 h, and it has a lower metabolic liability than 6-shogaol. These observations may explain the better anti-inflammatory activities of M13 compared to those of 6-shogaol.
The proportions of circulating CD4+ and CD8+ T lymphocytes are reportedly increased in IBD patients compared with healthy individuals, and the heightened chronic inflammation in IBD is associated with increased activation of circulating T lymphocytes [31]. M13 seems likely to be safe for healthy individuals, since it does not affect the proportions of resting CD4+ or CD8+ lymphocytes in vitro. The ability of M13 to inhibit the proliferation of IL-2-activated CD4+ and CD8+ lymphocytes suggests that M13 may be able to specifically modulate the activated T lymphocytes in IBD patients [32].
The Ames test, introduced by Bruce Ames in the early 1970s, is widely employed to assess the mutagenic/ carcinogenic potential of drug candidates [33]. This test revealed that M13 was not mutagenic up to a concentration of 5000 μg/plate. In vitro safety screening is an essential tool for predicting clinical adverse effects during drug discovery, enabling developers to address the possibility of clinical liabilities at an early stage of drug development. The results of our InVEST test demonstrated that M13 did not have any significant effect on the activities of 25 assorted targets from five target classes. In addition, a single-dose oral toxicity study of M13 showed that the maximum tolerated dose of M13 is > 1000 mg/kg in mice. These in vitro and in vivo results indicate that M13 can be viewed as a very safe drug candidate.
Some early studies showed that M13 exhibited bioactivity in human colon cancer cells (HCT-116) and lung cancer cells (H-1299), while showing low toxicity against human normal colon fibroblast cells (CCD-18Co) and normal lung cells (IMR-90) [9]. Here, we systematically compared the growth inhibitory effects of M13 in one human normal intestinal cell line (HIEC-6) and three human intestinal cancer cell lines (HCT-116, Caco-2/BBe, and HC-CSC). We found that M13 specifically eliminated intestinal cancer cells (IC50s of 16–40 μM) but had much less effect on the growth of normal intestinal cells (IC50>100 μM). It is well known that the natural manner of a solid tumor in vivo can be recapitulated in 3D culture, not 2D culture. Although the 2D cell culture model has been widely used to predict in vivo drug efficacy, it has many limitations, including the lack of tissue-specific structure, cell-to-cell interactions, cell-to-matrix interactions, biochemical cues, and mechanical cues [34]. 3D cell culture models in which cells mix and grow within an extracellular matrix gel established by 3D printing are believed to reflect more tissue-specific functions and better mimic the in vivo tumor microenvironment, compared to 2D culture [35]. Here, we printed Caco-2/BBe-hydrogel mixed-cell spheres to simulate the response of a tumor mimicking intestinal epithelium to M13 in vivo. Our data showed that M13 treatment is also active and dose-dependent in the 3D Caco-2/BBe sphere model.
Mechanistically, 6-shogaol is thought to act via induction of apoptosis and inhibition of proliferation in cancer [36,37]. Therefore, we speculated that M13 (as a tripeptide conjugate of 6-shogaol) might employ similar mechanisms in inhibiting colorectal cancer. We found that 22 cancer-related proteins and two cell cycle control/phosphorylation proteins were significantly down-regulated in Caco-2/BBe cells exposed to M13 or M13-NL. In colorectal cancer cells, EGFR is directly involved in regulating Ras/ERK- and PI3K/Akt signaling-mediated apoptosis [38]. The apoptosis-inhibiting proteins, survivin and BCL-x, are regulated by the Notch signaling pathway [39]. Recent studies demonstrated that down-regulation of survivin, EGFR, or BCL-x significantly inhibited tumor growth and induced apoptosis in colorectal cancer [27–29]. In addition, the serum level of Dkk-1 in patients with colorectal cancer liver metastasis was reported to be significantly increased, suggesting that down-regulation of Dkk-1 could inhibit colorectal cancer liver metastasis [26]. Down-regulation of EpCAM was reported to effectively inhibit the migration and proliferation of colorectal cancer cells, while the cyclins CDC25A and RAD52 are essential promoters of colorectal cancer cell proliferation [23,24]. On the basis of these previous findings and our present results, we conclude that M13 contributes to regulating multiple signaling pathways and various physiological and biochemical reactions, including the apoptosis, proliferation, migration, and metastasis of colorectal cancer cells in vitro. This study provides an important reference point for further elucidating the mechanism(s) by which M13 acts against intestinal tumors.
Regarding the utilized delivery system, GDNPs offer the advantages of colon-targeting properties, lack of toxicity, low immunogenicity, and ease of mass production [11]. In previous studies, we confirmed that oral delivery of GDNP-encapsulated M13 has good anti-inflammatory activity in the intestinal tract of mice with ulcerative colitis and can promote intestinal wound healing [13]. The current study demonstrated in vitro that M13-NL could down-regulate several cancer-related proteins and cell cycle proteins in Caco-2/BBe cells to a significantly greater degree than free M13, indicating that this encapsulation enhances the anti-tumor-cell activity of M13. Moreover, we confirmed that the bioavailability of orally administered M13-NL in mouse intestinal tissues was significantly increased relative to that of free M13. This action, which may reflect the previously reported ability of NL to target the colon, improved the uptake efficiency of intestinal cells for M13-NL in vivo and increased the concentration and residence time of M13 in colon tissues [13].
It is noteworthy to mention that NL exhibited antitumor effectiveness in vivo. Analysis of its lipid composition revealed the presence of monogalactosyldiacylglycerol (MGDG), digalactosyldiacylglycerol (DGDG), and phosphatidic acid (PA). [12,19]. Research has indicated that MGDG has the potential to enhance apoptosis rates in human cancer cell lines, while both DGDG and PA may augment the anti-cancer efficacy of chemotherapeutic drugs [40–43]. These findings suggest that in addition to NĽs primary function of drug delivery, its components may exert a synergistic effect when combined with M13. However, further investigation is necessary to determine the extent of this synergistic power.
The AOM/DSS-induced CAC mouse model is known as a reproducible and relatively inexpensive initiation-promotion model of CAC that is generated by chemical induction of DNA damage followed by repeated cycles of colitis [44,45]. This model has been widely used to explore the therapeutic effects of drug candidates on acute inflammation-induced colon cancer [46,47]. We first evaluated the anticancer activities of M13-NL in this established mouse model and found that orally administered M13-NL could effectively retard the growth of colon tumors in AOM/DSS-induced CAC mice by significantly down-regulating many proteins, including EGFR, survivin, BCL-x, Dkk-1, CDC25A, and RAD52. Furthermore, and consistent with our previous reports, the physiological and biochemical indexes observed in AOM/DSS-induced CAC mice support the biocompatibility of orally administered NL and M13-NL [12,13].
It is generally believed that IBD and CAC are related to compositional changes and metabolic dysbiosis of the intestinal microbiota [48,49]. Accumulating evidence indicates that disturbance of the intestinal microbiota can directly impact the progression of IBD and CAC [49,50]. Recently, we demonstrated that NL could target microorganisms and thus significantly accelerate the microbiome composition changes caused by NL-encapsulated 6-shogaol [51]. In the present study, we found that treatment with M13-NL modulated the gut microbiota by increasing its overall abundance and diversity in AOM/DSS-induced CAC mice. This treatment appeared to enrich phyla with anti-inflammatory functions (e.g., Firmicutes) while depleting phyla with pro-inflammatory functions (e.g., Verrucomicrobia), thereby generating a more balanced microbiota similar to that of healthy mice [52] [53]. This might be another factor contributing to the therapeutic effect of M13-NL on CAC.
5. Conclusion
In summary, we systematically investigated the physicochemical properties, anti-colon cancer activities, and underlying mechanisms of the drug candidate, “M13”, and conducted preclinical studies of M13-loaded nanoliposomes for the treatment of CAC in vitro and in vivo. Our study shows that M13 has good biopharmaceutical properties, is not mutagenic, and is safe in vitro and in vivo, and that ginger-derived nanoliposomes encapsulation of M13 improves its delivery efficiency, safety, and effectiveness in vivo. Furthermore, our work demonstrates that oral delivery of M13-NL has the potential as an effective agent for treating CAC.
Supplementary Material
Funding
This work was supported by grants from the Crohn’s and Colitis Foundation (689659), the National Institute of Diabetes and Digestive and Kidney Diseases (RO1-DK-116306), and the Department of Veterans Affairs (BX002526 and BX004476).
Abbreviations:
- AOM
azoxymethane
- CAC
Colitis-associated cancer
- DSS
dextran sodium sulfate
- EGFR
epidermal growth factor receptor
- FACS
Fluorescence-activated cell sorting
- FBS
fetal bovine serum
- GPCR
G-protein-coupled receptor
- HC-CSC
human colon cancer stem cells
- H&E
hematoxylin and eosin
- IBD
inflammatory bowel disease
- HPLC
high-performance liquid chromatography
- IL-2
interleukin-2
- IF
immunofluorescence
- IHC
immunohistochemistry
- InVEST
in vitro evaluation of safety and toxicity
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- MTD
Maximum tolerated dose
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide
- M13-NL
M13-loaded nanoliposomes
- NPs
nanoparticles
- NL
nanoliposomes
- PBMCs
peripheral blood mononuclear cells
- PBS
phosphate-buffered saline
- PCoA
principal coordinates analysis
- 2D
two-dimensional
- 3D
three-dimensional
- WGS
whole genome shotgun sequencing
Footnotes
Declaration of Interest
D Long is a recipient of the Research Fellowship Award from Crohn’s and Colitis Foundation (CCF Award # 689659). D Merlin is a recipient of a Senior Research Career Scientist Award (BX004476) from the Department of Veterans Affairs. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
References
- 1.Sussman D, Santaolalla R, Strobel S, et al. Cancer in inflammatory bowel disease: lessons from animal models. Current opinion in gastroenterology. 2012;28(4):327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ingersoll SA, Ayyadurai S, Charania MA, et al. The role and pathophysiological relevance of membrane transporter PepT1 in intestinal inflammation and inflammatory bowel disease. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2012;302(5):G484–G492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Long D, Merlin D. Micro-and nanotechnological delivery platforms for treatment of dysbiosis-related inflammatory bowel disease. Future Medicine; 2021. p. 1741–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Long AG, Lundsmith ET, Hamilton KE. Inflammation and colorectal cancer. Current colorectal cancer reports. 2017;13(4):341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kotelevets L, Chastre E, Desmaele D, et al. Nanotechnologies for the treatment of colon cancer: From old drugs to new hope. International journal of pharmaceutics. 2016;514(1):24–40. [DOI] [PubMed] [Google Scholar]
- 6.Chen S, Yueh M-F, Bigo C, et al. Intestinal glucuronidation protects against chemotherapy-induced toxicity by irinotecan (CPT-11). Proceedings of the National Academy of Sciences. 2013;110(47):19143–19148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hua S, Marks E, Schneider JJ, et al. Advances in oral nano-delivery systems for colon targeted drug delivery in inflammatory bowel disease: selective targeting to diseased versus healthy tissue. Nanomedicine: nanotechnology, biology and medicine. 2015;11(5):1117–1132. [DOI] [PubMed] [Google Scholar]
- 8.Alqahtani MS, Kazi M, Alsenaidy MA, et al. Advances in oral drug delivery. Frontiers in Pharmacology. 2021:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu Y, Warin RF, Soroka DN, et al. Metabolites of ginger component [6]-shogaol remain bioactive in cancer cells and have low toxicity in normal cells: chemical synthesis and biological evaluation. PLoS one. 2013;8(1):e54677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H, Lv L, Soroka D, et al. Metabolism of [6]-shogaol in mice and in cancer cells. Drug Metabolism and Disposition. 2012;40(4):742–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang C, Zhang M, Merlin D. Advances in plant-derived edible nanoparticle-based lipid nano-drug delivery systems as therapeutic nanomedicines. Journal of Materials Chemistry B. 2018;6(9):1312–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang M, Viennois E, Prasad M, et al. Edible ginger-derived nanoparticles: A novel therapeutic approach for the prevention and treatment of inflammatory bowel disease and colitis-associated cancer. Biomaterials. 2016;101:321–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang C, Zhang M, Lama S, et al. Natural-lipid nanoparticle-based therapeutic approach to deliver 6-shogaol and its metabolites M2 and M13 to the colon to treat ulcerative colitis. Journal of Controlled Release. 2020;323:293–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang M, Xiao B, Wang H, et al. Edible ginger-derived nano-lipids loaded with doxorubicin as a novel drug-delivery approach for colon cancer therapy. Molecular Therapy. 2016;24(10):1783–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang M, Wang X, Han MK, et al. Oral administration of ginger-derived nanolipids loaded with siRNA as a novel approach for efficient siRNA drug delivery to treat ulcerative colitis. Nanomedicine. 2017;12(16):1927–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang M, Xu C, Liu D, et al. Oral delivery of nanoparticles loaded with ginger active compound, 6-shogaol, attenuates ulcerative colitis and promotes wound healing in a murine model of ulcerative colitis. Journal of Crohn's and Colitis. 2018;12(2):217–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saal C, Petereit AC. Optimizing solubility: kinetic versus thermodynamic solubility temptations and risks. Eur J Pharm Sci. 2012. Oct 9;47(3):589–95. [DOI] [PubMed] [Google Scholar]
- 18.Sugiyama K, Yamada M, Awogi T, et al. The strains recommended for use in the bacterial reverse mutation test (OECD guideline 471) can be certified as non-genetically modified organisms. Genes Environ. 2016;38:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sung J, Yang C, Viennois E, et al. Isolation, Purification, and Characterization of Ginger-derived Nanoparticles (GDNPs) from Ginger, Rhizome of Zingiber officinale. Bio Protoc. 2019. Oct 5;9(19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yue Z, Li C, Voth GA, et al. Dynamic Protonation Dramatically Affects the Membrane Permeability of Drug-like Molecules. J Am Chem Soc. 2019. Aug 28;141(34):13421–13433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bennion BJ, Be NA, McNerney MW, et al. Predicting a Drug's Membrane Permeability: A Computational Model Validated With in Vitro Permeability Assay Data. J Phys Chem B. 2017. May 25;121(20):5228–5237. [DOI] [PubMed] [Google Scholar]
- 22.Dahlen R, Strid H, Lundgren A, et al. Infliximab inhibits activation and effector functions of peripheral blood T cells in vitro from patients with clinically active ulcerative colitis. Scand J Immunol. 2013. Sep;78(3):275–84. [DOI] [PubMed] [Google Scholar]
- 23.Shen T, Huang S. The role of Cdc25A in the regulation of cell proliferation and apoptosis. Anti-Cancer Agents in Medicinal Chemistry (Formerly Current Medicinal Chemistry-Anti-Cancer Agents). 2012;12(6):631–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barlow JH, Rothstein R. Timing is everything: cell cycle control of Rad52. Cell division. 2010;5(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang K-H, Tso H-C, Hung S-H, et al. Extracellular domain of EpCAM enhances tumor progression through EGFR signaling in colon cancer cells. Cancer letters. 2018;433:165–175. [DOI] [PubMed] [Google Scholar]
- 26.Sui Q, Zheng J, Liu D, et al. Dickkopf-related protein 1, a new biomarker for local immune status and poor prognosis among patients with colorectal liver Oligometastases: a retrospective study. BMC cancer. 2019;19(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai Y, Ma W, Huang X, et al. Effect of survivin on tumor growth of colorectal cancer in vivo. International journal of clinical and experimental pathology. 2015;8(10):13267. [PMC free article] [PubMed] [Google Scholar]
- 28.Gao S-J, Ren S-N, Liu Y-T, et al. Targeting EGFR sensitizes 5-Fu-resistant colon cancer cells through modification of the lncRNA-FGD5-AS1-miR-330–3p-Hexokinase 2 axis. Molecular Therapy-Oncolytics. 2021;23:14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramesh P, Di Franco S, Taboada LA, et al. BCL-XL inhibition induces an FGFR4-mediated rescue response in colorectal cancer. Cell Reports. 2022;38(7):110374. [DOI] [PubMed] [Google Scholar]
- 30.Asami A, Shimada T, Mizuhara Y, et al. Pharmacokinetics of [6]-shogaol, a pungent ingredient of Zingiber officinale Roscoe (Part I). Journal of natural medicines. 2010;64(3):281–287. [DOI] [PubMed] [Google Scholar]
- 31.Funderburg NT, Stubblefield Park SR, Sung HC, et al. Circulating CD 4+ and CD 8+ T cells are activated in inflammatory bowel disease and are associated with plasma markers of inflammation. Immunology. 2013;140(1):87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bachmann MF, Oxenius A. Interleukin 2: from immunostimulation to immunoregulation and back again. EMBO reports. 2007;8(12):1142–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ames BN, McCann J, Yamasaki E. Methods for detecting carcinogens and mutagens with the Salmonella/mammalian-microsome mutagenicity test. Mutat Res;(Netherlands). 1975;31. [DOI] [PubMed] [Google Scholar]
- 34.Pampaloni F, Reynaud EG, Stelzer EHK. The third dimension bridges the gap between cell culture and live tissue. Nature reviews Molecular cell biology. 2007;8(10):839–845. [DOI] [PubMed] [Google Scholar]
- 35.Samy KE, Levy ES, Phong K, et al. Human intestinal spheroids cultured using Sacrificial Micromolding as a model system for studying drug transport. Scientific reports. 2019;9(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim MO, Lee M-H, Oi N, et al. [6]-Shogaol inhibits growth and induces apoptosis of non-small cell lung cancer cells by directly regulating Akt1/2. Carcinogenesis. 2014;35(3):683–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang T, He Y, Chang Y, et al. 6-shogaol a active component from ginger inhibits cell proliferation and induces apoptosis through inhibition of STAT-3 translocation in ovarian cancer cell lines (A2780). Biotechnology and Bioprocess Engineering. 2019;24(3):560–567. [Google Scholar]
- 38.Koveitypour Z, Panahi F, Vakilian M, et al. Signaling pathways involved in colorectal cancer progression. Cell & bioscience. 2019;9(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajendran DT, Subramaniyan B, Ganeshan M. Role of Notch signaling in colorectal cancer. Role of transcription factors in gastrointestinal malignancies: Springer; 2017. p. 307–314. [Google Scholar]
- 40.Grabowska K, Galanty A, Koczurkiewicz-Adamczyk P, et al. Multidirectional anti-melanoma effect of galactolipids (MGDG-1 and DGDG-1) from Impatiens parviflora DC. and their synergy with doxorubicin. Toxicol In Vitro. 2021. Oct;76:105231. [DOI] [PubMed] [Google Scholar]
- 41.Abedin MR, Barua S. Isolation and purification of glycoglycerolipids to induce apoptosis in breast cancer cells. Sci Rep. 2021. Jan 14;11(1):1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akasaka H, Mizushina Y, Yoshida K, et al. MGDG extracted from spinach enhances the cytotoxicity of radiation in pancreatic cancer cells. Radiat Oncol. 2016. Nov 22;11(1):153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sliva D, Harvey K, Mason R, et al. Effect of phosphatidic acid on human breast cancer cells exposed to doxorubicin. Cancer Invest. 2001;19(8):783–90. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka T, Kohno H, Suzuki R, et al. A novel inflammation-related mouse colon carcinogenesis model induced by azoxymethane and dextran sodium sulfate. 2003;94(11):965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Robertis M, Massi E, Poeta ML, et al. The AOM/DSS murine model for the study of colon carcinogenesis: From pathways to diagnosis and therapy studies. Journal of carcinogenesis. 2011;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y, Wang B, Yuan X, et al. Vitexin prevents colitis-associated carcinogenesis in mice through regulating macrophage polarization. Phytomedicine. 2021;83:153489. [DOI] [PubMed] [Google Scholar]
- 47.Osman J, Savari S, Chandrashekar NK, et al. Cysteinyl leukotriene receptor 1 facilitates tumorigenesis in a mouse model of colitis-associated colon cancer. Oncotarget. 2017;8(21):34773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee M, Chang EB. Inflammatory bowel diseases (IBD) and the microbiome—searching the crime scene for clues. Gastroenterology. 2021;160(2):524–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khan I, Ullah N, Zha L, et al. Alteration of gut microbiota in inflammatory bowel disease (IBD): cause or consequence? IBD treatment targeting the gut microbiome. Pathogens. 2019;8(3):126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nature Reviews Gastroenterology & Hepatology. 2019;16(11):690–704. [DOI] [PubMed] [Google Scholar]
- 51.Yang C, Long D, Sung J, et al. Orally Administered Natural Lipid Nanoparticle-Loaded 6-Shogaol Shapes the Anti-Inflammatory Microbiota and Metabolome. Pharmaceutics. 2021;13(9):1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sartor RB, Mazmanian SK. Intestinal microbes in inflammatory bowel diseases. The American journal of gastroenterology supplements. 2012;1(1):15. [Google Scholar]
- 53.Lin C-H, Chen C-C, Chiang H-L, et al. Altered gut microbiota and inflammatory cytokine responses in patients with Parkinson’s disease. Journal of neuroinflammation. 2019;16(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.