Abstract
Purpose
NRL is an influential transcription factor and central to animal modeling in ophthalmology. Disrupting NRL abrogates rod development and produces an excess of S-cones (also known as “UV cones” or “short-wavelength-sensitive1 [SWS1] cones”). Strikingly, mutations in zebrafish tbx2b produce the exact opposite phenotypes (excess rods and loss of SWS1 cones). We sought to define what genetic relationship exists, if any, between these transcription factors. We also infer whether these two phenotypes (altered rod abundance and altered SWS1 cone abundance) are independent versus inter-related.
Methods
Zebrafish mutants were bred to disrupt nrl and tbx2b in concert. Rods and SWS1 cones were quantified and characterized at ultrastructural and transcriptional levels.
Results
Considering single mutant zebrafish, we confirmed previously established phenotypes and noted that the number of rods lost in nrl−/− mutants is reflected by a concomitant increase in SWS1 cone abundance. The tbx2b−/− mutants present the opposite phenotype(s) but exhibit a similar trade-off in cell abundances, with lots of rods and a concomitant decrease in SWS1 cones. Double mutant nrl−/−;tbx2b−/− zebrafish recapitulate the nrl−/− mutant phenotype(s).
Conclusions
The tbx2b is thought to be required for producing SWS1 cones in zebrafish, but this can be over-ridden when nrl is absent. Regarding the altered cell abundances observed in either tbx2b−/− or nrl−/− mutants, the alterations in rod and SWS1 cones appear to not be two separate phenotypes but are instead a single intertwined outcome. The tbx2b and nrl are in an epistatic relationship, with nrl phenotypes dominating, implying that tbx2b is upstream of nrl in photoreceptor cell fate determination.
Keywords: visual system development, gene regulatory network, specification
The vertebrate retina is a complex structure comprised of numerous cell types working in concert to produce the sense of vision. Of the retinal cells, the photoreceptors are responsible for capturing light and translating it into a biological signal for the brain to interpret,1 and they are divided into two classes of cells: rods and cones.2 Rod cells are extremely sensitive, capable of being stimulated by a single photon,3 and providing vision in low-light conditions. Cone cells are less sensitive, but have adaptations to limit the wavelengths of light to which they respond maximally, allowing for the distinction of colors.4 Damage or degradation of these photoreceptors is a common cause of vision loss. Mutations that disrupt photoreceptor biochemistry cause a variety of dystrophies, such as cone-rod dystrophy and retinitis pigmentosa (RP) which effect 1 in 30,000 to 40,000 and 1 in 3000 to 4000 individuals, respectively.5,6 In RP and various other retinal degenerative diseases, vision loss begins with night blindness as rod cells are lost first, followed by a reduction in overall visual acuity as cone photoreceptors degenerate in the following decades. Current treatments of many of these retinal degenerative diseases often focus primarily on the preservation of photoreceptors,7,8 however, there is little that can be done currently to recover photoreceptor function once it has been lost. Applying stem cells to treat blindness has great potential and progress has been impressive,9,10 but the outcomes have largely focused on producing rod photoreceptors.11,12 A major hurdle is tuning stem cells to produce cones, in order to restore a patient's daytime color and high-acuity vision. The genes nrl, nr2e3, and perhaps tbx2b investigated herein, lie at the heart of this photoreceptor specification. Our work aims to elucidate the specific molecular mechanisms that govern the development of photoreceptors in general, and rod versus cone cells in particular. To this end, we have focused our attentions on a transcription factor in the MAF gene family known as neural retina leucine-zipper (NRL).
NRL is a critical gene for the development and maintenance of rod photoreceptors, and mutations in NRL are linked to multiple retinal degenerative diseases (OMIM: 162080). Nrl is required for the development of rods in mice,13 but is also known to be sufficient to drive developing cells to take on a rod fate. For example, overexpression of nrl in developing xenopus eye primordia is sufficient to drive the rod fate, at the direct expense of the cone fate.14 Similarly, transgenic expression of nrl in cone precursor cells is sufficient to drive the rod fate in cells otherwise fated to develop as short wavelength sensitive-cones, or S-cones.15,16 The correlation between rod development and short-wavelength cone development is consistent with observations that whereas nrl mutant animal models, and, in some instances, human patients, fail to develop rods, they also produce an excess of S-cones.17,18 Phenotypes that are shared with a condition known as Enhanced S-Cone syndrome, often associated with mutations in NR2E319,20 (OMIM: 604485), which is a principle transcriptional target of NRL.
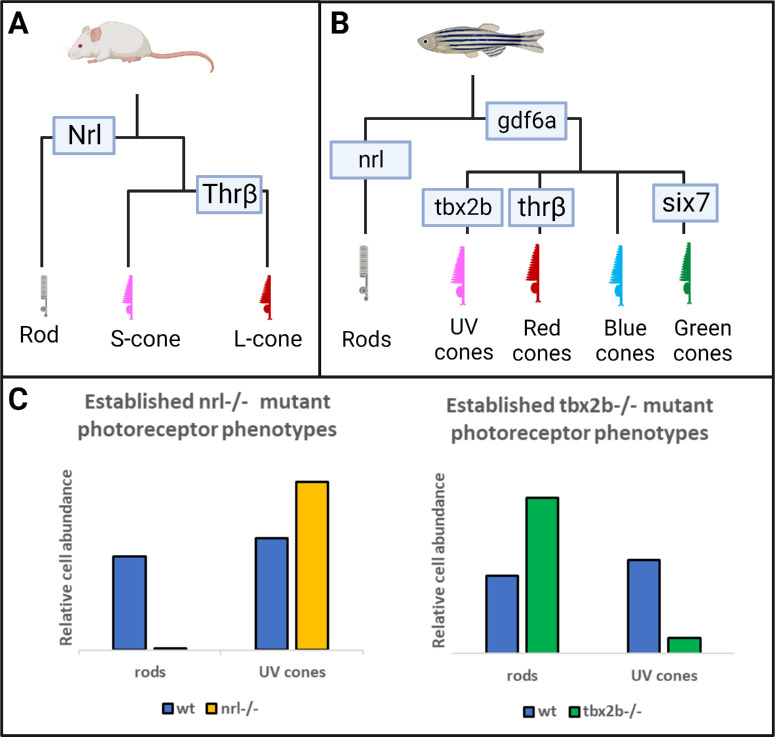
Much of the foundational work on nrl has been done in the mouse model, where the loss of rods and increased S-cone phenotypes are often used to study cone specific development and pathologies.21 Notably, whereas the S-cone in mice is equivalent to the blue cone in humans, expressing the OPN1SW opsin, the homologous cell in the zebrafish model is the UV sensitive cone (expressing sws1 or opn1sw1), rather than the zebrafish's own blue cone (sws2). Appreciating the development of mouse photoreceptors has the advantage of a simpler control system compared to humans, having only two cone types whose developmental control requires only two genes, Nrl to specify rods from cones, and thrB to specify L-cones from S-cones (Fig. 1A). Unfortunately, the mouse model has fundamental limitations in the study of photoreceptors due to its rod dominant retina with a paucity of cone photoreceptors that occur only at low density.
Figure 1.
The nrl and tbx2b mutants have strikingly opposite effects on rod and sws1 cone developmental phenotypes. (A, B) Schematics highlighting that the role of Nrl in patterning the rod fate in mouse models is recapitulated and expanded upon in the zebrafish model. (C) Previous research has established that nrl mutants exhibit a dramatic loss of rod cell development and a concomitant increase of short-wavelength cones (mice and zebrafish),13,15 whereas the opposite is observed in tbx2b mutants (zebrafish).25 We seek to resolve whether this relation between the rod and UV cone fates represents a single fate determination mechanism, or two independent mechanisms governing each cell fate.
In this regard, zebrafish provide a tractable model for the study of photoreceptor specification. Zebrafish photoreceptors are developed in a highly organized mosaic pattern22 and have consistent ratios of UV-, blue-, red-, and green-sensitive cones throughout.23 The additional complexity of producing four cone subtypes requires a more extensive regulatory network to control their development, and previous work has implicated another gene, the t-box transcription factor tbx2b, as a core regulatory element required for Sws1 cone development (Fig. 1B).24,25 TBX2 homologs have been understudied in the retina, presumably because mouse Tbx2 mutants have heart defects and die early in development; zebrafish offer an advantage in this regard because tbx2b mutants are viable (perhaps due to partial redundancy with paralog tbx2a). The tbx2b has been shown to be a critical factor in diverse processes, such as parapineal development,26 liver regeneration27 and cardiac development,28 but of particular interest here is that disruptions to tbx2b also result in the retinal phenotype that exactly mirrors the phenotype when disrupting nrl. The tbx2b mutant fish fail to produce UV cones but also develop an excess of rods (Fig. 1C).25 It is unclear whether the consistent fluidity between rod and sws1 cone development represents two paired but independent mechanisms (i.e. two phenotypes), wherein tbx2b enforces the UV cone fate similarly to how nrl enforces the rod fate, or whether these two transcription factors interact in some way within a single genetic mechanism controlling a developmental switch between the two fates (i.e. one phenotype with a trade-off between rod and sws1 cone abundances). To investigate these possibilities, we analyzed a line of nrl;tbx2b double mutants to assess whether an epistatic relation exists between these genes.
We predict that the abundance of rods and UV cones within these double mutants will be indicative of the genetic relationship of nrl and tbx2b. In the event that the two genes each control separate mechanisms and thereby produce two independent phenotypes, we expect to find that the double mutants produce some mix of the two single mutant's phenotypes. If each gene is required for rod and UV cone development, respectively, we may see a double mutant with neither rods nor UV cones. However, if these genes are involved in two competing mechanisms, then the absence of them both could well result in a high abundance of both cell types. Such complex genetic interactions could also result in disorganized photoreceptor abundance, disrupting the zebrafish's neatly organized photoreceptor mosaic. Alternatively, if these two genes are both involved in a single genetic mechanism wherein rods and UV cones are presented as mutually exclusive developmental end points of a single precursor cell then we predict that the nrl−/−;tbx2b−/− double mutants to recapitulate one of the single mutant phenotypes. If one of these genes acts upstream of the other in order to regulate its activity, then the absence of them both is expected to only produce a single phenotype, likely representant of the effects of the downstream gene's function.
Considering the use of Nrl−/− mutant mice as “cone-rich” ophthalmology models, the potential of NRL as a gene therapy target,29 and NRL’s importance in the study of photoreceptor specification, the elucidation of the role of tbx2b in rod versus cone development is an exciting opportunity to expand upon the foundational principles of photoreceptor specification.
Methods
Animal Ethics
All individuals working with, and protocols involving zebrafish husbandry or experimentation were approved by institutional ethics Committees at the University of Alberta under protocol # AUP00000077. All ethics compliance and protocols overseen by the Canadian Council on Animal Care and the work adhered to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Animal Genotyping
The presence of the nrlua5009 mutant allele (ZFin ID: ZDB-ALT-210805-1) in both the nrl−/− and nrl−/−;tbx2b-/- lines was assessed using the presence of a Taq1 restriction enzyme cut site present only in the mutant. PCR of nrl was performed using the nrl Genotyping Primers listed in Supplementary Table S1 and the following parameters: 96°C 2 minutes, 35 times [56°C 15 seconds, 72°C 1 minutes, 96°C 15 seconds], 72°C 5 minutes. PCR product was subsequently treated with Taq1 enzyme. Mutant sequences were cleaved into two products, whereas wildtype nrl sequence remains uncut.
The presence of the tbx2blor allele (ZFin ID: ZDB-ALT-080920-1; lor = “lots of rods”) was confirmed in both the single mutants and nrl−/−;tbxb2−/− mutants via sequence analysis of a known single nucleotide polymorphism (SNP).25 Sequencing primers are listed in Supplementary Table S1.
Immunostaining and Confocal Imaging
Larvae were anaesthetized in MS-222 before fixation with 4% paraformaldehyde (PFA) in 0.1 M phosphate buffer with 5% sucrose pH 7.4; fixation was at room temperature for at least 2 hours, or overnight at 4°C. Fixed larvae were washed of PFA with phosphate buffered saline pH 7.4 with 0.1% Tween20 (“PBSTw”), then prepared for immunohistochemistry. Fish were washed in water 5 minutes, −20°C acetone for 7 minutes, rinsed out of acetone with PBSTw + 1.0% DMSO, and then blocked for at least 30 minutes in 10% normal goat serum (ThermoFisher) in PBSTw. Blocking solution was removed, and primary antibody was applied. Antibody incubations were mixed 1:100 for primary, and 1:1000 for secondary antibodies, each in PBSTw with 2% goat serum, and incubated at 4°C at least overnight. Larvae were rinsed twice and washed twice in PBSTw, and then transferred to 60% glycerol in PBSTw.
For mounting whole retinas, glycerol-equilibrated larvae were relieved of their lenses using electrolysis-sharpened tungsten wire needles, produced as described by Conrad, G.W., et al.30 After de-lensing, the corneal and scleral tissues covering the retina were cut and folded back, and the retina was removed from the socket with the tungsten needles using a scooping maneuver. The retinas were then positioned vitreal-side down upon slides, and cover slips positioned upon the scleral side. Imaging was performed using an Olympus Fluoview FV3000 confocal microscope and analyzed through the FluoView and CellSens software packages. Images were exported and analyzed using ImageJ software. (https://imagej.nih.gov/ij/index.html, Wayne Rasband, National Institutes of Health, Bethesda, MD, USA). A 100 um × 100 um area was outlined, using the optic nerve as a reference point to ensure consistent placement and total quantity of rods and UV cones within this area were then counted using the ImageJ Cell Counter Plugin.
Rods were detected using both zpr-3 (antibody ID: ZDB-ATB-081002-45, AB_10013805) and 4C12 (antibody ID: ZDB-ATB-090506-2) antibodies. Antibody labeling of tissues in nrl−/− and nrl−/−;tbx2b−/− larvae was determined to be successful, despite lack of detectable rods, because the antibodies also label the pineal gland in larval fish. UV cones were detected using 10C9.1 antibody (antibody ID: ZDB-ATB-140728-2).
Electron Microscopy
Samples were fixed overnight in a solution of 2.5% glutaraldehyde and 2% PFA in 0.1 M phosphate buffer (pH 7.2-7.4). Samples were washed of fixative 3 times with 0.1 M phosphate buffer for 15 minutes, before being incubated for 1 hour in 1% Osmium Tetroxide, followed by 3 more 15-minute phosphate buffer washes. Samples were then dehydrated via sequential 25-minute washes in 50%, 70%, and 90% ethanol, and 3 washes in 100% ethanol. Spurr Resin was introduced via wash in 1:1 mixture of 100% ethanol and Spurr resin, followed by an overnight wash in pure Spurr resin. Spurr resin was replaced twice the following day before samples were placed in block molds and embedded in fresh Spurr and cured in a 70°C oven overnight. Excess resin was then trimmed away with razor blades to expose embedded tissues for ultrathin sectioning on an Ultracut E ultramicrotome (Reichert-Jung). Sections were placed on a copper grid and treated with uranyl acetate for 20 minutes before being rinsed and treated with lead citrate for 7 minutes and rinsed. Stained sections were imaged using a Phillips FEI Morgagni 268 Transmission Electron Microscope, operating at 80 kV. Images were captured with a Gatan Orius CCD camera.
RNA Extraction and Quantitative PCR
RNA samples were extracted from 5-day post fertilization zebrafish embryos using a Qiagen RNeasy minikit (Qiagen: 74104) and quality was verified using an Agilent RNA Nano RNA chip in a 2100 bioanalyzer. All samples had an RNA Integrity Number (RIN) of 9 or higher. Once RNA quality and concentration were validated, sample concentration was standardized to 50 ng/ul. The cDNA was synthesized from extracted RNA using QuantaBio qScript cDNA supermix (QuantaBio 95048-100) in preparation for quantitative PCR.
The quantitative PCR was performed with Applied Biosystems 7500 Fast Real-Time PCR system. The 2X QPCR Mastermix (*Dynamite*) used in this study is a proprietary mix developed and distributed by the Molecular Biology Service Unit (MBSU), in the Department of Biological Science at the University of Alberta, Edmonton, Alberta, Canada. It contains Tris (pH 8.3), KCl, MgCl2, Glycerol, Tween 20, DMSO, dNTPs, ROX as a normalizing dye, SYBR Green (Molecular Probes) as the detection dye, and an antibody inhibited Taq polymerase. Then, 5 ul of quantitative PCR mastermix was used in conjunction with 2.5 ul of 3.2 uM primer mix per 2.5 ul of cDNA sample. Each genotype was analyzed in triplicate with three technical replicates of each sample. All primers were validated via dilution series to produce a standard curve with a minimum r2 of 0.97 and melt curve analysis confirming a single amplified product.
All genotypes were compared against wildtype and each primer pair assessed against primers for b-actin as an endogenous control31 in order to perform ΔΔ-Ct analysis and assign each sample an RQ value. The RQ values were analyzed within the Applied Biosciences 7500 software and used in order to determine proportional transcript abundance, reported as a percentage of wildtype abundance.
Statistical Analysis of Cell Counts and Quantitative PCR
Statistical analyses were performed in GraphPad Prism version 10.0.0 for Windows (GraphPad Software, Boston, MA, USA, www.graphpad.com). Analysis of cell count data used unpaired t-tests. Analysis of quantitative PCR data used Brown-Forscythe and Welch 1-way ANOVA with Dunnett T3’s multiple comparison tests. All quantitative PCR data are presented as mean expression ± standard error of the mean (SEM). Values determined as percentages relative to wildtype transcript abundance. Significance markers are denoted in each respective figure legend.
Results
Nrl−/−;tbx2b−/− Homozygous Double Mutants Recapitulate Single nrl Homozygous Mutant Phenotype: Overabundant UV Cones and No Detectable Rods
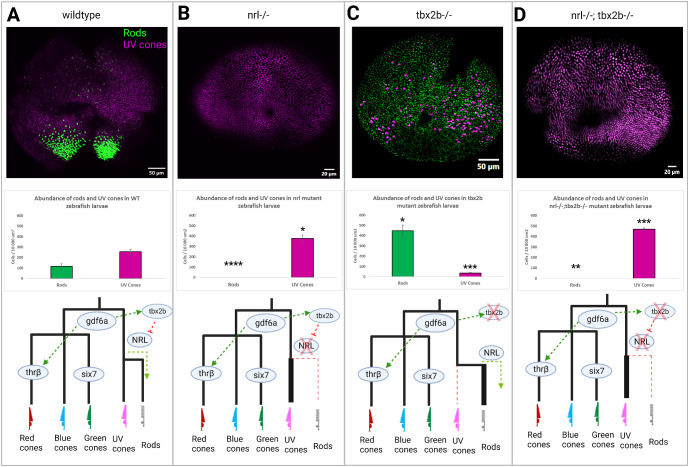
We produced a line of zebrafish with mutations in both nrl and tbx2b (nrl−/−;tbx2b−/−) and compared them against nrl−/− homozygous mutants and against tbx2b−/− homozygous mutants. Immunohistochemistry on 4 days post fertilization (4dpf) larval retina demonstrates that nrl homozygous mutants fail to generate rods while UV cone abundance is significantly increased (Fig. 2B). In exact contradistinction to this, tbx2b−/− single mutants have an excess of rods, but have reduced abundance of UV cones (Fig. 2C; Supplementary Fig. S1). The nrl−/−;tbx2b−/- mutants recapitulate the single nrl mutant phenotype, failing to generate rods, whereas increasing abundance of UV cones (Fig. 2D).
Figure 2.
The nrl and tbx2b share an epistatic relationship governing rod versus UV cone fate. Schematic of putative interaction of tbx2b repressing activation of nrl in otherwise UV cone fated cells. (A) In wildtype zebrafish, nrl is sufficient to induce the rod fate but is repressed by tbx2b in UV fated cells. (B) Loss of nrl expression leads to failure to develop rods and increase in UV cone production (N = 4), as cells fail to adopt the rod fate and instead take on a default UV cone program. (C) Loss of tbx2b releases nrl from repression, driving an excess of rod development in otherwise UV cone fated cells (N = 3). (D) Double mutants recapitulate the single nrl mutant phenotype, producing excess UV cones at the expense of rods (N = 5) as tbx2b is not needed to act on an already non-functional nrl. Together these results indicate that tbx2b is not required for UV cone development, but instead likely prevents nrl expression from imposing the rod fate on UV fated cells. Rods and UV cones detected with antibodies 4C12 and 10C9.1, respectively, label an unknown rod epitope and Sws1 opsin. * = P < 0.05, ** = P < 0.005, *** = P < 0.0005 significance as determined by unpaired t-test. Significance markers denote differences in abundance of each photoreceptor cell type relative to wildtype.
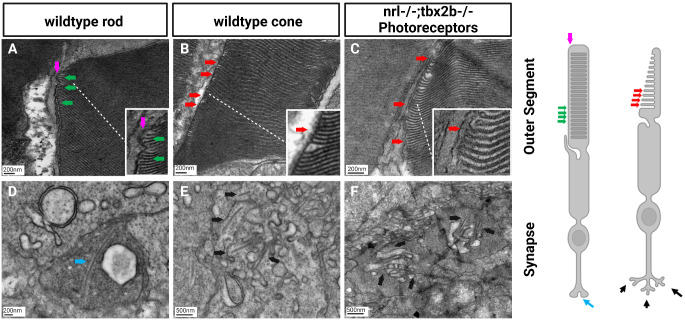
Absence of rods in the nrl−/−;tbx2b−/− mutants was confirmed by ultrastructural examination of developing photoreceptors through transmission electron microscopy. Rod photoreceptors are identifiable in wildtype animals by the distinct morphologies of their outer segments (OS) and synaptic terminals. Rod cells produce outer segment disks with a distinct “hairpin loop” structure, with all the disks being completely enclosed by the OS membrane (Fig. 3A) as opposed to cones’ OS disks being comprised of invaginations of the OS membrane itself (Fig. 3B). Rod synapses are distinguishable by their electron dense terminals, appearing darker in transmission electron microscopy imaging than neighboring cone synapses, and possessing a single synaptic ribbon rather than several ribbons in cones synapses (Figs. 3D, 3E). By these criteria we were not able to confidently identify a single rod cell within the nrl−/−;tbx2b−/− zebrafish, despite examination of eyes from six individual larvae. Contrasting these results to wildtype zebrafish, in which rods were easily identified in all six individuals, supports the conclusion that the necessity of nrl to produce rods supersedes the promotion of the rod fate by loss of tbx2b.
Figure 3.
Electron microscopy reveals lack of rod-like outer segments or synapses in nrl−/−; tbx2b−/− double mutants. Using the metrics described below, no nrl−/−;tbx2b−/− double mutant larvae was found to have a cell that could confidently be described as a rod, despite assessing six individual larvae. In wild types, all six larvae examined had obvious rod photoreceptors. This reaffirms that although mutation of tbx2b promotes a rod-like phenotype, it is insufficient to rescue loss of nrl. Rod cells can be identified morphologically using EM from key distinctive features. Rod outer segments (A) are fully enclosed by the outer segment membrane (magenta arrow) and contain discrete stacked disks which can be identified by characteristic hairpin loops at their ends (green arrows). Cone outer segments (B, C) result from invaginations of the outer segment membrane, producing disk layers made of continuous membrane that are exposed to the extracellular matrix (red arrows). Rod synapses (D) are electron dense structures, appearing darker than the surrounding cone synapses, and contain a single, relatively large, synaptic ribbon (blue arrow). Cone synapses (E, F) are identifiable by an abundance of comparatively shorter synaptic ribbons (black arrows).
Transcript Abundance of Related Photoreceptor Genes Support the nrl and tbx2b Interaction Models
Further connections between these two transcription factors were probed by analysis of transcript abundance through quantitative PCR. Overall transcript abundance of assayed genes was consistently similar between the nrl mutants and the double mutants (Fig. 4). The exception to this trend is abundance of sws1 transcript, which differed significantly between the two, although neither differed significantly from wildtype. Of particular note, was that the abundance of nrl transcript was significantly upregulated in all three mutant lines, consistent with an autoregulatory role for nrl as has been proposed in our previous work, and thus possibly a similar repressive relation between tbx2b and nrl. Abundance of rod specific factors, such as nr2e3 and rh1, were notably downregulated in both the nrl−/− and nrl−/−;tbx2b−/− mutant larvae, consistent with the observed lack of rods from immunohistochemistry. Unexpectedly, the abundance of these genes was not increased in the tbx2b−/- despite their observed increase in rod cells, raising further questions regarding the functionality of these fate shifted photoreceptors.
Figure 4.
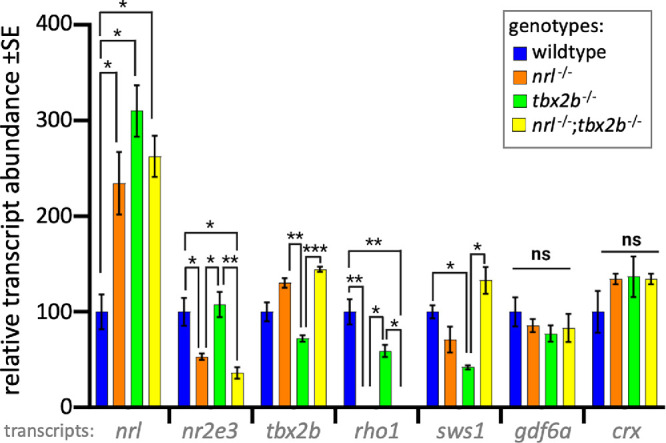
Photoreceptor transcripts in larval zebrafish show that nrl−/−; tbx2b−/− mutants recapitulate phenotypes of nrl−/− mutants. The nrl mutant zebrafish larvae have significantly increased levels of mRNA abundance compared to wildtype, consistent with the notion that nrl has auto-regulatory properties. Similarly, tbx2b mutants also have high nrl transcription, suggesting a repressive interaction. The nrl−/− and nrl−/−;tbx2b−/− mutants share consistently similar transcription profiles across most other assayed genes. Examination of the opsin genes expressed in rods (rho1) and UV cones (sws1) reveals disruption to expected levels of opsin relative to observed abundance of each cell type. Despite their observed overabundance of rods, tbx2b mutants have slightly lower levels of rho1 transcription. Similarly, despite nrl and nrl−/−/tbx2b−/− mutants having increased UV cone abundance, sws1 mRNA abundance does not show similar increases. Error bars represent standard error of the mean. N = 3 to 6 biological replicates. * = P < 0.05, ** = P < 0.005, *** = P < 0.0005 as determined by Brown-Forscythe and Welch 1-way ANOVA with Dunnett T3’s multiple comparison tests.
Discussion
Seminal discoveries in the study of nrl and tbx2b each produced intriguing phenotypes13,16,25 that precisely mirror one another. But those discoveries occurred in distantly related taxa (mice and zebrafish, respectively) that have distinct complements of photoreceptors, and this limited their obvious comparability. Our modest recent contribution confirmed nrl mutant phenotypes in zebrafish,15 and also highlighted the sufficiency of nrl for producing rods in zebrafish,15 prompting the recognition of the strikingly mirrored pattern of rod versus UV cone abundances between genotypes within a single species. Having both genotypes now accessible in the same model organism, the comparison between the two phenotypes became particularly pertinent.
Considering that the single mutant phenotypes suggest that nrl is necessary for rod development, and that tbx2b is necessary for UV cone development, we anticipated the outcome of the double mutant would be informative. If each gene is in fact necessary for the development of their respective photoreceptor, the double mutants would be expected to keep the two phenotypes separate, either failing to produce any rods or UV cones at all (or losing some kind of mutual repression, allowing each cell type to develop). Instead, we observed a complete lack of rods, and retention of UV cones, suggesting that rather than tbx2b being always necessary for UV cone production, tbx2b may instead be involved in an epistatic relation with nrl governing a rod versus UV cone fate decision. Because the double mutants recapitulate the single nrl mutant phenotype, we propose that tbx2b acts upstream of nrl within this genetic mechanism in order to repress nrl’s expression within otherwise UV cone fated cell progenitors (see Fig. 2). With such an interaction, loss of nrl disrupts the cell's ability to take on the rod fate, allowing the UV cone fate to predominate as a default state. Loss of tbx2b thus merely frees nrl to pattern all UV cone fated cells into rods, rather than losing the ability to pattern UV cones themselves.
This potential repressive relationship is further supported by quantitative PCR analysis of these mutants’ transcripts. As has been previously shown, nrl mutants have significantly increased nrl transcript abundance despite their lack of functional Nrl protein,32 suggesting an autoregulatory role with Nrl controlling its own transcription.15 Similarly, the increased abundance of nrl transcript in the tbx2b mutant line suggests a role of tbx2b counteracting the effects of nrl expression within UV cone fated cells. It is, however, worth considering possible confounding explanations. This increase in nrl transcript abundance could potentially be related to extracting total RNA from whole larvae rather than isolating only the retinal tissue, although nrl is only appreciably expressed within eye tissue so complication due to excess tissue is not expected to dramatically influence nrl transcript abundance. Alternatively, the excess nrl transcript could also be impacted by the tbx2b mutant's increased production of rod-like cells, which may over-represent nrl transcription due merely to the increased abundance of cells expected to express nrl. However, the abundance of nr2e3 transcript does not support this notion: if the increased nrl transcript due simply to an excess of rods in tbx2b mutants, we would expect a concomitant increase in nr2e3 transcript as well.
The data demonstrated that tbx2b mutants display a similar increase in nrl transcript abundance, and as such we suspect it plays a similar role in repressing expression of nrl in UV fated cells. Such a developmental relation draws new light to the importance of tbx2b and its homologs in photoreceptor fate determination. Rather than being a critical regulatory element governing the UV cone fate at the expense of potential rod development, tbx2b may instead acts as an anti-nrl factor to prevent the initiation of the rod fate in otherwise UV cone fated cells. These results suggest a pathway wherein the UV cone fate appears as a default state unless acted upon by nrl to divert the developing cell toward the rod fate.
These mutant photoreceptors also demonstrate disruptions to transcription of rod and UV cone opsin in unexpected ways. The mRNA transcript abundance of Rh1 and Sws1, the rod and UV cone opsins, respectively, do not follow the trends expected based on the observed abundance of each photoreceptor type. Surprisingly, tbx2b mutants had significantly decreased transcript abundance of Rh1, despite producing a large population of rods. Likewise, nrl and nrl−/−;tbx2b−/− double mutants do not have the increased sws1 opsin transcript abundance expected based on their increased UV cone production compared to wildtype, although they do differ significantly from one another. These discrepancies open new questions regarding the necessity of these transcription factors to produce phototransduction machinery rather than strictly initiating the patterning of cell identity. Future experiments would be needed to assess whether these disruptions to gene transcription result in measurable electrophysiological or behavioral disturbances in response to light stimuli to determine whether these fate shifted photoreceptors can perform their proper function.
Taken together, this work has begun an exciting investigation into new possibilities in the study of photoreceptor development. By considering tbx2b and short-wavelength cones within the same developmental paradigm as nrl and rods we believe that this work will ultimately offer many novel insights into the developmental and possibly evolutionary history of these cell types.
Future work to investigate the exact nature of tbx2b’s role in this fate determination mechanism is highly warranted. More comprehensive transcriptomics would identify additional details of great interest regarding what additional aspects of rod and UV cone are altered by this transcriptional network, but would likely require single-cell RNA-Seq or similar considering that the questions here are only about a subset of retinal cells, whereas nrl and tbx2b likely have some expression in various cell types. We believe that a zebrafish model expressing tbx2b within developing rod cells would mirror our existing study and further elucidate the potential repressive effects of txb2b on nrl and the rod fate in general. Further, it would be interesting to explore any interactions between nrl and tbx2b in adult zebrafish, where the rod photoreceptor specification appears intact, but their maintenance may not be.15 Additionally, extending these insights to mammalian models would be greatly informative. A retina-specific conditional knockout of Tbx2 in mice would help to solidify our understanding of this genetic interaction and could inform future efforts in cell programming for applications in stem cell therapies.
Supplementary Material
Acknowledgments
The authors thank Phil Oel for discussions regarding conceptualization and early technical assistance. Jim Fadool kindly provided 4C12 antibody. The Zebrafish International Resource Center supplied antibody zpr-3. We would like to thank Arlene Oatway and Kacie Norton of the University of Alberta Biological Sciences Microscopy Unit for their assistance with TEM and confocal imaging, and Troy Locke of the University of Alberta Molecular Biology Service Unit for assistance with RNA work, qPCR troubleshooting and assistance with reagents.
Funding: G.J.N. was supported by an Alberta Graduate Excellence Scholarship and the Alberta Vision Net Studentship via the Cook Family Endowment. Operating funds to W.T.A. were provided by the Natural Sciences and Engineering Research Council of Canada.
Disclosure: G.J. Neil, None; K.H. Kluttig, None; W.T. Allison, None
References
- 1. Fain GL, Hardie R, Laughlin SB.. Phototransduction and the evolution of photoreceptors. Curr Biol. 2010; 20: R114–R124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schultz M. Archiv für mikroskopische Anatomie. Berlin, Germany: J. Springer [etc.]; 1865. [Google Scholar]
- 3. Pugh EN. The discovery of the ability of rod photoreceptors to signal single photons. J Gen Physiol. 2018; 150: 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hunt DM, Carvalho LS, Cowing JA, Davies WL.. Evolution and spectral tuning of visual pigments in birds and mammals. Philos Trans R Soc Lond B Biol Sci. 2009; 364: 2941–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boulanger-Scemama E, El Shamieh S, Démontant V, et al.. Next-generation sequencing applied to a large French cone and cone-rod dystrophy cohort: mutation spectrum and new genotype-phenotype correlation. Orphanet J Rare Dis. 2015; 10: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Verbakel SK, van Huet RAC, Boon CJF, et al.. Non-syndromic retinitis pigmentosa. Prog Retin Eye Res. 2018; 66: 157–186. [DOI] [PubMed] [Google Scholar]
- 7. Charish J, Shabanzadeh AP, Chen D, et al.. Neogenin neutralization prevents photoreceptor loss in inherited retinal degeneration. J Clin Invest. 2020; 130: 2054–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sahaboglu A, Barth M, Secer E, et al.. Olaparib significantly delays photoreceptor loss in a model for hereditary retinal degeneration. Sci Rep. 2016; 6: 39537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Osakada F, Ikeda H, Mandai M, et al.. Toward the generation of rod and cone photoreceptors from mouse, monkey and human embryonic stem cells. Nat Biotechnol. 2008; 26: 215–224. [DOI] [PubMed] [Google Scholar]
- 10. Singh MS, Maclaren RE.. Stem cells as a therapeutic tool for the blind: biology and future prospects. Proc R Soc B: Biol Sci. 2011; 278: 3009–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou L, Wang W, Liu Y, et al.. Differentiation of induced pluripotent stem cells of swine into rod photoreceptors and their integration into the retina. Stem Cells. 2011; 29: 972–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhong X, Gutierrez C, Xue T, et al.. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat Commun. 2014; 5: 4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mears AJ, Kondo M, Swain PK, et al.. Nrl is required for rod photoreceptor development. Nat Genet. 2001; 29: 447–452. [DOI] [PubMed] [Google Scholar]
- 14. McIlvain VA, Knox BE.. Nr2e3 and Nrl can reprogram retinal precursors to the rod fate in Xenopus retina. Dev Dyn. 2007; 236: 1970–1979. [DOI] [PubMed] [Google Scholar]
- 15. Oel AP, Neil GJ, Dong EM, Balay SD, Collett K, Allison WT.. Nrl is dispensable for specification of rod photoreceptors in adult zebrafish despite its deeply conserved requirement earlier in ontogeny. Iscience. 2020; 23(12): 101805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oh ECT, Khan N, Novelli E, Khanna H, Strettoi E, Swaroop A.. Transformation of cone precursors to functional rod photoreceptors by bZIP transcription factor NRL. Proc Natl Acad Sci USA. 2007; 104: 1679–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim JW, Yang HJ, Oel AP, et al.. Recruitment of rod photoreceptors from short-wavelength-sensitive cones during the evolution of nocturnal vision in mammals. Dev Cell. 2016; 37: 520–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Littink K, Stappers P, Riemslag F, et al.. Autosomal recessive NRL mutations in patients with enhanced S-Cone syndrome. Genes. 2018; 9: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yzer S, Barbazetto I, Allikmets R, et al.. Expanded clinical spectrum of enhanced S-cone syndrome. JAMA Ophthalmol. 2013; 131: 1324–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garafalo AV, Calzetti G, Cideciyan AV, et al.. Cone vision changes in the enhanced S-cone syndrome caused by NR2E3 gene mutations. Invest Ophthalmol Vis Sci. 2018; 59: 3209–3219. [DOI] [PubMed] [Google Scholar]
- 21. Samardzija M, Caprara C, Heynen SR, et al.. A mouse model for studying cone photoreceptor pathologies. Invest Ophthalmol Vis Sci. 2014; 55: 5304–5313. [DOI] [PubMed] [Google Scholar]
- 22. Raymond PA, Colvin SM, Jabeen Z, et al.. Patterning the cone mosaic array in zebrafish retina requires specification of ultraviolet-sensitive cones. PLoS One. 2014; 9: e85325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Allison WT, Barthel LK, Skebo KM, Takechi M, Kawamura S, Raymond PA.. Ontogeny of cone photoreceptor mosaics in zebrafish. J Comp Neurol. 2010; 518: 4182–4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. DuVal MG, Allison WT.. Photoreceptor progenitors depend upon coordination of gdf6a, thrβ, and tbx2b to generate precise populations of cone photoreceptor subtypes. Invest Ophthalmol Vis Sci. 2018; 59: 6089–6101. [DOI] [PubMed] [Google Scholar]
- 25. Alvarez-Delfin K, Morris AC, Snelson CD, et al.. Tbx2b is required for ultraviolet photoreceptor cell specification during zebrafish retinal development. Proc Natl Acad Sci USA. 2009; 106: 2023–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Snelson CD, Santhakumar K, Halpern ME, Gamse JT.. Tbx2b is required for the development of the parapineal organ. Development. 2008; 135: 1693–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Choi TY, Khaliq M, Tsurusaki S, et al.. Bone morphogenetic protein signaling governs biliary-driven liver regeneration in zebrafish through tbx2b and id2a. Hepatology. 2017; 66: 1616–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chi NC, Shaw RM, De Val S, et al.. Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes Dev. 2008; 22: 734–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yu W, Mookherjee S, Chaitankar V, et al.. Nrl knockdown by AAV-delivered CRISPR/Cas9 prevents retinal degeneration in mice. Nat Commun. 2017; 8: 14716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Conrad GW, Bee JA, Roche SM, Teillet MA.. Fabrication of microscalpels by electrolysis of tungsten wire in a meniscus. J Neurosci Methods. 1993; 50: 123–127. [DOI] [PubMed] [Google Scholar]
- 31. Fleisch VC, Leighton PL, Wang H, et al.. Targeted mutation of the gene encoding prion protein in zebrafish reveals a conserved role in neuron excitability. Neurobiol Dis. 2013; 55: 11–25. [DOI] [PubMed] [Google Scholar]
- 32. Liu F, Qin Y, Huang Y, et al.. Rod genesis driven by mafba in an nrl knockout zebrafish model with altered photoreceptor composition and progressive retinal degeneration. PLoS Genet. 2022; 18: e1009841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.