Abstract
Streptolysin S (SLS) is a potent cytolytic toxin produced by nearly all group A streptococci (GAS). SLS-deficient Tn916 insertional mutants were generated from two clinical isolates of GAS, MGAS166s and T18Ps (M serotypes 1 and 18, respectively), by transposon mutagenesis using Tn916 donor strain Enterococcus faecalis CG110. Representative nonhemolytic transconjugants SBNH5 and SB30-2 each harbored a single Tn916 insertion in identical loci. The insertion in SBNH5 was located in the promoter region of an open reading frame, designated sagA, rendering it transcriptionally inactive. Protease, streptolysin O, and DNase activities and the production of M protein remained the same in the nonhemolytic mutants and the wild-type strains, as did the growth rates and exoprotein profiles. Transconjugants were evaluated in an established murine model by injecting the organisms subcutaneously and monitoring the mice for alterations in weight and the development of necrotic lesions. Animals infected with SBNH5, compared to those infected with MGAS166s, gained weight during the first 24 h (+1.15 versus −1.16 g; P < 0.05) and had fewer necrotic lesions (0 versus 7; P = 0.0007). Animals infected with SB30-2, compared to those infected with T18Ps, also gained weight within the first 24 h (+0.54 versus −0.66 g; P < 0.05) and produced fewer necrotic lesions (1 versus 8; P = 0.001). Revertants of the mutants in which Tn916 had been excised regained the hemolytic phenotype and the virulence profile of the wild-type strains. This study demonstrates that SLS-deficient mutants of GAS, belonging to different M serotypes and containing identical Tn916 mutations, are markedly less virulent than their isogenic parents.
Streptolysin S (SLS) is produced by virtually all strains of group A streptococci (GAS) and has a direct cytopathic effect on a broad range of cell types (5, 10, 14). SLS is an oxygen-stable, nonimmunogenic cytotoxin which causes a zone of beta-hemolysis observed on the surface of blood agar. This property, used routinely in the clinical laboratory to identify GAS, distinguishes SLS activity from streptolysin O (SLO). SLO is an oxygen-labile, immunogenic hemolysin which does not cause beta-hemolysis on the surface of blood agar plates (14).
The cytolytic spectrum of SLS is broad, including not only erythrocytes of all tested eukaryotes but also lymphocytes, polymorphonuclear leukocytes, platelets, several tissue culture cell lines, tumor cells, bacterial protoplasts, and L forms of bacteria as well as intracellular organelles such as mitochondria and lysozomes (14). By weight, it is one of the most toxic cytolytic agents known (2, 22, 24). The impressive biological activity of SLS is based primarily upon in vitro assays. In vivo evaluations have used impure preparations of SLS, animals of heterogeneous lineage, SLS of various doses, and various routes of administration of the toxin that may not represent routes by which humans are commonly infected by GAS (3, 15, 17, 34). Thus, the role of SLS in the production of tissue damage in vivo has not been precisely elucidated.
Two previous investigations were concerned with generating SLS-deficient mutants of GAS. Owens et al. used nonspecific chemical mutagenesis to create SLS-deficient strains which showed reduced virulence when injected intraperitoneally into mice (29). However, the SLS mutant exhibited growth differences compared to the wild-type parent, suggesting that the expression of multiple genes may have been altered. By Tn916 insertion mutagenesis, Nida and Cleary generated isogenic mutants of an M-type 12 GAS which showed no SLS expression (27). These mutants, however, were not evaluated in an animal model of streptococcal infection, and the chromosomal region associated with loss of SLS production was not characterized.
In this investigation, SLS-deficient Tn916 mutants were generated from two strains of GAS. Each of the wild types was associated with severe streptococcal disease in humans (26, 32). The mutants were compared to the wild types in a murine model of cutaneous streptococcal infection, and a gene associated with SLS production, designated sagA (for SLS-associated gene), was identified.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Strains used in this investigation are listed in Table 1. Gram-positive bacteria were grown in Todd-Hewitt broth (Oxoid, Basingstoke, England) or on Columbia agar (Oxoid) plates containing 5% defibrinated sheep blood (Woodlyn Laboratories, Guelph, Ontario, Canada). When antibiotic selection was required, 2,000 μg of streptomycin (Sigma Laboratories, St. Louis, Mo.) per ml and 5 μg of tetracycline (Sigma) per ml were added to the appropriate media. Escherichia coli was propagated by using Luria-Bertani broth (Difco, Detroit, Mich.). When appropriate, 25 μg of tetracycline per ml and/or 50 μg of chloramphenicol per ml was added to the media. Strains T18P, MGAS166, and CG110 were kindly provided by Patrick Schlievert (University of Minnesota, Minneapolis), James Musser (Baylor College of Medicine, Houston, Tex.), and Don Clewell (University of Michigan, Ann Arbor), respectively. E. coli RN6851(pRN6680) contains a 2.2-kb tetM gene from Tn551 cloned into pBS-bluescript and was kindly provided by Barry Krieswirth (New York Public Health Research Institute, New York).
TABLE 1.
Bacterial strains and relevant properties
| Strain | Relevant phenotypea | Reference | Comments |
|---|---|---|---|
| S. pyogenes | |||
| T18P | M18 Sts Tcs SLS+ | 32 | Isolate associated with rheumatic fever |
| MGAS166 | M1 Sts Tcs SLS+ | 26 | Invasive clinical isolate |
| T18Ps | M18 Str Tcs SLS+ | This paper | Spontaneous Strr derivative of T18P |
| MGAS166s | M1 Str Tcs SLS+ | This paper | Spontaneous Str derivative of MGAS166 |
| SB30-2 | M18 Str Tcr SLS− | This paper | Nonhemolytic derivative of T18Ps |
| 30-2rev | M18 Str Tcs SLS+ | This paper | Hemolytic derivative of SB30-2 |
| SBNH5 | M1 Str Tcr SLS− | This paper | Nonhemolytic derivative of MGAS166s |
| NH5rev | M1 Str Tcs SLS+ | This paper | Hemolytic derivative of SBNH5 |
| ATCC 21547 | SLS+ SLO+ | NRb | Hemolytic control strain |
| ATCC 27762 | SLS− SLO+ | 14 | Nonhemolytic control strain |
| E. faecalis CG110 | Sts Tcr | 11 | Tn916 donor strain |
| E. coli | |||
| RN6851 | Tcr | NR | Contains pRN6680 |
| DH5αMCR | mcrA φ80dlacΔZM15 | Gibco BRL | Library efficiency competent cells |
| SL-1 | Tcs Cmr | This paper | Contains pACYC184 with a 3.8-kb insert |
Str, streptomycin resistant; Sts, streptomycin sensitive; Tcs, tetracycline sensitive; Tcr, tetracycline resistant; Cmr, chloramphenicol resistant.
NR, no reference.
M typing and quantitation.
Serotyping of recipients and nonhemolytic transconjugants was conducted by the National Reference Center for Streptococci (Edmonton, Alberta, Canada) in a blinded fashion according to standard techniques (16). M protein was quantitated by Western blotting with monoclonal antibody to the constant region of the M6 protein (kindly performed by Vincent Fischetti, Rockefeller University) according to published methods (9).
Generation of transconjugants.
Strains MGAS166 and T18P were made resistant to streptomycin by plating each strain on Columbia blood agar containing streptomycin and selecting a colony which became spontaneously resistant to streptomycin. Tn916 was mobilized from Enterococcus faecalis CG110 to MGAS166s and T18Ps by a variation of a method described by Nida and Cleary (27). Cells of the recipient and the donor were added to a nonselective Columbia blood agar plate in a ratio of 1:1, which corresponded to 107 CFU of each strain, and the entire plate was cross-streaked with a sterile loop. After overnight incubation at 37°C in 5% CO2, the bacterial mat was replica plated onto selective media containing tetracycline and streptomycin by use of Acutran sterile replicators (Schleicher and Schuell, Keene, N.H.). Nonhemolytic transconjugants which were devoid of a beta-hemolytic phenotype were chosen and were passaged at least 10 times on selective media to ensure stability of the mutant phenotype. Lancefield grouping was conducted for the nonhemolytic transconjugants (Prolab, Richmondhill, Ontario, Canada) as outlined by the manufacturer.
Southern hybridization analysis.
A probe specific for the tetM gene of Tn916 was used to identify the transposon insertion in the transconjugants. The tetM determinant was amplified from pRN6680 by PCR using T3 and T7 universal primers (Stratagene Cloning Systems, La Jolla, Calif.) and parameters recommended by the manufacturer. The PCR product was confirmed by its size on a 0.7% agarose gel and was purified from the gel with a Qiaex II gel extraction kit (Qiagen, Chatsworth, Calif.). The purified product was labeled by using the enhanced chemiluminescence (ECL) direct labeling system (Amersham, Oakville, Ontario, Canada) as outlined by the manufacturer. Genomic DNA was isolated from GAS by cell lysis (28) followed by DNA purification (36). DNA was digested with HindIII (Boehringer-Mannheim, Laval, Quebec, Canada), subjected to 0.7% agarose gel electrophoresis, transferred to Hybond N+ nylon membranes (Amersham), and probed with the ECL-labeled tetM-specific probe as indicated by the manufacturer.
Cloning and sequencing.
Genomic DNA from MGAS166s was digested with HindIII, ligated into the HindIII site of pACYC184 (New England Biolabs, Mississauga, Ontario, Canada), and transformed into E. coli DH5αMCR high-efficiency competent cells (Gibco BRL, Burlington, Ontario, Canada) by standard techniques (13). Plasmid DNA from transformants was isolated by alkaline lysis (31) and dot blotted onto Hybond N+ membranes by vacuum suction. In order to identify transformants harboring the sequence disrupted by Tn916 in SBNH5, a probe based on the sequence flanking the transposon in SBNH5 was generated by partial-inverse PCR as follows (30). Genomic DNA from SBNH5 was digested with HindIII, self-ligated, and used as a template with outward-reading primers based on the ends of Tn916. The resulting amplicon consequently consisted of the sequence flanking Tn916 and was used as a probe for identifying transformants from MGAS166s harboring the sequence associated with SLS production. Sequencing was done commercially (Mobix, Inc., Hamilton, Ontario, Canada) with an automated sequencer (Applied Biosystems, Oakville, Ontario, Canada) according to the manufacturer’s guidelines. Analysis of sequence data was done by using the Wisconsin Genetics Computer Group sequence analysis software as well as the FASTA algorithm and BLAST search engines of the National Biotechnology Institute.
Northern analysis.
Total RNA was extracted by using Trizol (Gibco BRL) according to the manufacturer’s directions. RNA was isolated from bacteria at mid-log phase (optical density at 550 nm [OD550] = 0.6 to 0.8) and then every 2 h thereafter for a maximum of 10 h. Total RNA was standardized spectrophotometrically and resolved by using 1.9% formaldehyde–agarose gels. RNA electrophoresis and Northern blot transfer were performed according to standard techniques (13). DNA probes were labeled with [α-32P]dCTP by using Ready-to-Go DNA labeling beads (Pharmacia Biotech, Baie d’Urfé, Quebec, Canada) according to the manufacturer’s instructions. The integrity of the RNA was checked simultaneously by probing all samples with a conserved 16S rRNA sequence.
Excision of Tn916.
Phenotypic revertants were produced in a manner similar to that described by Nida and Cleary (27). Briefly, 106 CFU, determined by an OD550 of 1.0 to 1.2, of a late-log-phase culture of nonhemolytic transconjugants was inoculated into 50 ml of nonselective Todd-Hewitt broth. After overnight incubation, 108 CFU was plated onto a single nonselective blood agar plate. Following overnight incubation, zones of hemolysis were identified within the bacterial mat, and colonies within the hemolytic zones were subcultured on nonselective media to isolate the hemolytic revertants. Tetracycline resistance was determined by growth on Columbia blood agar plates containing tetracycline (5 μg/ml).
Growth rate analysis.
To determine the growth rates of wild-type and mutant GAS, 10 ml of Todd-Hewitt broth was inoculated with a single colony. Mutants were grown in the presence of tetracycline. After overnight growth, 106 CFU was used to inoculate 50 ml of Todd-Hewitt broth. OD550 readings with a Beckman spectrophotometer (Beckman Instruments Inc., Fullerton, Calif.) were taken at the time of inoculation and every hour subsequently for 12 h. The actual CFU at each time point were confirmed by serial dilutions and plating on Columbia blood agar.
Hemolysis assays.
To confirm that SLO was still being produced by the nonhemolytic mutants, assays similar to those previously described by Smyth and Duncan (33) were employed. Late-log-phase cultures (OD550 = 1.0 to 1.2) of GAS were centrifuged to pellet the bacteria. Culture supernatants (750 μl) were reduced by adding l-cysteine to a final concentration of 20 mM and incubating the mixture at ambient temperature for 10 min. An equal volume of a 5% solution of sheep erythrocytes, washed three times in 0.15 M sodium phosphate buffer (pH 6.8) (PBS) and resuspended in the same buffer, was added to the culture supernatants, and samples were incubated at 37°C for 60 min. After centrifugation, the OD540 was measured to determine the release of hemoglobin. An equivalent amount of lysed erythrocytes suspended in sterile Todd-Hewitt broth was used as a control to represent 100% hemolysis, and sample values were recorded as a fraction of this value. Trypan blue (Sigma), at a final concentration of 13 μg/ml, and cholesterol (Sigma), at a final concentration of 0.5 mg/ml, were used as inhibitors of SLS and SLO, respectively. ATCC 21547 (SLO+ SLS+) and 27762 (SLO+ SLS−) were used as control strains (Table 1).
SLS activity was also measured during early, mid-, and late log phase by the method described above. Overnight broth cultures of MGAS166s and SBNH5 were subcultured in Todd-Hewitt broth, and samples were withdrawn hourly for 8 h and immediately frozen at −70°C. Samples were thawed, and the bacteria were pelleted by centrifugation. Serial dilutions of culture supernatant were added to PBS-washed 5% rabbit erythrocytes, and the mixtures were incubated at 37°C for 60 min. Cells were removed by centrifugation, and the optical density was determined as described above.
Preparation of extracellular and cell-associated proteins.
Bacteria were grown in 200 ml of Todd-Hewitt broth, and samples were collected at either mid-log phase (OD550 = 0.6) or late log phase (OD550 = 1.0 to 1.2). Bacteria were pelleted by centrifugation at 10,000 × g for 15 min at 4°C. Ammonium sulfate was added to the culture supernatants gradually with constant shaking at 4°C until the solution reached 80% saturation. After gentle mixing overnight at 4°C, the tubes were centrifuged at 10,000 × g, and the supernatant was discarded. The ammonium sulfate precipitate was dissolved in 2 ml of 0.01 M ammonium bicarbonate (pH 7.0) and dialyzed against the same solution by using Slide-A-Lyzer dialysis cassettes (Pierce Chemical Co., Arlington Heights, Ill.) overnight at 4°C. Dialysate samples were boiled for 5 min in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (23), resolved with an SDS–10% polyacrylamide gel, and stained with Coomassie brilliant blue R.
For analysis of cellular proteins, bacteria were grown in 10 ml of Todd-Hewitt broth and were pelleted when they reached mid- or late log phase. The supernatants were discarded, and the pellets were resuspended in 15 μl of 0.1% Triton X-100 (Sigma)–25 mM PBS (pH 7.2) and vortexed briefly. After the cells were incubated at 37°C for 30 min, 15 μl of SDS-PAGE loading buffer was added and samples were resolved as described above.
Production of caseinase and DNase.
Caseinase activity was determined by the method of Wheeler et al. (35). DNase production was determined by using commercial media (Difco). In the two assays, equivalent inocula of late-log-phase organisms were spotted onto assay plates. The plates were incubated anaerobically overnight, and zones of opacity or clearing were measured to determine caseinase or DNase activity, respectively. SBNH5 and SB30-2 were tested with and without 5 μg of tetracycline per ml in the media.
Quantitation of hyaluronic acid.
Bacteria were grown in 150 ml of Todd-Hewitt broth to an OD550 of 0.6 to 0.8. Mutants were grown in the presence of tetracycline. Aliquots of the cultures were removed, serially diluted, and subcultured to confirm the exact number of CFU. The bacterial pellet was harvested by centrifugation and washed once with sterile distilled water. The pellet was resuspended in 1.5 ml of water, an equal volume of chloroform was added, and the solution was mixed vigorously and incubated at room temperature for 1 h. The mixture was centrifuged to separate the aqueous phase from the chloroform. The aqueous phase was used in the carbazole method of uronic acid quantitation as described by Knutson and Jeanes (21). Human umbilical hyaluronic acid (Sigma) was used as a standard.
Dermonecrotic mouse model.
Virulence of GAS strains was determined by using a dermonecrotic mouse model as previously described (6). Organisms were grown to mid-log phase (OD550 = 0.6 to 0.8) in Todd-Hewitt broth with appropriate antibiotic selection. A 100-μl volume of mid-log-phase organisms was mixed with an equal volume of sterilized Cytodex beads (Sigma) suspended in PBS at a concentration of 20 μg/ml. The 200-μl Cytodex-bacterium suspension was injected subcutaneously into the right flank of hairless, 4-week-old male crl:SKH1(hrhr)Br mice (Charles River, Wilmington, Mass.) weighing 15 to 20 g by using a 1-ml tuberculin syringe. Nine animals were injected for each strain examined. Viable counts were performed for all cultures to confirm the exact number of CFU injected. Animals were weighed immediately prior to inoculation and every 24 h subsequently for a total of 5 days. The length and width of the lesions were measured daily by an observer blinded to the identity of the infecting strain. The wound area (A) was determined by A = π (L × W)/2, where L is the longest axis and W is the shortest axis.
Culturing of necrotic lesions and histopathology.
To determine the phenotypes of the organisms in the lesions, a single animal was randomly chosen from each group 24 and 120 h after infection and euthanized. The wounds were excised from the euthanized animals and divided equally. One half of each lesion was cultured, and the other half was used for the preparation of histological specimens. Tissue for culture was suspended in 1 ml of sterile PBS and then ground in a sterile tissue homogenizer. Aliquots of the PBS-tissue homogenate were serially diluted and inoculated on either selective or nonselective Columbia blood agar plates and scored for beta-hemolysis. Histologic sections were prepared by immersion in 10% buffered formalin and embedded in paraffin. Sections were stained with hematoxylin and eosin or tissue Gram stain (Brown-Benn stain) and examined by light microscopy by a pathologist blinded to the source of the biopsies.
Statistics.
Statistical analysis was conducted as described previously (6). Group means for weight loss and lesion size were compared by analysis of variance (ANOVA). Post hoc tests were done by using Fisher’s protected least significant difference (Fisher’s PSLD). P values reported refer to the ANOVA tests unless stated otherwise. Significant differences between pairs of groups were reported if P was <0.05. Fisher’s exact test was used to compare counts of dermonecrotic lesions.
RESULTS
Generation of nonhemolytic transconjugants.
Nonhemolytic tetracycline-resistant transconjugants resulted from mating GAS strains MGAS166s and T18Ps with the Tn916 donor strain E. faecalis CG110 at a frequency of 10−4 for both recipients. Transconjugants maintained the nonhemolytic phenotype after subculture on selective media. When Tn916 excision assays were conducted with nonhemolytic transconjugants derived from T18Ps and MGAS166s, the wild-type, beta-hemolytic phenotype was restored and detected as a zone of beta-hemolysis within a confluent mat of nonhemolytic bacteria. The frequencies of excision of Tn916 were on the order of 10−8 and 10−7 for SBNH5 and SB30-2, respectively. Because of the low frequency of Tn916 excision, it was necessary to screen for hemolytic revertants on a confluent mat of bacteria.
Genetic characterization of the nonhemolytic transconjugants and hemolytic revertants.
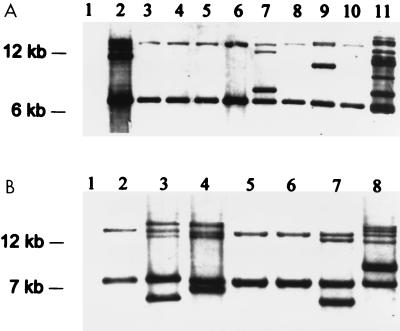
The Tn916 probe used spanned the only HindIII restriction site within Tn916. Cleavage at this site divides the transposon into two fragments of approximately 6 and 12 kb (7). After HindIII digestion, each copy of Tn916 which has integrated into the chromosome of the recipient strains yields two bands that hybridize with the probe. Two HindIII fragments, approximately 14 and 7.8 kb, from each of the nonhemolytic mutants derived from MGAS166s hybridized with the tetM probe. For each nonhemolytic mutant derived from T18Ps, HindIII fragments of 14 and 6.5 kb hybridized with the tetM probe. This pattern was seen even in nonhemolytic transconjugants which possessed more than one insertion (Fig. 1). Two nonhemolytic transconjugants derived from T18Ps, designated SB30-2, and one from MGAS166s, designated SBNH5, were chosen for further study.
FIG. 1.
Southern hybridization analysis of HindIII restriction digests of genomic DNA probed with tetM. (A) Hemolytic wild-type T18P (lane 1) does not hybridize with the tetM probe. Nonhemolytic transconjugants SB1-4, SB2-9, SB30-2, SB1-9, SB5-9, SB6-9, SB7-9, SB1-1, and SB8-2 (lanes 2 to 10, respectively) all contain at least one copy of Tn916 and hybridize with the tetM probe. Isolates in lanes 2, 7, and 9 possess more than one Tn916 insertion. All lanes possess two bands hybridizing with the tetM probe corresponding to approximately 6.5 and 14 kb. The Tn916 donor strain CG110 (lane 11) contains several copies of Tn916. (B) Hemolytic wild-type MGAS166s (lane 1) does not hybridize with the tetM probe. The nonhemolytic transconjugants SBNH1, SBNH3, SBNH4, SBNH5, SBNH6, SBNH7, and SBNH8 (lanes 2 to 8, respectively) all possess at least one copy of Tn916. Isolates in lanes 3, 4, 7, and 8 possess more than one Tn916 insertion. Isolates in all lanes possess two bands of similar size, approximately 14 and 7.5 kb. The migration of molecular size standards (1-kb ladder) is indicated on the left.
Excision of Tn916 from SB30-2 and SBNH5 was permitted by growth in the absence of tetracycline and confirmed by detecting tetracycline-susceptible hemolytic revertants. Restoration of the wild-type phenotype is consistent with previous reports that Tn916 is capable of precise excision (12). Two revertants, NH5rev and 30-2rev (derived from SBNH5 and SB30-2, respectively), were selected for further analysis. Neither revertant hybridized with the tetM-specific probe, and excision of Tn916 was precise, since it resulted in restoration of the hemolytic phenotype.
Analysis of the Tn916 insertion site.
In order to identify the wild-type region into which Tn916 inserted, a genomic library of MGAS166s was generated by using the low-copy-number plasmid pACYC184. Clones containing the wild-type region corresponding to the insertion site of Tn916 were identified by using a 2.2-kb partial-inverse-PCR product which was generated with Tn916-derived outward-reading primers. Three clones containing a 3.8-kb fragment which hybridized with the Tn916 flanking-region probe were identified. A single clone, SL-1, was chosen for further analysis. It was confirmed that the 3.8-kb insert in pSL-1 corresponded to the region interrupted by Tn916 insertion in the wild type by probing HindIII-digested genomic DNA from both MGAS166s and SBNH5 with the 3.8-kb insert. A single band at 3.8 kb was detected in MGAS166s, while two bands at approximately 14 and 7.8 kb were detected in SBNH5.
The entire 3.8-kb insert in pSL-1 was sequenced in both directions, yielding a fragment of exactly 3,732 bp. Analysis of the sequence with the Wisconsin Genetics Computer Group computer program identified several putative open reading frames (ORFs). However, only a single ORF, designated sagA, demonstrated nearly all of the conserved elements of a functional transcript (Fig. 2). A consensus Shine-Dalgarno sequence (AGGAGG) is located exactly 10 bp upstream of the ATG start codon. Approximately 150 bp upstream of this site is the −10 promoter (TATAAT), and 167 bp upstream of this lies the putative −35 promoter region sequence TTTACA. The sagA ORF appears to code for a peptide of 53 amino acids which is devoid of a signal sequence. Note the unusual presence of several cysteine residues near the amino terminus: seven cysteines, five consecutive, followed by two tyrosines, followed by two more cysteine residues. Analysis of the sequence of sagA by FASTA and BLAST searches failed to detect significant homology with other known sequences. Furthermore, the sequence of sagA was not found in the Oklahoma GAS genomic sequence database.
FIG. 2.
Nucleotide sequence and protein translation of sagA. A 390-bp region of genomic DNA from MGAS166s corresponding to the chromosomal point of insertion of Tn916 (▿) is indicated. The putative consensus elements of the sagA ORF and the putative 53-amino-acid translation product are shown. S.D., Shine-Dalgarno consensus sequence.
To determine the exact insertion site of Tn916, PCR products were generated by using primers based on the known 3.8-kb sequence coupled with outward-reading primers from Tn916. The PCR products were sequenced and allowed precise determination of the Tn916 insertion point which was midway within the putative promoter region of sagA, 11 bp downstream of the −35 element and 6 bp upstream of the −10 TATA box.
Sequence analysis of PCR products of the genomic DNA flanking Tn916 derived from SB30-2 confirmed that the Tn916 insertion was in exactly the same locus. Furthermore, Tn916 was oriented in the same direction as it was in SBNH5.
Expression of sagA.
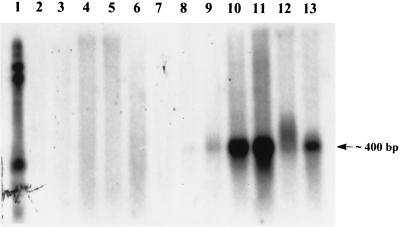
To determine if sagA was transcribed, RNA was isolated from MGAS166s and SBNH5 and probed with DNA corresponding only to sagA (Fig. 3). A transcript in RNA isolated from MGAS166s which gave a maximal signal at 4 to 6 h post-mid-log phase was detected. The transcription product corresponded to a size of approximately 400 bp, which was in keeping with the expected size of an mRNA product from sagA. No detectable transcript was observed from RNA isolated from SBNH5 at any time point. Probing of the same membranes with the 16S rRNA probe did not yield any differences between RNAs from MGAS166s and SBNH5.
FIG. 3.
Total RNA extracted from mutant SBNH5 (lanes 2 to 7) and wild-type MGAS166s (lanes 8 to 13) was quantified, standardized, blotted, and probed with a PCR amplicon of sagA labeled with α-32P. Lane 1, 0.16- to 1.77-kb RNA standard; lane 2, SBNH5 RNA harvested at mid-log phase; lanes 3 to 7, SBNH5 RNA at 2, 4, 6, 8, and 10 h post-mid-log phase, respectively; lane 8, MGAS166s RNA harvested at mid-log phase; lanes 9 to 13, MGAS166s RNA at 2, 4, 6, 8, and 10 h post-mid-log phase, respectively. The mutant strain is devoid of any transcripts from sagA, while the wild type contains sagA transcripts at all time points.
M typing of mutants.
M typing of nonhemolytic transconjugants confirmed that M protein was produced, and both SB30-2 and SBNH5 had the same M-protein phenotypes as their M18 and M1 parent strains, respectively. No difference in quantity of M protein was seen between MGAS166s and SBNH5 by Western blotting using a monoclonal antibody to the constant region of M6 protein.
Hemolytic activity.
The nonhemolytic mutants SBNH5 and SB30-2 showed no beta-hemolysis on blood agar, indicating that SLS activity had been ablated. Their hemolysis profiles were identical to that of ATCC 27762, which does not produce SLS but does produce SLO (4). An assay specific for SLO conducted under reducing conditions showed continued SLO production in all strains of GAS tested. Hemolysis was detected in the presence of the SLS inhibitor trypan blue but not in the presence of both trypan blue and the SLO inhibitor cholesterol (Table 2). SLS production peaked at late log phase for MGAS166s, whereas there was no detectable SLS activity for SBNH5 at any time point measured. These results confirm that SLO was not affected by the insertion of Tn916 and that the absence of beta-hemolysis was attributable to the loss of SLS activity, a profile similar to that of the SLS-deficient Tn916 insertion mutants described by Nida and Cleary (27).
TABLE 2.
SLO activities of wild-type and mutant streptococci
| Assay contents | Fraction of complete lysisa exhibited by bacterial strain:
|
|||||
|---|---|---|---|---|---|---|
| T18Ps | SB30-2 | MGAS166s | SBNH5 | ATCC 27762 | ATCC 21547 | |
| Supernatant | 0.57 | 0.48 | 0.56 | 0.45 | 0.62 | 0.78 |
| Supernatant with trypan blueb | 0.64 | 0.50 | 0.48 | 0.39 | 0.58 | 0.69 |
| Supernatant with trypan blue and cholesterolc | 0.05 | 0.09 | 0.10 | 0.07 | 0.11 | 0.04 |
Complete lysis was determined by lysing 750 μl of 5% washed sheep erythrocytes in hypotonic saline and adding the mixture to an equal volume of sterile Todd-Hewitt broth.
The concentration of trypan blue was 13 μg/ml.
The concentration of cholesterol was 0.5 mg/ml.
Growth rate comparisons.
To determine if the mutation conferred by Tn916 insertion had affected growth in addition to ablating SLS activity, the growth rates of the mutants were compared to those of their parent strains. No difference in the growth rate of either SB30-2 or SBNH5 was found.
Protein and hyaluronic acid capsule production.
There was no difference in production of cell-associated and extracellular proteins, resolved by SDS-PAGE, between the nonhemolytic mutants and their parent strains, suggesting that the Tn916 insertion had no gross pleiotropic effect. In addition, hyaluronic acid production was measured, and the strains were tested for DNase and caseinase activity. Both nonhemolytic insertion mutants retained their wild-type activities in all three assays (Table 3).
TABLE 3.
Phenotypic comparisons between hemolytic and nonhemolytic GAS
| Strain | Assay result
|
||
|---|---|---|---|
| Caseinase (mm)a | DNase (mm)a | Hyaluronic acid (fg/CFU) | |
| MGAS166s | 12.8 ± 1.3 | 15.1 ± 0.9 | 2 |
| SBNH5 | 12.2 ± 1.1 | 16.3 ± 1.3 | 3.1 |
| T18Ps | 0 | 16.1 ± 1.6 | 68 |
| SB30-2 | 0 | 17.4 ± 2.0 | 54 |
Results are diameters of zones surrounding the inoculum after overnight anaerobic incubation of assay plates at 37°C. Measurements are means ± SD of three experiments.
Reduced virulence of SLS-deficient transconjugants.
Reproducible, nonlethal lesions were generated following injection of 106 CFU of MGAS166s and 107 CFU of T18Ps subcutaneously into mice. The different inoculum sizes needed to produce the same virulence profile are likely due to inherent differences in virulence between the M1 and M18 serotypes of GAS.
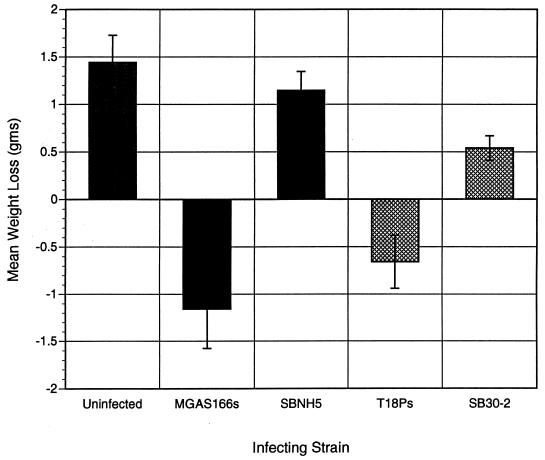
Nonhemolytic transconjugants SB30-2 and SBNH5 showed virulence which was markedly reduced compared to that of their wild-type counterparts. Mice infected with 106 CFU of wild-type strain MGAS166s exhibited a mean weight loss (± standard deviation [SD]) of −1.16 ± 0.42 g, compared to mice which received either SBNH5 (+1.15 ± 0.2 g) or sterile Cytodex alone (1.44 ± 0.29 g) (P < 0.05; Fisher’s PSLD) (Fig. 4). Similarly, mice injected with 107 CFU of the wild-type hemolytic T18Ps exhibited a significant mean weight loss (−0.66 ± 0.28 g [± SD]) in the first 24 h after infection, in contrast to the weight gain observed in mice which received the same infective dose of the nonhemolytic SB30-2 (+0.54 ± 0.13 g) (P < 0.05; Fisher’s PSLD).
FIG. 4.
Comparisons of mean weight changes 24 h after infection with wild-type strains (MGAS166s and T18Ps) and the respective isogenic nonhemolytic mutants (SBNH5 and SB30-2). Animals infected with nonhemolytic mutants of each wild type gained weight, in contrast to the marked weight loss caused by infection with the parent strains.
None of the nine mice which received the nonhemolytic transconjugant SBNH5 developed a necrotic lesion, while seven of the nine mice (78%) which received the wild-type MGAS166s developed necrotic lesions (P = 0.0007; Fisher’s exact test). Similarly, of the nine mice which received SB30-2, only one (11%) developed a necrotic lesion, compared to eight of the nine mice (89%) which developed necrotic lesions when injected with the wild-type T18Ps (P = 0.001; Fisher’s exact test). Data for the M1 and M18 strains were similar to each other in two separate experiments.
Two phenotypic revertants (30-2rev and NH5rev) from which Tn916 had been excised (derived from SB30-2 and SBNH5, respectively) were compared to the wild types, T18Ps and MGAS166s. The number of necrotic lesions and weight changes were not significantly different from those produced by the wild type in each case.
Gross and histological characterization of infected tissue.
For mice infected with MGAS166s, initial examination of the lesions revealed indurated zones surrounded by edema. The indurated zones subsequently progressed, yielding centralized ulceration and necrosis which did not penetrate the underlying musculature (Fig. 5). MGAS166s produced a maximum mean necrotic lesion size of 90.4 mm2. No necrotic lesions were observed in animals infected with SBNH5, though some animals did develop slight localized edema within 24 h of infection, similarly to the mice which received sterile Cytodex. Animals infected with the M18 strains, T18Ps and SB30-2, showed similar patterns when the wild types were compared with the nonhemolytic mutants. The maximum mean necrotic lesion area was 31 mm2 in animals infected with T18P. For the single animals which were infected with SB30-2 and developed lesions in two separate experiments, the maximum area was 10 mm2.
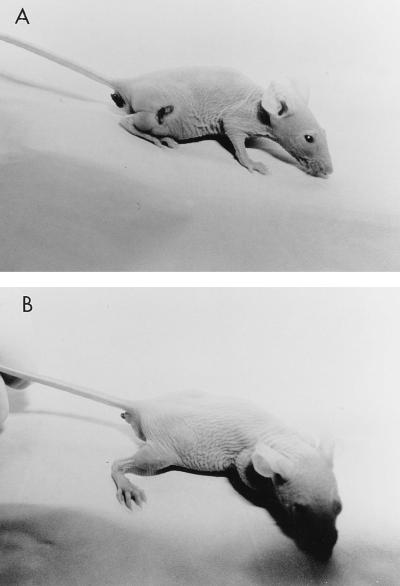
FIG. 5.
Photographs of hairless SKH1 mice 24 h after infection with 106 CFU of either the SLS-producing wild-type MGAS166s (A) or the SLS-deficient Tn916 mutant SBNH5 (B). A well-demarcated zone of induration with centralized necrosis is visible on the right flank of the mouse infected with MGAS166s. No necrosis was seen in mice infected with SBNH5.
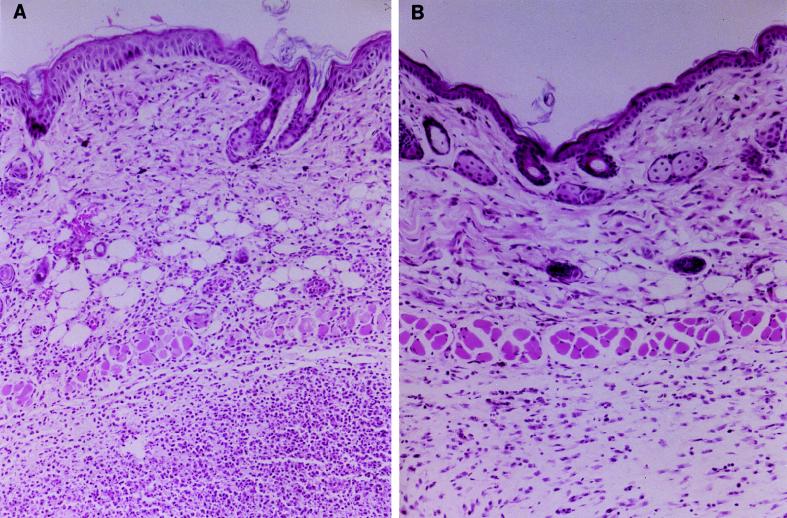
Twenty-four hours postinfection, biopsies of tissue from animals which had been infected with MGAS166s differed histologically from those from SBNH5- or sterile-Cytodex-inoculated animals. Sections of tissue from mice which received MGAS166s demonstrated evidence of profuse acute inflammation with dense infiltration of neutrophils and tissue necrosis. Biopsies from mice which received SBNH5 did not show evidence of acute inflammation, and no tissue damage was evident (Fig. 6). Gram staining of the sections revealed gram-positive cocci distributed throughout the tissue obtained from mice infected with MGAS166s, while tissue from mice which received SBNH5 failed to demonstrate any bacteria in all fields scanned. Examination of hematoxylin-and-eosin-stained or Gram-stained tissue sections from mice which received SBNH5 did not show an appreciable difference compared with tissue from mice which received a sterile-Cytodex injection.
FIG. 6.
Tissue biopsies from euthanized mice which were infected with 106 CFU of either the SLS-producing wild-type MGAS166s (A) or the SLS-deficient Tn916 mutant SBNH5 (B). The tissue section in panel A demonstrates acute inflammation with edema and tissue necrosis. The tissue depicted in panel B does not show evidence of necrosis, and the inflammation is markedly reduced compared with that in panel A. Tissue samples were stained with hematoxylin and eosin, and the final magnification is approximately ×17.
Culturing of lesions.
To determine if the phenotype of the infecting strains had remained the same as that of the injected organisms, lesions from animals which had received either MGAS166s or SBNH5 were cultured after 1 and 5 days. As there were no necrotic lesions on mice infected with SBNH5, the erythematous injection site, comparable in size to the lesion on the mice which received sterile Cytodex, was excised for culturing. All lesions from animals which received MGAS166s grew tetracycline-susceptible hemolytic GAS. However, no organisms grew from tissue cultured at either 1 or 5 days from mice which had received SBNH5. In two separate experiments, one of nine mice infected with SB30-2 developed necrotic lesions from which hemolytic tetracycline-susceptible GAS were cultured. Animals infected with SB30-2 received an inoculum of 107 CFU, probably sufficient to permit the emergence of revertants from which Tn916 had been excised. Growth of the revertants may explain the production of the small necrotic lesion in animals infected with the nonhemolytic SB30-2 in two separate experiments.
DISCUSSION
Despite the fact that SLS activity is one of the defining phenotypes of GAS, there has been a lack of research into the association between SLS production and the virulence of GAS. In this investigation, two distinct strains of GAS belonging to different M serotypes which were each isolated from human GAS infections (26, 32) were rendered SLS deficient by Tn916 mutagenesis. All nonhemolytic Tn916 mutants generated in this investigation belonging to the same M serotype harbored a Tn916 insertion in an identically sized HindIII chromosomal fragment, suggesting that mutation of a common region is responsible for loss of the SLS phenotype. Comparison of the Tn916 insertions in SB30-2 and SBNH5 confirmed that the insertion site, an AT-rich region which is a preferred site for Tn916 integration (25), was identical in these two mutants. Because of this finding and the fact that the SLS hemolytic phenotype was restored upon excision of Tn916, it is unlikely that loss of the SLS phenotype was due to an independent spontaneous mutation. Furthermore, because phenotypic reversion was demonstrated with the excision of Tn916, loss of the hemolytic phenotype was not due to deletions in the chromosomal regions flanking Tn916 as has been reported by other investigators (8).
The ORF identified in this study as being associated with SLS production, sagA, was transcriptionally active in MGAS166s but not in SBNH5 (Fig. 3). Identical results were obtained by Northern analysis of T18Ps and SB30-2 RNAs (results not shown). Although sagA is transcriptionally active and its Shine-Dalgarno sequence suggests that it is translated, it is not possible at this point to conclude whether sagA corresponds to the structural gene for SLS. However, the small size of the gene, with an estimated translational product size of 53 amino acids, is in keeping with the estimated small size of SLS (24). The sequence of sagA appears to be novel, since it lacks homology with any other known determinant, whether regulatory or structural. In addition, it is of interest that the sequence corresponding to sagA is absent from the Oklahoma database representing more than 95% of the M1 streptococcal genome.
Interestingly, the putative translational product of sagA contains several cysteine residues. Early work done to determine the amino acid composition of purified SLS (24) failed to detect any cysteine. Akao et al. (1) demonstrated that a peptide essential for SLS production was rich in cysteine-mediated disulfide bridges. Whether there is any functional relationship between the essential peptide described by Akao et al. and the translational product of sagA is not known. The possibility that sagA may correspond to a transcriptional or functional regulator of SLS activity must be considered; however, the fact that sagA fails to show homology with other characterized transcriptional regulators makes this appear unlikely.
Both SLS-deficient mutants, SBNH5 and SB30-2, were shown to have markedly reduced virulence compared to that of their wild types in a murine model of subcutaneous GAS infection. Weight loss and the development of necrotic lesions were used in this investigation as indicators of virulence when mice infected with different GAS were compared (6, 20). Such nonlethal models may be more capable of detecting subtle differences in virulence than are lethal models (19). The ability of this investigation to demonstrate such a marked difference in virulence between nonhemolytic GAS and their wild-type parents may be due to the animal model we employed. We used a model of cutaneous infection which facilitates close contact between streptococcal cells and eukaryotic subcutaneous tissue. Also, subcutaneous injection of GAS may more closely resemble the mechanism by which humans become infected by GAS, in contrast to intraperitoneal injection, as was used by previous investigators (29). Furthermore, as SLS often exists in a cell-bound state (14), direct inoculation of the bacteria into tissue may facilitate a mechanism whereby SLS could mediate its pathogenic effect in a localized manner (18).
No bacteria could be detected in the tissue from mice infected with SBNH5, either by direct microscopic examination or by culturing. It is possible that SLS is directly involved in preventing clearance of the GAS from the host or that other factors that were not detected in vitro are involved. It is notable that other investigators have documented that SLS is cytotoxic to numerous cells, including lymphocytes and polymorphonuclear leukocytes (10, 14). This cytotoxic activity of SLS may consequently impair clearance of GAS and could account for the rapid clearance of the SLS-deficient mutant SBNH5 from the mice.
A previous investigation demonstrated decreased virulence of a chemically mutagenized SLS-deficient derivative of GAS (29). However, the nonhemolytic mutant in that study demonstrated not only SLS deficiency but also a decreased growth rate in comparison to the wild-type strain. This suggested that more than SLS production had been affected, making the contribution of SLS to virulence difficult to assess. Unlike the SLS-deficient mutants described previously (29), the Tn916 nonhemolytic mutants generated in this investigation did not differ from their wild-type parents in growth rate. Furthermore, there was no evidence that the SLS-deficient mutants, SBNH5 and SB30-2, exhibited any pleiotropic alteration compared to their respective wild types, since the results of specific assays for hyaluronic acid production, caseinase, DNase, SLO, and M-protein quantitation remained unaltered. Also, no difference was found by SDS-PAGE analysis of cell-associated and extracellular proteins.
As is always the case with mutagenesis experiments, the possibility that subtle alterations affecting undetected determinants may have occurred or that such differences, although not manifested in vitro, are expressed in vivo cannot be ruled out. Indeed, polar effects conferred by the transposon could affect more than SLS expression alone. Direct polar effects of Tn916 appear unlikely, as Northern analysis using nonoverlapping probes constructed from the 3,729-bp fragment failed to detect any transcriptional activity in either the wild type or the mutant (results not shown). Furthermore, the idea that Tn916 has disrupted a pleiotropic regulator, although unlikely on the basis of phenotypic and DNA homology comparisons, must also be entertained. Neither of these potential effects can be definitively ruled out on the basis of negative data.
In conclusion, this investigation suggests that Tn916 insertion into a common putative ORF in two distinct strains of GAS results in an SLS-deficient phenotype. The mechanism by which SLS expression is affected is unclear. However, this phenotype is associated, either directly or indirectly, with decreased virulence in a murine model of cutaneous infection. These initial efforts at identification and characterization of this novel region should facilitate the ongoing elucidation of the elusive virulence determinants of GAS.
ACKNOWLEDGMENTS
We thank Vincent Fischetti for conducting our M-protein quantitation assays; the Department of Pathology, Ann Arbor Department of Veterans’ Affairs, for preparation and interpretation of histologic specimens; and James Talbot and Margaret Lovgren for M typing.
This work was supported by a grant from the Canadian Bacterial Diseases Network.
REFERENCES
- 1.Akao T, Takahashi T, Kobashi K. Purification and characterization of a peptide essential for formation of streptolysin S by Streptococcus pyogenes. Infect Immun. 1992;60:4777–4780. doi: 10.1128/iai.60.11.4777-4780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alouf J E. Streptococcal toxins (streptolysin O, streptolysin S, erythrogenic toxin) Pharmacol Ther. 1980;11:661–717. doi: 10.1016/0163-7258(80)90045-5. [DOI] [PubMed] [Google Scholar]
- 3.Barnard W G, Todd E W. Lesions in mouse produced by streptolysins O and S. J Pathol Bacteriol. 1940;51:43–47. [Google Scholar]
- 4.Bernheimer A W. Streptococcal infections. New York, N.Y: Columbia University Press; 1954. p. 19. [Google Scholar]
- 5.Bernheimer A W. Hemolysins of streptococci: characterization and effects on biological membranes. In: Wannamaker L W, Matson J M, editors. Streptococci and streptococcal diseases. New York, N.Y: Academic Press; 1972. pp. 19–31. [Google Scholar]
- 6.Bunce C, Wheeler L, Reed G, Musser J, Barg N. A murine model of cutaneous infection with gram-positive cocci. Infect Immun. 1992;60:2636–2640. doi: 10.1128/iai.60.7.2636-2640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clewell D B, Flannagan S E. The conjugative transposons of gram-positive bacteria. New York, N.Y: Plenum Press; 1993. pp. 369–393. [Google Scholar]
- 8.Dougherty B A, Van de Rijn I. Molecular characterization of a locus required for hyaluronic acid capsule production in group A streptococci. J Exp Med. 1992;175:1291–1299. doi: 10.1084/jem.175.5.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischetti V A, Jones K F, Scott J R. Size variation of the M protein in group A streptococci. J Exp Med. 1985;161:1384–1401. doi: 10.1084/jem.161.6.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freer J H, Arbuthnott J P. Biochemical and morphological alterations of membranes by bacterial toxins. In: Bernheimer A W, editor. Mechanisms in bacterial toxinology. New York, N.Y: John Wiley and Sons; 1976. pp. 170–193. [Google Scholar]
- 11.Gawron-Burke C, Clewell D B. A transposon in Streptococcus faecalis with fertility properties. Nature (London) 1982;300:281–284. doi: 10.1038/300281a0. [DOI] [PubMed] [Google Scholar]
- 12.Gawron-Burke C, Clewell D B. Regeneration of insertionally inactivated streptococcal DNA fragments after excision of transposon Tn916 in Escherichia coli: strategy for targeting and cloning of genes from gram-positive bacteria. J Bacteriol. 1984;159:214–221. doi: 10.1128/jb.159.1.214-221.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilman M. Preparation of RNA from eukaryotic and prokaryotic cells. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1997. pp. 4.1.1–4.1.23. [Google Scholar]
- 14.Ginsburg I. Streptolysin S. In: Montie T C, Kadis S, Ajl S J, editors. Microbial toxins. New York, N.Y: Academic Press; 1970. pp. 99–171. [Google Scholar]
- 15.Ginsburg I, Bentwich Z, Zeiri N, Silberstein Z, Lav S. Mechanism of action of streptolysin S. In: Caravano R, editor. Current research on group A streptococci. Amsterdam, The Netherlands: Excerpta Medical Foundation; 1968. pp. 239–254. [Google Scholar]
- 16.Griffith F. Serological classification of Streptococcus pyogenes. J Hyg (London) 1934;34:542–584. doi: 10.1017/s0022172400043308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hare R. Heat-labile toxin of hemolytic streptococci. J Pathol Bacteriol. 1937;44:71–90. [Google Scholar]
- 18.Jelijaszwicz J, Szmigielski S, Hryniewicz W. Biological effects of staphylococcal and streptococcal toxins. In: Jelijaszewicz J S, Wadstrom T, editors. Bacterial toxins and cell membranes. New York, N.Y: Academic Press; 1978. pp. 185–227. [Google Scholar]
- 19.Ji Y, McLandsborough L, Kondagunta A, Cleary P P. C5a peptidase alters clearance and trafficking of group A streptococci by infected mice. Infect Immun. 1996;64:503–510. doi: 10.1128/iai.64.2.503-510.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinsman O S, Arbuthnott J P. Experimental staphylococcal infections in newborn mice: inhibition of weight gain as an index of virulence. J Med Microbiol. 1980;13:281–290. doi: 10.1099/00222615-13-2-281. [DOI] [PubMed] [Google Scholar]
- 21.Knutson C A, Jeanes A. A new modification of the carbazole analysis: application to heteropolysaccharides. Ann Biochem. 1968;24:470–481. doi: 10.1016/0003-2697(68)90154-1. [DOI] [PubMed] [Google Scholar]
- 22.Koyama J, Egami F. Biochemical studies on streptolysin S formed in the presence of yeast ribonucleic acid. I. The purification and some properties of the toxin. J Biochem (Japan) 1963;53:147–154. doi: 10.1093/oxfordjournals.jbchem.a127670. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Lai C Y, Wang M T, De Faria J B, Akao T. Streptolysin S: improved purification and characterization. Arch Biochem Biophys. 1978;191:804–812. doi: 10.1016/0003-9861(78)90423-x. [DOI] [PubMed] [Google Scholar]
- 25.Lu F, Churchwood G. Tn916 target DNA sequences bind the C-terminal domain of integrase protein with different affinities that correlate with transposon insertion frequency. J Bacteriol. 1995;177:1938–1946. doi: 10.1128/jb.177.8.1938-1946.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Musser J, Kanjilal S, Shah U, Musher D, Barg N, Nelson K, Selander R K, Johnson K, Schlievert P, Henrichsen J, Gerlach D, Rakita R, Tanna A, Cookson B, Huang J. Geographic and temporal distribution and molecular characterization of two highly pathogenic clones of Streptococcus pyogenes expressing allelic variants of pyrogenic exotoxin A (scarlet fever toxin) J Infect Dis. 1993;167:337–346. doi: 10.1093/infdis/167.2.337. [DOI] [PubMed] [Google Scholar]
- 27.Nida K, Cleary P P. Insertional inactivation of streptolysin S expression in Streptococcus pyogenes. J Bacteriol. 1983;155:1156–1161. doi: 10.1128/jb.155.3.1156-1161.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Connor S P, Cleary P P. In vivo Streptococcus pyogenes C5a peptidase activity: analysis using transposon and nitrosoguanidine-induced mutants. J Infect Dis. 1987;156:495–504. doi: 10.1093/infdis/156.3.495. [DOI] [PubMed] [Google Scholar]
- 29.Owens W, Henley F, Baridge B D. Hemolytic mutants of group A Streptococcus pyogenes. J Clin Microbiol. 1978;7:153–157. doi: 10.1128/jcm.7.2.153-157.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pang K M, Knecht D A. Partial inverse PCR: a technique for cloning flanking sequences. BioTechniques. 1997;22:1046–1048. doi: 10.2144/97226bm07. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. pp. 1.21–1.31. [Google Scholar]
- 32.Schlievert P M, Bettin K M, Watson D W. Purification and characterization of group A streptococcal pyrogenic exotoxin type C. Infect Immun. 1977;16:673–679. doi: 10.1128/iai.16.2.673-679.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smyth C J, Duncan J L. Thiol-activated (oxygen-labile) cytolysins. In: Jelijaszewicz J S, Wadstrom T, editors. Bacterial toxins and cell membranes. New York, N.Y: Academic Press; 1978. pp. 129–183. [Google Scholar]
- 34.Weld J T. Toxic properties of serum extracts of hemolytic streptococci. J Exp Med. 1934;59:83–95. doi: 10.1084/jem.59.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wheeler M C, Roe M H, Kaplan E L, Schlievert P M, Todd J K. Outbreak of group A streptococcus septicemia in children. JAMA. 1991;266:533–537. [PubMed] [Google Scholar]
- 36.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1994. pp. 2.4.1–2.4.5. [DOI] [PubMed] [Google Scholar]