Abstract
Exosomes, a class of extracellular vesicles of endocytic origin, play a critical role in paracrine signaling for successful cell-cell crosstalk in vivo. However, limitations in our current understanding of these circulating nanoparticles hinder efficient isolation, characterization, and downstream functional analysis of cell-specific exosomes. In this work, we sought to develop a method to isolate and characterize keratinocyte-originated exosomes () from human chronic wound fluid. Furthermore, we studied the significance of in diabetic wounds. LC-MS-MS detection of KRT14 in and subsequent validation by Vesiclepedia and Exocarta databases identified surface KRT14 as a reliable marker of . dSTORM nanoimaging identified KRT14+ extracellular vesicles () in human chronic wound fluid, 23% of which were of exosomal origin. An immunomagnetic two-step separation method using KRT14 and tetraspanin antibodies successfully isolated from the heterogeneous pool of EV in chronic wound fluid of 15 non-diabetic and 22 diabetic patients. Isolated (Ø75–150nm) were characterized per EV-track guidelines. dSTORM images, analyzed using online CODI followed by independent validation using Nanometrix, revealed Ø as 80–145nm. The abundance of was low in diabetic wound fluids and negatively correlated with patient HbA1c levels. The isolated from diabetic wound fluid showed a low abundance of small bp RNA (<200 bp). Raman spectroscopy underscored differences in surface lipids between non-diabetic and diabetic Uptake of by monocyte-derived macrophages (MDM) was low for diabetics versus non-diabetics. Unlike from non-diabetics, the addition of diabetic to MDM polarized with LPS and INFγ resulted in sustained expression of iNOS and pro-inflammatory chemokines known to recruit macrophage (mϕ) This work provides maiden insight into the structure, composition, and function of from chronic wound fluid thus providing a foundation for the study of exosomal malfunction under conditions of diabetic complications such as wound chronicity.
Keywords: Extracellular vesicles, keratinocyte-originated exosomes, Diaexosomes, macrophage, resolution of inflammation, wound healing, dSTORM microscopy, Nanometrix
Graphical Abstract
Biological body fluids are enriched in nanosized membrane-bound extracellular vesicles (EVs; Ø 50–1000 nm) which play a critical role in cell-to-cell communication via a paracrine mechanism.[1] EVs are abundant in most body fluids such as blood, urine, saliva, and milk.[2–4] EVs are known to be heterogeneous, differing in physical characteristics (size, density, composition), cellular origin, molecular compositions, and/or function.[5] Traditionally, it is common to classify EVs as exosomes and membrane-originated particles such as ectosomes, microparticles, and apoptotic bodies.[6] Unlike other EVs of membrane origin, exosomes (Ø 50–150 nm) are of endocytic origin that selectively package a distinctive repertoire of cargo such as mRNA/miRNAs, protein, and anti-microbial peptides (AMPs).[7–9] Presence of such a well-coordinated sorting mechanism led to the notion that the paracrine effects of exosomes are an active process and is distinct from other shedding membrane vesicles.[10, 11] The role of exosomes as a mediator of intercellular communication within the tissue milieu is of outstanding interest.[12–14]
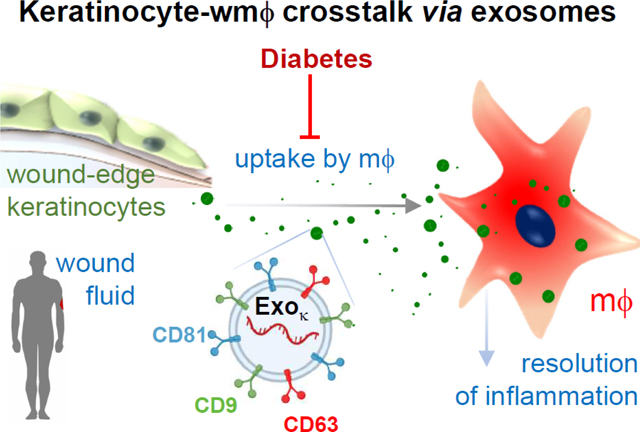
The molecular composition of exosomes reflects the (patho)physiological state of the cell of origin.[15] Thus, exosomes are known to serve as powerful markers providing diagnostic and prognostic cues.[16] As sophisticated work on well-characterized exosomes has started to emerge,[7] it is important to acknowledge that much of the earlier literature on exosomes and data interpretation is clouded and sometimes misleading.[17] Specifically, the study of cell-specific exosomes is scanty, making interpretation of data derived from a heterogeneous pool of EV[18], challenging.[17, 19, 20] Adding to the challenge are other factors such as incomplete reporting of standards and methodological details.[17, 21] The human wound fluid serves as an excellent reservoir of EVs that are causatively linked to healing outcomes.[22] Our previous work has demonstrated that EVs isolated from wound fluid of healing chronic wound patients are more effective in transdifferentiation of macrophages (mϕ) to a pro-healing phenotype when compared to that from non-healing diabetic patients.[22] It is widely accepted that keratinocytes in cutaneous wound healing and inflammation.[23] Emerging evidence positions exosomes as a major functional component of the wound microenvironment.[24–29] The resolution of wound inflammation relies on successful crosstalk between wound-site keratinocytes and mϕ via exosomal gene transfer.[30] Based on our previous studies, here we test the hypothesis that under diabetic conditions, the cross-talk between the resident keratinocyte and blood-borne wound mϕ is compromised resulting in persistent inflammation at the wound site. However, unlike in murine studies, where isolation of cell-specific exosomes is feasible using a genetic approach,[30] isolation of high-purity keratinocyte-originated exosomes from the human wound is more challenging. Furthermore, concerns with inappropriate separation techniques of different nanovesicles of similar size limit the ability to delineate specific mechanisms in human subjects.[21, 31, 32] In this work, we report a technique to isolate exosomes of keratinocyte origin from human chronic wound fluids that may provide critical insight into diabetic wound chronicity.
RESULTS AND DISCUSSION
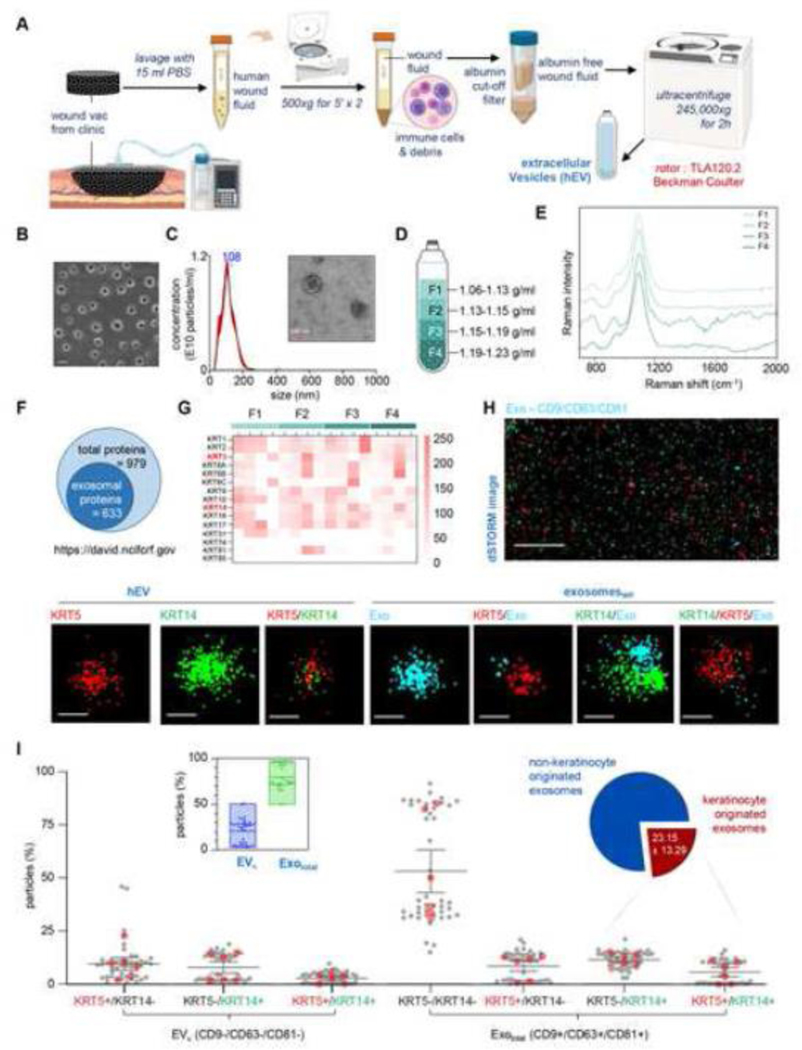
Identification of keratinocyte-derived exosomes in human wound fluid.
Negative Pressure Wound Therapy (NPWT) is widely known to promote wound closure by a number of mechanisms including improvements of the wound microenvironment as a result of removal of wound fluid and influx of fresh bodily fluids into the affected site.[33–37] The discarded NPWT sponge dressing is thus rich in EVs released from wound-edge cells. This work reports the first approach to quantitatively identify the presence of keratinocyte-originated exosomes () in chronic human wound fluid. Clinical wound fluid EVs were isolated using differential ultracentrifugation, a process that is regarded as the “golden standard” (Fig. 1A).[38–40] The morphology and concentration of the exosomes were analyzed by scanning electron microscopy (SEM), transmission electron microscopy (TEM), and nanoparticle tracking analysis (NTA) (NanoSight™) (Fig. 1B-C). Using sequential four discontinuous one-step density gradient ultracentrifugation, four density (ρ) ranges of EVs (1.06–1.13 g/mL, 1.13–1.15 g/mL, 1.15–1.19 g/mL and 1.19–1.23 g/mL) were isolated from chronic human wound fluid (Fig. 1D). Raman spectra of the EVs derived from all four ρ ranges showed distinct differences in the intensity of peaks that were characteristic for proteins and lipids within the ranges determined by the Variable Importance on Projection (VIP) values (Fig. 1E). The average ρ of exosomes (Ø 50–150nm) is known to be in – the range of 1.15 – 1.19 g/mL.[41–43] Thus, fractions 2 and 3 (F2 and F3) were expected to be exosomal fractions. However, the spectral resemblance of F1 and F2 led us to the notion that F2 is likely to be of non-endosomal origin including particle size comparable to that of exosomes. Proteomic analysis of the four ρ ranges of EVs revealed a total of 979 proteins across all samples. Six hundred and thirty-three of these proteins were of exosomal origin as determined by DAVID bioinformatics resources (Fig. 1F).[44] All four ρ ranges of EVs showed abundance of keratin proteins (Fig. 1G). The path to this successful identification of exosomal proteins of keratinocyte origin taught us that the use of anti-pan-keratin conjugated to superparamagnetic Dynabeads™ is inadequate as non-keratinocyte cells did express some keratin proteins (Fig. S1A-C).[45–48] High keratin 14 (KRT14) abundance is specific to keratinocytes (Fig. S2).[49–51] Thus, a combination of the use of anti-K14 conjugated Dynabeads™ and tetraspanins exosome markers CD9, CD63, and CD81 conjugated Dynabeads™ is necessary. Direct Stochastic Optical Reconstruction Microscopy (dSTORM) of EVs (Fig. 1H, Fig. S3) using anti-keratin 5-Alexa 647 (KRT5647) and KRT14-Atto 488(KRT14488) together with cyan conjugated tetraspanins exosome markers CD9, CD63 and CD81 (Exocy3) validated the presence of KRT14 and its counterpart intermediate filament KRT5 in human chronic wound fluid (Fig. 1H). Image analyses using the Collaborative Discovery (CODI, ONI, UK) online platform demonstrated a lower abundance of compared to the total exosome pool (Fig. 1I). Based on the presence of KRT14 and KRT5 on exosomes, nearly one-fourth (23.15±13.29%) of the total exosomes in human chronic wound fluid were of keratinocyte origin (Fig. 1I). In the interest of additional rigor, it is important to note that in a subset of keratinocytes, KRT14 is co-expressed with KRT5 (type II, 58kDa), which forms a parallel coiled-coil heterodimer to support the dynamic cytoskeleton.[50, 52–54] dSTORM imaging thus identifies a KRT14+/KRT5+ subset alongside an all KRT14+ marker (Fig. 1H).
Figure 1: Identification of keratinocyte-derived exosomes in human wound fluid.
A, Schematic diagram showing enrichment of EVs in human chronic wound fluid isolated from the dressings of patients undergoing NPWT by using differential ultracentrifugation. B, Representative SEM image of EVs. Scale, 200nm. C, Representative size distribution of EVs. (n=10). The inset shows a representative zoomed TEM image of EVs. Scale, 100nm. D, Schematic diagram showing the four density (ρ) ranges of the EVs. E, Normalized Raman spectra of the EVs in the four different fractions (F). F, Schematic diagram showing the proportions of exosomal proteins to total proteins identified by LC-MS from all the four fractions. G, Heat map showing the abundance of keratin proteins identified from four different ρ ranges of EVs. (n=4). H, Representative super-resolution dSTORM immunofluorescence images of EVs showing localization of tetraspanins markers (CD9, CD63, CD81) on exosome (cyan) and keratin markers KRT14 (green) and KRT5 (red). Scale, 5um. Zoomed images of EV showing the abundance of single, double, and triple markers. Scales, 50nm. I, Localization of tetraspanins and keratins on EVs were analyzed using CODI online analysis platform and plotted graphically. Each grey dot corresponds to one ROI, the red dots correspond to the mean of each biological replicate. At least 6 ROI per sample. (n=6). The box plot represent abundance of Keratinocyte-originated EVs ()(CD9-/CD63-/CD81-) and total exosomes (CD9+/CD63+/CD81+). Each dot represents an individual ROI taken from all six biological replicates. The pie chart represents the abundance of exosomes of keratinocytes () and non-keratinocyte-origin. Data in I were shown as mean ± SEM and were analyzed by one-way ANOVA with the post-hoc Bonferroni’s multiple comparison test. Figures 1A and D were created with BioRender.com.
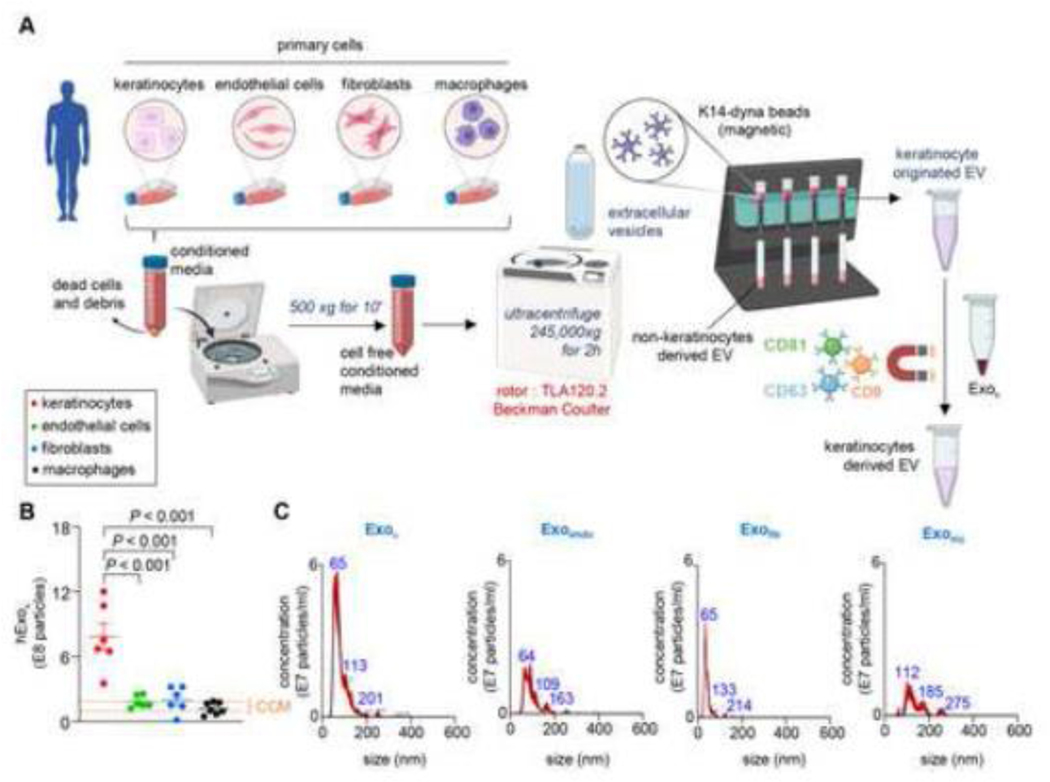
Validation of the KRT14 immunomagnetic separation method to specifically isolate exosomes of keratinocyte origin.
Abundant KRT14 (type I, 50kDa) is a unique characteristic of the keratinocytes (Fig. S2).[49–51] Thus, KRT14 immunomagnetic separation (IMS) is useful to isolate . Next, using Dynabeads™ conjugated with three tetraspanins exosome markers (CD9/CD63/CD81)[55] as reported in EVPedia[56] and Exocarta,[57] can be specifically isolated from chronic human wound fluid. To test the specificity of such isolation, human primary keratinocytes, dermal fibroblasts, dermal microvesicular endothelial cells, and monocyte-derived macrophages (MDM) were independently cultured in media supplemented with 10% exosome depleted serum. The corresponding conditioned media were collected. A three-step method was performed to isolate for rigor. First, EVs were isolated from the cell cultured-conditioned media using differential ultracentrifugation (Fig. 2A). Second, the heterogenous EVs were incubated with superparamagnetic Dynabeads™ conjugated with antibodies against KRT14 (Fig. 2A). Successful conjugation of anti-KRT14 antibody on the superparamagnetic Dynabeads™ was tested using flow cytometry and FITC conjugated anti-KRT14 antibody demonstrating a clear shift in green fluorescence (Fig. S4A-F). From the heterogenous EV pool, only those exosomes expressing KRT14 were trapped allowing the non-keratinocyte EVs in the flow-through to pass (Fig. 2A). Finally, was incubated with superparamagnetic Dynabeads™ conjugated with antibodies for CD9, CD63, and CD81 to specifically isolate (Fig. 2A). The concentration of KRT14+ exosomes was analyzed by NTA (NanoSight™). The abundance of KRT14+ exosomes was significantly higher in primary human keratinocytes compared to the background signal (comparable to particles isolated from media with 10% exosome depleted serum resuspended in with PBS, Fig. 2B, C, Fig. S4G) and from dermal microvascular endothelial cells, dermal fibroblast, and mϕ in the cultured conditioned media. To eliminate the possibility of non-specific binding of exosomes to the superparamagnetic Dynabeads™, exosomes were labeled with Vybrant™ DiO. Incubation of the DiO-labeled exosome with superparamagnetic Dynabeads™ did not result in any shift in green fluorescence thus ruling out any non-specific binding to the superparamagnetic Dynabeads™ via say adsorption (Fig. S4A, B, D, F). These findings established the specificity of the reported three-step KRT14 IMS method for isolating .
Figure 2: Intermediate filament protein keratin 14 are specific for vesicles of keratinocyte origin.
A, Schematic diagram showing enrichment of KRT14 expressing exosomes in primary human adult epidermal keratinocytes, primary human dermal microvascular endothelial cells, primary human adult dermal fibroblasts, and monocytes derived macrophages. B-C, Quantitative KRT14+ exosome abundance (B) and representative NTA images showing the size distribution of KRT14 expressing exosomes (C) in primary human adult epidermal keratinocytes, primary human dermal microvascular endothelial cells, primary human adult dermal fibroblasts, and monocytes derived mϕ (n=6). The orange dashed line shows the range of particles obtained from cell culture media (CCM) containing 10% exosome-depleted serum. Data in B were shown as mean ± SEM and were analyzed by one-way ANOVA with the post-hoc Bonferroni’s multiple comparison test. Figure A was created with BioRender.com.
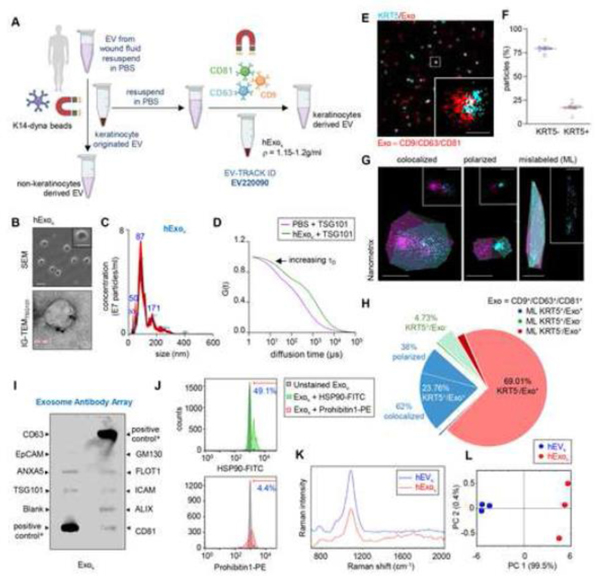
Isolation and characterization of keratinocyte-derived exosomes in human wound fluid.
The isolation strategy described above was applied to isolate from albumin-free human chronic wound fluid (Fig. 3A). The ρ of was determined to be in the range of 1.15 −1.2g/ml. The size, concentration, and morphology of were analyzed using SEM, immunogold transmission electron microscopy (IG-TEM) of TSG101, and NTA (NanoSight™) (Fig. 3B, C). Additionally, the binding ability of exosome-specific PE-tagged TSG101 antibody to the isolated was examined by fluorescence correlation spectroscopy (Fig. 3D). A shifted autocorrelation curve confirmed the successful binding of TSG101-PE to the isolated (Fig. 3D). Super-resolution dSTORM image analysis using built-in CODI demonstrated that one-fifth of the isolated were KRT5+ (Fig. 3E-F, Fig. S5A). Analysis of the dSTORM images was also performed on a separate platform using the Nanometrix software’s batch EV identification and characterization pipeline (Fig. 3G, Fig. S5B). Automated repeated measurements across datasets demonstrated that 69.01% of the were KRT5- (Fig. 3G, Table 1). Colocalization analyses demonstrated that 23.76% were KRT5+ (Fig. 3G, Table 1). Of these, 62% of the KRT5+ exosomes were colocalized (with overlap) while 38% were polarized (no overlap) (Fig. 3H, Table 1). An estimated 1.49% of the were detected as mislabeled and did not fit the criteria of circularity (Fig. 3G, Table 1). The three-step IMS method, as reported here, yielded 4.73% of the fraction that was negative for the three tetraspanins CD9, CD63, and CD81 (Fig. 3G, Table 1). This objectively addresses the limitation of our approach and may be viewed as impurities.
Figure 3: Isolation and characterization of keratinocyte-originated exosome from human wound fluid.
A, Schematic diagram showing enrichment of in human wound fluid isolated from the sponge dressings of patients undergoing NPWT by using differential ultracentrifugation. This method was reported in EV-track. (EV220090). B, Representative SEM image of showing cup-shaped morphology. Scale, 200nm. Inset showing a zoomed morphology of . Scale, 100nm. Representative immunogold TEM showing TSG101 on isolated from human chronic wound fluids. Scale, 100nm. C, Representative nanoparticle tracking analysis showing the size distribution of isolated from human wound fluid. (n=10). D, Binding of TSG-101 PE with the human keratinocyte-originated exosome was tested by autocorrelation curves as determined by fluorescence correlation spectroscopy (FCS). shows the increase in diffusion time. E, Representative super-resolution dSTORM immunofluorescence images of keratinocyte originated exosome showing the abundance of tetraspanins markers (CD9, CD63, CD81) on exosome (red) and keratin markers KRT5 (cyan). Scale, 50nm. F, Quantification of KRT5- and KRT5+ . Each dot corresponds to one quantified ROI, except the blue and red dots, which correspond to the mean of biological replicates. At least 6 ROI per sample. (n=4). G, The colocalization of exosome markers and KRT5 markers was analyzed using the Nanometrix software showing colocalized, polarized and mislabeled (ML) exosomes. Scale,100nm H, Pie diagram showing analysis of dSTORM image datasets using Nanometrix. ML, mislabeled. I, Antibody array of keratinocyte originated exosome from human wound fluid. PC, labeled positive control for HRP detection; GM130, cellular contamination marker. ALIX, TSG101, CD9, CD63, CD81, FLOT1, exosomal marker; EpCAM, epithelial cell adhesion molecule; ANXA5, Annexin 5. * Positive control for HRP detection derived from human serum exosomes. J, Flow cytometric analysis of conjugated with super magnetic dynabeads™ functionalized with CD63, CD9, and CD81 showing binding of HSP90-FITC (exosome marker) and Prohibitin1-PE (contaminants associated with exosome) antibodies. The histogram demonstrates the shift in FITC and PE fluorescence after binding with HSP90. The mean percentage of beads with exosomes showing FITC and PE fluorescence was mentioned over the marker bar. K, Raman fingerprint of and isolated from chronic wound fluid. L, PC2 vs PC1 2D-score plots for and isolated from chronic wound fluid constructed from principal component analysis of Raman spectra. (n=3) plot. Figure 3A was created with BioRender.com.
Table 1:
Physical characteristics of keratinocytes originated exosomes
| Characteristics | Channel 1 | Channel 2 | Overlay | Total |
|---|---|---|---|---|
| Exosome internal properties | ||||
| Median localization/exosome | 51 | 53 | 111 | 61 |
| Density (loc/nm2) | 0.02 | 0.01 | 0.01 | 0.02 |
| Average distance from all localization | 20.09 | 24.77 | 37.55 | 25.00 |
| Morphology | ||||
| Convex area (nm2) | 2530.35 | 5688.21 | 8476.95 | 3690.16 |
| Perimeter (nm) | 209.46 | 300.42 | 370.31 | 251.96 |
| Size (nm) | 82.14 | 113.93 | 142.96 | 99.01 |
| Circularity | 0.52 | 0.58 | 0.53 | 0.52 |
| Distribution | ||||
| Median distance to closest exosomes (nm) | 681.44 | 669.21 | 715.92 | 687.88 |
| Colocalization | ||||
| Overlapping channel 1 & channel 2 | - | - | 62% | 15% |
| Distance between channel 1 & channel 2 | - | - | 47.40 | - |
| Median channel 1 localization / exosome | - | - | 82 | - |
| Median channel 2 localization / exosome | - | - | 16 | - |
| Channel 1 photon intensity | - | - | 1828.91 | - |
| Channel 2 photon intensity | - | - | 455.78 | |
| Polarized channel 1 & channel 2 | - | - | 38% | - |
This work categorically tested all criteria set forward by EV-track for transparent reporting.[21] The isolated was characterized by an abundance of exosome markers reported in Exocarta such as FLOT1, ALIX, CD81, ICAM1, TSG101, and ANXA5 (Fig. 3I). The absence of GM130 validated the acceptable purity of isolation per EV-track guidelines.[17, 21] Additional quantitative flow cytometry analysis of anti-KRT14 conjugated superparamagnetic Dynabeads™ bound following incubation with FITC conjugated HSP90 (HSP90-FITC; marker) and phycoerythrin (PE) conjugated prohibitin (Prohibitin -PE, contaminants) antibodies rigorously validated the purity of (Fig. 3J). These observations validated that within the limits of acceptable purity, the three-step isolation process was successful in separating from the heterogenous EV pool in human chronic wound fluid. Raman spectroscopic analyses demonstrated a spectral difference across the averaged Raman spectra of and after normalization and baseline correction (Fig. 3K). These data were acquired in the range of 700−2000 cm−1 (fingerprint region). Unlike , displayed characteristic Raman bands of nucleic acids (725–836, 1200–1260, 1455–1460 cm−1). The peaks associated with phospholipids (780–830 cm−1), proteins (925–945 cm−1), phenylalanine (1005 cm−1), and lipids and proteins (1090 and 1100 cm−1) were present in both and .[58–60] To reduce the initial complexity of data set, the original components were transformed into principal components (PC). A multivariate principal component analysis (PCA) technique was carried out using OriginPro PCA spectroscopy app for Raman spectra investigation. Here, the principal component depends on the percentage of variance, and in the case of EVs and exosomes, the first component (PC1) showed the highest percentage i.e., 99.5% of total spectral variance compared to PC2 (0.4%) (Fig. 3L). The separation of EVs and exosomes is shown by a 2D score plot constructed from PC1 and PC2. Principal component 3 was not used to construct the score plot as it had no significant effect on the separation of and (Fig. S5D).
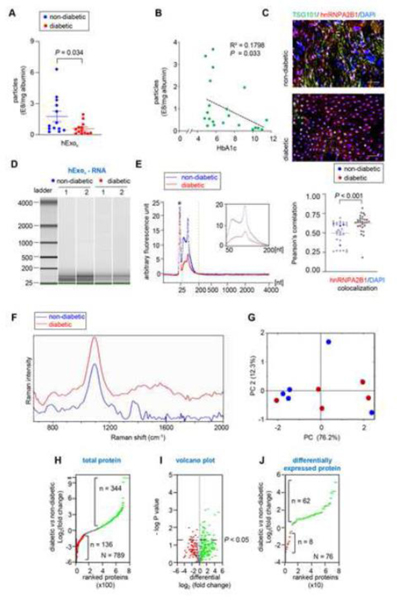
Low abundance of keratinocyte-derived exosomes in diabetic wound fluid.
The isolation of from albumin-free human chronic wound fluid in non-diabetic and diabetic patients (Table 2) revealed a significantly higher number of in non-diabetic patients compared to those from patients with diabetic wound (Fig. 4A). Interestingly, the abundance of was negatively correlated with the HbA1c level (Fig. 4B). TSG101, a core component of the ESCRT-I complexes, plays a critical role in the release of exosomes.[61–63] Wound-edge biopsies isolated from chronic non-diabetic and diabetic patients showed evidence of TSG101 depletion in the epidermis (Fig. 4C, Fig. S6A-B). Our previous work in the murine model reported that the silencing of heterogeneous nuclear ribonucleoproteinA2B1 (hnRNPA2B1), the key protein responsible for recognizing the EXOmotif sequence in nucleic acid packaging and loading them in multivesicular bodies,[7] causes persistent inflammation primarily characterized by non-resolving wound mϕ.[30] In this work, hnRNPA2B1 was observed to be more localized in the nucleus in diabetic wound-edge keratinocytes (Fig. 4C). Because the packaging of miRNAs is known to occur within cytosolic MVBs, nuclear localization of hnRNPA2B1 is expected to compromise the loading of small RNA in exosomes. Indeed, high-resolution automated electrophoretic analyses demonstrated significantly lower abundance of small RNA (50 – 200 bp) in isolated from patients with diabetic ulcers (Fig. 4D-E). The average Raman spectra of in non-diabetic and diabetic wound fluid showed a striking contrast in the intensity of characteristic bands of lipids and proteins (Fig. 4F-G). The from diabetic wound fluid were deficient in distinct peaks for lipids and proteins (1200–1300, 1450, 1465, 1606, 1615, 1600–1700 cm−1) as determined by the VIP values (Fig. 4F-G, Table 3). There were 789 proteins identified in and their intensity was plotted based on the log base 2 of protein intensity for diabetic over non-diabetic (Fig. 4H). A total of 77 unique proteins were identified in diabetic leaving 712 proteins in common between both the groups (Fig. 4H). Out of these, 344 proteins were enriched in diabetic and 136 are enriched in non-diabetic (Fig. 4H). These data were tested more rigorously in the volcano plot where only proteins with a p value < 0.05 are above the threshold line (Fig. 4I). Considering proteins that were significantly different between the non-diabetic and diabetic , 76 proteins were found to be significantly different leaving the remaining 713 proteins that were not comparable (Fig. 4J). Out of these 76 proteins, 62 and 8 were identified in diabetic and nondiabetic respectively (Fig. 4J, Table 4).
Table 2:
Demographics and clinical characteristics of chronic wound patients
| Characteristics | Non-Diabetic (n=15) | Diabetic (n=22) |
|---|---|---|
| Age § | 41 (22–69) | 57 (40–77) |
| Race † | ||
| White | 10 | 18 |
| African American | 5 | 3 |
| Others | 1 | |
| Ethnicity † | ||
| Hispanic | 0 | 0 |
| Non-hispanic | 15 | 22 |
| Sex | ||
| Male | 8 | 12 |
| Female | 7 | 10 |
| Weight (lbs) § | 212.96 (141.3 – 547.8) | 237.435 (127.16 – 382.4) |
| Smoking | ||
| Former | 1 | 6 |
| Current | 2 | 6 |
| Never | 12 | 10 |
| HbA1c § | 5.3 (4.8 – 5.7) | 7.3 (5.8 – 16.4) |
| Wound Etiology | ||
| Surgical | 13 | 12 |
| Non-Surgical | 2 | 10 |
| Wound Infection | ||
| Infected | 13 | 20 |
| Non-infected | 2 | 2 |
| Wound status | ||
| Healing | 10 | 15 |
| Non-healing | 4 | 6 |
| Unknown * | 1 | 1 |
Wound fluid was collected from these patients undergoing NPWT
Data reported as median;
Race and ethnicity was reported by patients;
Patient didn’t show up for visit 2 for assessment of wound status
Figure 4: Human chronic wound fluid of diabetic subject has a low abundance of Keratinocyte-derived exosomes with compromised cargo loading:
A, Quantification of from chronic non-diabetic and diabetic human wound fluid normalized with albumin. B, Regression analysis of the abundance of keratinocyte-originated exosomes with HbA1C. C, Representative fluorescence images of human non-diabetic and diabetic wound-edge epidermis showing immunofluorescence staining of hnRNPA2B1(red) and TSG101 (green) with DAPI counterstaining in. Scale, 50 μm. D, High-resolution automated electrophoresis of RNA in isolated from human chronic wound fluid of non-diabetic and diabetic subjects shows more abundance of RNA in non-diabetic human wound fluid compared to diabetic human wound fluid. E, Comparison of bioanalyzer generated electropherograms of RNA in isolated from human chronic wound fluid of non-diabetic and diabetic subjects. F, Representative Raman spectra of isolated from non-diabetic and diabetic chronic wound fluid. G, 2D-score plot constructed from principal component analysis of Raman spectra. H, Ranked protein plot showing intensity of proteins in isolated from human chronic wound fluid of non-diabetic and diabetic subjects. I, Volcano plot showing proteins with a p value <0.05 are above the threshold line. Note that only significant proteins with a p < 0.05 have a -log10(.05) > 1.3. J, Ranked plot of the proteins that were significantly different between t isolated from human chronic wound fluid of nondiabetic and diabetic subjects. Data in A and C were shown as mean ± SEM and analyzed by Student’s t-test.
Table 3:
Observed Raman peaks
| Wavenumber (cm-1) | Non-diabetic exosomes | Diabetic exosomes | Origin | ||
|---|---|---|---|---|---|
| 790 | Yes | Yes | Yes | No | O-P-O stretching in nucleic acids |
| 824 | No | Yes | Yes | Yes | Phosphodiester |
| 1003 | Yes | Yes | Phenylalanine ring | ||
| 1087 | Yes | Yes | Phosphodioxy | ||
| 1200–1300 | No | Yes | Yes | No | Amide III |
| 1450 | No | Yes | Yes | No | Nucleic acids, proteins and lipids |
| 1465 | Yes | No | Lipids | ||
| 1600–1700 | No | Yes | Yes | No | Amide I |
| 1615 | Yes | No | Protein | ||
| 1606 | Phe, Tyr ring vibrations |
Table 4:
Proteomics output file for proteins enriched in diaexosomes (in black) and (in italics)
| S.No. | Accession | Description | Enrichment* | −log (P) |
|---|---|---|---|---|
| 1. | H0YA55 | Albumin | 1.53 | 1.57 |
| 2 | D6RHD5 | Albumin | 1.55 | 1.44 |
| 3. | H7C0L5 | Inter-alpha-trypsin inhibitor heavy chain 4 | 1.71 | 1.55 |
| 4. | P0C0L5 | Complement C4-B | 1.93 | 1.72 |
| 5. | A0A140TA44 | Complement C4-A | 1.93 | 1.72 |
| 6. | A0A0G2JL54 | Complement C4-B | 1.93 | 1.72 |
| 7. | F5GXS0 | Complement C4-B | 1.93 | 1.72 |
| 8. | P0C0L4 | Complement C4-A | 1.96 | 1.72 |
| 9. | O75083 | WD repeat-containing protein 1 | 1.96 | 1.72 |
| 10. | D6RD66 | WD repeat domain 1 | 1.96 | 1.72 |
| 11. | A0A140TA32 | Complement C4-A | 1.97 | 1.74 |
| 12. | A0A140TA29 | Complement C4-B | 1.97 | 1.74 |
| 13. | P04114 | Apolipoprotein B-100 | 1.98 | 2.04 |
| 14. | A0A0G2JPR0 | Complement C4-A | 2.10 | 1.74 |
| 15. | A0A140TA49 | Complement C4-A | 2.10 | 1.74 |
| 16. | D6RF20 | Vitamin D-binding protein | 2.13 | 1.84 |
| 17. | Q14624 | Inter-alpha-trypsin inhibitor heavy chain H | 2.17 | 2.32 |
| 18. | B7ZKJ8 | ITIH4 protein | 2.17 | 2.32 |
| 19. | A0A3B3ISA6 | Complement C4B | 2.18 | 2.00 |
| 20. | P14780 | Matrix metalloproteinase-9 | 2.26 | 1.81 |
| 21 | G3V5M4 | Actinin alpha 1 | 2.32 | 1.36 |
| 22. | G3V2E8 | Actinin alpha 1 | 2.32 | 1.36 |
| 23. | P06312 | Immunoglobulin kappa variable 4–1 | 2.37 | 1.70 |
| 24. | D6RBJ7 | Vitamin D-binding protein | 2.57 | 1.98 |
| 25. | P17661 | Desmin | 2.59 | 1.31 |
| 26. | G3V2N5 | Actinin alpha 1 | 2.63 | 1.46 |
| 27. | G3V2X9 | Actinin alpha 1 | 2.63 | 1.46 |
| 28. | A0A087WSZ2 | Actinin alpha 3 | 2.63 | 1.46 |
| 29. | Q08043 | Alpha-actinin-3 | 2.63 | 1.46 |
| 30. | M0R0Q9 | Complement C3 | 2.73 | 2.33 |
| 31. | P02760 | Protein AMBP | 2.94 | 1.94 |
| 32. | A0A1W2PNV4 | Actin-related protein 2/3 complex subunit 1A | 2.99 | 1.54 |
| 33. | C9JEY1 | Actin-related protein 2/3 complex subunit 1B | 2.99 | 1.54 |
| 34. | C9K057 | Actin-related protein 2/3 complex subunit 1B | 2.99 | 1.54 |
| 35. | O15143 | Actin-related protein 2/3 complex subunit 1B | 2.99 | 1.54 |
| 36. | J3KS17 | Apolipoprotein H | 3.11 | 1.55 |
| 37. | P02774 | Vitamin D-binding protein | 3.12 | 3.01 |
| 38. | D6RF35 | Vitamin D-binding protein | 3.12 | 3.01 |
| 39. | B0YJC5 | Vimentin | 3.21 | 1.52 |
| 40. | B0YJC4 | Vimentin | 3.35 | 1.34 |
| 41. | P08670 | Vimentin | 3.35 | 1.34 |
| 42. | P00734 | Prothrombin | 4.41 | 1.73 |
| 43. | C9JV37 | Coagulation factor II, thrombin | 4.41 | 1.73 |
| 44. | E9PIT3 | Prothrombin | 4.41 | 1.73 |
| 45. | P02749 | Beta-2-glycoprotein 1 | 5.16 | 3.06 |
| 46. | A0A087X232 | Complement C1s subcomponent | 5.55 | 1.61 |
| 47. | P09871 | Complement C1s subcomponent | 5.55 | 1.61 |
| 48. | H0Y5D1 | Complement C1s | 5.55 | 1.61 |
| 49. | Q05639 | Elongation factor 1-alpha 2 | 5.55 | 1.59 |
| 50. | A0A2U3TZH3 | Elongation factor 1-alpha | 5.55 | 1.59 |
| 51. | A096LPE2 | SAA2-SAA4 readthrough | 6.46 | 2.13 |
| 52. | P35542 | Serum amyloid A-4 protein | 6.46 | 2.13 |
| 53. | A0A0G2JH38 | C3/C5 convertase | 6.85 | 1.59 |
| 54. | P19827 | Inter-alpha-trypsin inhibitor heavy chain H1 | 10.10 | 1.34 |
| 55. | C9JV77 | Alpha-2-HS-glycoprotein | 15.19 | 1.58 |
| 56. | P02765 | Alpha-2-HS-glycoprotein | 15.19 | 1.58 |
| 57. | V9GYE3 | Apolipoprotein A-II | 17.51 | 2.74 |
| 58. | V9GYM3 | Apolipoprotein A-II | 17.51 | 2.74 |
| 59. | P02652 | Apolipoprotein A-II | 17.51 | 2.74 |
| 60. | V9GYG9 | Apolipoprotein A-II | 17.51 | 2.74 |
| 61. | V9GYS1 | Apolipoprotein A-II | 35.86 | 2.88 |
| 62. | V9GYC1 | Apolipoprotein A-II | 35.86 | 2.88 |
| 63. | P07357 | Complement component C8 alpha chain Keratin, type I cytoskeletal 17 (Fragment) | 0.14 | 1.98 |
| 64. | K7EPJ9 |
Neutrophil elastase
Keratin, type II cytoskeletal 73 (Fragment) |
0.18 | 1.35 |
| 65. | P08246 | Azurocidin | 0.26 | 1.40 |
| 66. | H0YIC5 | Hemoglobin subunit beta Immunoglobulin lambda variable 3–10 | 0.29 | 1.77 |
| 67. | P20160 | 0.33 | 1.47 | |
| 68. | A0A2R8Y7R2 | 0.47 | 1.56 | |
| 69. | A0A075B6K4 | 0.64 | 1.88 |
enrichment = diabetic / healthy
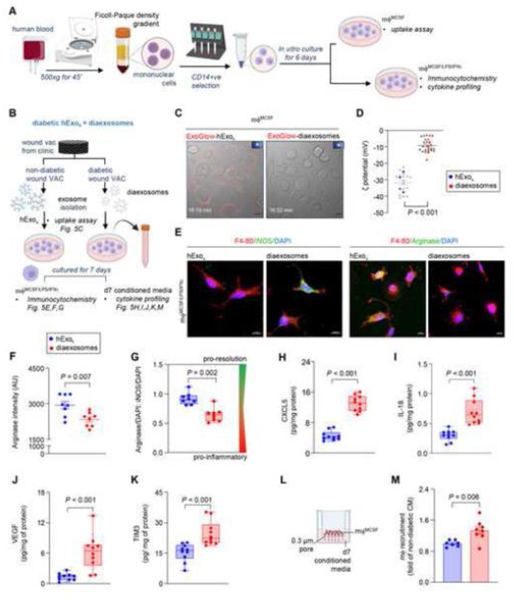
Diaexosomes compromised the resolution of wound inflammation.
Here, isolated from diabetic chronic wound fluid with compromised cargo and specific lipid-protein fingerprints are designated as diaexosomes. To test the significance of diaexosomes in the resolution of wound inflammation, blood-derived monocytes were isolated and differentiated in mϕ (Fig. 5A). The effect of and diaexosomes were tested by mϕ uptake assay, expression of inflammatory markers, and cytokine profiling (Fig. 5B). Because mycoplasma contamination in mϕ culture is likely to have a significant bearing on cellular responses,[64, 65] the presence of mycoplasma contamination in differentiated mϕ was ruled out using semi-quantitative PCR (Fig. S7A-B). Incubation of non-diabetic mϕ with non-diabetic caused overt phenotypic changes of mϕ (Fig. S7C). Such mϕ were not responsive to diaexosomes (Fig. S7C). Such response was functionally significant. Unlike that for non-diabetic , the uptake of diaexosomes by healthy non-diabetic mϕ was severely compromised (Fig. 5C, Fig. S8A-B). The surface charge of exosomes is known to influence cellular uptake.[66, 67] The Zeta potential (ζ) of were thus quantified. The ζ of diaexosomes was significantly higher than that of from non-diabetic wound patients (Fig. 5D). When exposed to from non-diabetic patients, mϕ exhibited induction of pro-resolution markers such as arginase (Fig. 5E-G, Fig. S9A-B). In contrast, addition of diaexosomes to mϕ maintained a persistent pro-inflammatory state as is known to be common in patients with diabetic ulcers (Fig. 5E-G, Fig. S9A-B). Human cytokine profiling in response to exposure of mϕ to diaexosomes further characterized the persistent pro-inflammatory state as evident by induction of angiogenin, CXCL5, IL-18, CXCL11, VEGF, and TIM3 (Fig. S9C-D). Quantitative ELISA showed significant elevated levels of CXCL5, IL-18, VEGF and TIM3 (Fig. 5H-K). Interleukin-18, also known as interferon-gamma inducing factor, is a pro-inflammatory cytokine critical for the release of chemokines recruiting inflammatory cells.[68] Similar to IL-18, CXCL11 is also induced by interferon-gamma and is critical for directing the temporal and spatial migration of activated T cells and natural killer cells.[69] CXCL5 is critical for recruiting neutrophils during inflammation.[70] Furthermore, in vitro mϕ invasion assay (Fig. 5L) demonstrated that the diaexosomes exposed conditioned media recruited significantly higher number of mϕ compared to non-diabetic exposed conditioned media (Fig. 5M). TIM3 is expressed on monocytes/macrophages and is involved in the regulation of inflammatory responses. TIM3 can synergize with lipopolysaccharide resulting in the production of the pro-inflammatory cytokine tumor necrosis factor (TNF) and it directly induces the expression of pro-inflammatory nuclear factor-κB (NF-κB).[71,72] These data demonstrated that the condition of diabetes brings for changes in wound-fluid exosomes in a way that opposes the resolution of inflammation and supports persistent inflammation. Taken together, this work provides critical insight that positions as a major target for the rescue from the diabetic complication of persistent inflammation which is not only implicated in wound chronicity but also in other diabetic inflammatory disorders.[73–75]
Figure 5: Diaexosomes failed to mount the inflammatory response in mf required for timely resolution of inflammation.
A, Schematic diagram showing the isolation of blood monocyte derived mϕ using MCSF and priming them to pro-inflammatory state using LPS and INFγ. B, Schematic diagram showing the experimental design to test the significance of and diaexosomes in human chronic wound fluid on mϕ. C, Live-cell confocal images showing compromised uptake of and diaexosomes by day 6 mϕ . Scale, 10 μm. and diaexosomes were stained with DiD before imaging.  Indicate movies in the supplement. D, Zeta potentials of and diaexosomes at physiological pH (pH-7.4). Each grey dot corresponds to one technical replicate, and the blue and red dots correspond to the mean of each biological replicates. (n=4). E, Representative immunofluorescence staining of F 4–80 (red) in mϕ () with either iNOS (green, pro-inflammatory mϕ marker) or Arginase (green pro-resolution mϕ marker) with DAPI counterstaining at day 7. Scale, 10 μm. F-G, Quantification of Arginase intensity (F) and the ratio of Arginase to iNOS intensity (G) in at day 7. H-K, Quantitative abundance of CXCL5 (H), IL-18 (I), VEGF (J), and TIM3 (K) in mϕ conditioned media exposed to either or diaexosomes for 7 days. L, Schematic diagram showing the experimental set up for mϕ recruitment assay. M, mϕ recruitment in response to the conditioned media exposed to either or diaexosomes for 7 days. Data in D, F, G, H, I, J, K and M were shown as mean ± SEM and analyzed by Student’s t-test. Figure A and B were created with BioRender.com.
Indicate movies in the supplement. D, Zeta potentials of and diaexosomes at physiological pH (pH-7.4). Each grey dot corresponds to one technical replicate, and the blue and red dots correspond to the mean of each biological replicates. (n=4). E, Representative immunofluorescence staining of F 4–80 (red) in mϕ () with either iNOS (green, pro-inflammatory mϕ marker) or Arginase (green pro-resolution mϕ marker) with DAPI counterstaining at day 7. Scale, 10 μm. F-G, Quantification of Arginase intensity (F) and the ratio of Arginase to iNOS intensity (G) in at day 7. H-K, Quantitative abundance of CXCL5 (H), IL-18 (I), VEGF (J), and TIM3 (K) in mϕ conditioned media exposed to either or diaexosomes for 7 days. L, Schematic diagram showing the experimental set up for mϕ recruitment assay. M, mϕ recruitment in response to the conditioned media exposed to either or diaexosomes for 7 days. Data in D, F, G, H, I, J, K and M were shown as mean ± SEM and analyzed by Student’s t-test. Figure A and B were created with BioRender.com.
CONCLUSION
In the complex landscape of in vivo systems biology wherein interaction between the components determines outcomes at the tissue, organ, and organismal levels, exosomes are emerging as nanocarriers of molecular signals that enable intercellular crosstalk – both paracrine and within the microenvironment.[76, 77] Such signals, often represented by functional proteins, metabolites and nucleic acids, are packed as cargo by well defined molecular processes.[43, 78] These cargobearing nanocarriers are decorated at the surface such that they are likely to be taken up by specific recipient cells with higher affinity. For example, mannose-decorated exosomes are preferentially taken up by macrophages and modify inflammation outcomes.[30] Taken together, these observations support the emergence of a paradigm wherein exosomes originating from specific cell types are loaded with specific cargo for delivery to specific recipient cells to achieve changes in functional outcomes. The current patient-based work reports on exosomal malfunction due to diabetes at all four levels i.e. cargo packing, exosomal release, exosomal uptake by recipient cell, and functional inflammation outcomes. This work provides key insight into the mechanistic underpinnings of impaired resolution of inflammation, known to cause wound chronicity in diabetic patients. These findings are enabled by the development of an unprecedented approach, supported by an array of methodological tools. Exosomes originating from human keratinocytes, and present in clinical wound fluid, have been isolated and characterized. The power of super-resolution dSTORM nanoscale imaging has been harnessed to characterize exosomal surfaces thus identifying exosomal subsets and their functional significance. This approach identified mislabeled exosomes thus adding to investigational rigor in an unprecedented manner. Raman spectroscopic-based lipid fingerprinting recognized clear differences between exosomes originating from diabetic versus non-diabetic chronic wound patients. The rigorous nanoscopic characterization of cell-specific single exosome in clinical samples presented in this work provides critical insight into a major diabetic complication as presented clinically and paves the path to other such clinical studies that address other disease processes.
METHODS
Human samples.
Human wound fluid samples were obtained from the dressing (sponges) of chronic wound patients undergoing Negative Pressure Wound Therapy (NPWT) at Indiana University CWC (Comprehensive Wound Center). The wound dressing was lavaged with 10 ml of saline solution supplemented with 0.1% gentamycin. The collected wound fluid was centrifuged at 500g for 5 min to remove debris and dead cells and passed through a 0.22 μm filter before storing them in aliquots at −80°C. Human wound biopsy samples were obtained from chronic wound patients from The Ohio State University (OSU) Comprehensive Wound Center (CWC). All human studies were approved by The Ohio State University and Indiana University Institutional Review Board. Declaration of Helsinki protocols was followed, and patients gave their written informed consent.
Cells and cell culture.
Primary human dermal microvascular endothelial cells (Cat PCS-110–010, Lot #80326190), primary human adult epidermal keratinocytes (Cat PCS-200–011, Lot # 70024770), and primary human adult dermal fibroblasts (Cat PCS-201–012, Lot #70017605) were procured from ATCC and cultured as per manufactures’ s instruction.
Human peripheral blood monocytes were isolated from fresh blood leucocyte source packs (American Red Cross, Columbus, OH) using Ficoll-Paque PLUS media (Cytiva [formerly GE Healthcare] Marlborough, MA) using density gradient centrifugation as described previously.[79, 80] Positive selection for monocytes was accomplished using CD14 antibody conjugated to magnetic microbeads (Miltenyi Biotec, Auburn, CA). The purity and recovery of these preparations of monocytes were greater than 90% as determined by FACS analysis using the CD14 antibody. Cell viability was tested using Trypan blue exclusion method and was found to be greater than 98%. Differentiation of these cells to macrophages (monocyte-derived macrophages or MDMs) was performed by culturing them with macrophage colony-stimulating factor (MCSF) for 6 days.[81] Macrophages were polarized to M1 phenotype using LPS (1 μg/ml) and IFNγ (20 ng/ml).[22] The cells were maintained in a standard culture incubator with humidified air containing 5% CO2. All experiments involving isolation or uptake of exosomes were performed with exosome depleted FBS (Gibco, Catalog # A2720803).
Detection of Mycoplasma contamination in Macrophages.
Mycoplasma contamination of MDM was detected using Universal Mycoplasma Detection Kit (ATCC, Catalog Number 30–1012K) as per the manufacturer’s instructions. Briefly, MDM isolated from human blood were scraped using 500ul PBS and transferred to a new sample lysis tube. The lysis tube was centrifuged at 13000 rpm for 3 min at 4°C. The supernatant was discarded, and the cell pellet was resuspended in 50μL of lysis buffer by vortexing. The resuspended cells were incubated at 37°C for 15 min to lyse the cells. Following incubation, the samples were heated at 95°C for 10 min to inactivate the protease. The cell lysate was collected by centrifuging at 13000 rpm for 5 min at 4°C. Polymerase chain reaction (PCR) was performed using the cell lysate as a template. For PCR reaction, the cell lysate was mixed with the reaction mixtures prepared in PCR tubes as per kit instructions. The PCR product was loaded on 3% agarose gel and electrophoresed until the tracking dye migrated 80% the length of the gel. The gel was stained with ethidium bromide and viewed over UV illumination in a gel imager (Azure, c600).
Conjugation of KRT14 antibody with superparamagnetic Dynabeads™.
Superparamagnetic Dynabeads™ were coupled with KRT14 antibody (# 905301, BioLegend, San Diego, CA) by using Dynabeads Antibody coupling kit (14311D, Invitrogen) according to the manufacturer’s protocol. Briefly, 5mg of superparamagnetic Dynabeads™ were washed with the wash buffer and incubated overnight with 20 μl antibody on a roller at 37°C. The next day, superparamagnetic Dynabeads™ were collected by using a magnet, and the supernatant was discarded. The excess antibody was removed using washed buffers. The beads were incubated for 15 min with SB buffer at room temperature. Beads were resuspended in 500μl SB buffer and stored at 4°C.
Isolation of extracellular vesicles and exosome.
Extracellular vesicles and exosomes were isolated from cell culture conditioned media supernatant using differential ultracentrifugation (Beckman Coulter Optima Max-XP Ultracentrifuge, rotor TLA120.2) following protocol as described previously in EV track (EV-TRACK ID: EV190103).[30] Extracellular vesicles were collected from the human wound fluid following the same protocol as described previously with modifications.[30] Albumin was removed from human wound fluid by centrifuging at 14000rpm for 15 min at 4°C using 100KDa centrifugal filters (Amicon Ultra centrifugal filters).
For isolation of keratinocyte-originated exosomes, the pellet post-ultracentrifugation was resuspended in PBS and incubated overnight at 4°C with magnetic Dynabeads conjugated with keratin 14 (KRT14) antibody. The keratinocyte-specific EVs () attached to the beads were magnetically separated from flow-through using magnetic MS column (MS columns from Miltenyi Biotec MACS, 130–042-201) after incubating with elution buffer (ExoFlow Exosome Elution Buffer, System Biosciences, EXOFLOWBUFR-2) for 2h at RT. Isolated were incubated overnight with superparamagnetic CD9, CD63 and CD81 Dynabeads™ (Invitrogen, Cat # 10614D, 10606D, 10616D). The attached to the beads were magnetically separated and re-pelleted for further analysis following differential ultracentrifugation at 245,000g for 1.5h. This method was submitted to EV track (EV-TRACK ID: EV220090) and received a preliminary score of 100%.
For flow cytometry, the exosomes were not removed from the beads. For NTA or TEM imaging, exosomes were eluted from the beads using elution buffer as per the manufacturer’s protocol. The in wound fluid was normalized against albumin concentration quantified using Human Albumin ELISA Kit (abcam, ab227933) as per manufacture’s protocol.
The density of isolated extracellular vesicles.
The wound fluid filtered through a 0.22 μm filter was defatted by centrifugation at 300 × g for 10 min at 4 °C. The supernatant was isolated and centrifuged at 2,000 × g for 10 min. at 4 °C to remove cells and other debris. The macromolecular proteins were isolated by centrifugation at 10,000 × g at 4 °C for 30 min. The resulting supernatant, a mixture of proteins, lipids, and other species, including EVs, is loaded above a series of sucrose cushions to isolate EV density ranges. The supernatant was diluted to 10 mL and the resulting supernatant is transferred to an open-top thin wall ultra-clear tube (Beckman Coulter 13.2 mL) and floated over 1 mL of a 1.19 g/mL sucrose solution with 10 mM tris in water. The density of sucrose solutions was calculated from readings with a brix refractometer. The ultracentrifuge tube was spun down at 210,000 g for 70 min at 4 C in an Optima XPN, Ultracentrifuge to pellet EVs greater than 1.19 g/mL. The top 1 mL was removed, and the remaining supernatant was combined with the cushion. The combined supernatant was transferred to a new ultracentrifuge tube and floated over 1 mL of a 1.15 g·mL−1 sucrose solution with 10 mM tris in water. The ultracentrifuge process was repeated to collect the lower density fraction of 1.15–1.19 g/mL. Again, the top 1 mL of the supernatant is discarded, and the rest is floated above a 1.13 g/mL sucrose solution with 10 mM tris in water to collect the 1.13–1.15 g/mL density fraction under the same ultracentrifuge conditions. The lowest density fraction, less than 1.13 g/mL, was collected by transferring the resultant supernatant to a new ultracentrifuge tube and was spun down at 210,000 g for 70 min at 4 °C with no cushion. All resulting pellets were resuspended with 200 μL of 100 mM ammonium acetate and used for analysis.
The density of isolated keratinocyte-originated exosomes.
The isolated pellet was redispersed in 150 μL of PBS and were loaded on top of a discontinuous sucrose step-gradient ranging from 0.5–2.5M sucrose (S5–500, Sigma Aldrich). The gradient was ultracentrifuged at 100000 ×g for 12 h at 4 °C (Optima MAX-XP Ultracentrifuge, Beckman Coulter). The density of exosomes was calculated to be in the range of 1.15 to 1.19 g/mL.
Preparation of samples for proteomics analyses.
Samples of EVs that were expected to be enriched in exosomes were also subjected to liquid chromatography (LC)-MS-database search proteomics analysis to assess what proteins are present in the samples. For these analyses, aliquots of each sample were dried and dissolved in 8 M urea. Protein disulfide bonds from concentrated samples were reduced with tris 2-carboxyethyl phosphine hydrochloride (2 mM, Sigma Aldrich, St. Louis, MO) for 1h at 56 ˚C. Reduced proteins were alkylated with iodoacetamide (4 mM, Sigma Aldrich, St. Louis, MO) for 45 min in the dark at room temperature. The urea concentration was then diluted to 1M using 100 mM ammonium bicarbonate (pH 7.5, Sigma Aldrich, St. Louis, MO) and trypsin (modified sequencing-grade trypsin from Promega, Madison, WI) was added at a 1:100, w:w protein ratio. Trypsin digestion was allowed to progress overnight at 37 ˚C. After digestion, peptides were desalted using a C-18 preparative Sep-Pak cartridge (Waters UK, Elstree, Herts, UK).
LC-MS-database search proteomics characterization.
Desalted peptides were resolubilized in buffer A (0.1% formic acid in water, Fisher Scientific, Hampton, NH) and loaded onto a reverse-phase trap column (Acclaim PepMap 100, 75μm × 2 cm, nano-viper, C18, 3 μm, 100 Å, ThermoFisher, Waltham, MA) by an easyNanoLC 1200 (ThermoFisher, Waltham, MA) at a flow rate of 5 μL∙min−1 for 10 μl. Peptides were resolved using an analytical reversed-phase column (Acclaim PepMap RSLC, 75 μm dia., 25 cm in length, using 2 μm diameter beads with an average pore size of 100 Å, ThermoFisher, Waltham, MA) over a 120 min. A linear gradient from 7% to 38% buffer B (0.1 % formic acid, 80% acetonitrile, 20% H2O, Fisher Scientific, Hanover, NH). Peptides eluting from the analytical column were electrosprayed into a Fusion Lumos Tribrid (Thermo Fisher, Waltham, MA). Precursor ions were monitored with a resolving power of 120,000 (@ 200 m/z). Individual precursors (z = 2 to 7) were selected using the quadrupole offset from the monoisotopic mass by 0.5 m/z with a window of 2 m/z. Peptides were subjected to higher energy collision dissociation (30 ± 5 %) and fragment ion masses were measured with a resolving power of 15,000 (@ 200 m/z). The AGC target was set to 2.0 × 105 or a fill time of 200 ms. Peptide database searching was completed using Proteome Discoverer 2.5 (Thermo Fisher, Waltham, MA) against the UniprotKB Bos Taurus database (Download Date: 2017-Nov-11) with a precursor mass accuracy of 10 ppm and a product ion mass accuracy of 0.02 Da. Modifications include methionine oxidation, N-terminal pyro-glutamate, N-terminal acetylation, N-terminal methionine loss, and cysteine carbamidomethylation. Percolator scoring was used with a 0.01 strict and 0.05 relaxed FDR, based on the q-value. Label-free protein quantification was based on the top three most abundant peptides from the identified protein.
Raman Spectroscopy.
EVs and exosomes were characterized by Raman spectroscopy and spectra were recorded on a commercial Raman microscope (Horiba Xplora Plus). The instrument was calibrated using a Si reference sample. The EVs and exosomes measurements were recorded by placing 70 μL of the sample (dispersed in PBS) in a cavity glass slide. To avoid contamination and evaporation, the cavity was covered with a coverslip (0.22 μm). The measurements were done with a 532 nm laser, 10x objective lens, 1800 (450–850 nm) grating, 100 μm entrance slit, and a pinhole of 300μm. All spectra were recorded in the range of 100–3000cm−1. The data was pre-processed to remove the background signals from glass and other deviations to minimize the insignificant variations. Baseline correction and noise of spectra were reduced using the Labspec6 software. The data were normalized using OriginPro software.
Nanoparticle tracking analysis.
The average size (mean particle diameter) and concentration of extracellular vesicles and exosomes were measured using Nanoparticle tracking analysis (Nanosight NS300) equipped with a SCMOS camera (Malvern, Worcestershire, UK) and 532 nm laser. Prior to measurement, the instrument was calibrated using 100 nm standard latex spheres (dilution 1:1000) in Milli-Q. All measurements were taken using sample dilution 1:100–1:500 to achieve 32–100 particles/frame and analyzed using 7 runs of the 30s collecting 25 frames per second (749 frames per run). The syringe pump speed was adjusted to 60, the detection threshold was set at 5, and the camera level was typically set at either 14 or 15. Data were analyzed by NTA 3.0 software (Malvern Instruments).[30, 82, 83]
Electron microscopy.
Electron Microscopy (EM) measurements were performed on a 300-μm mesh of carbon-coated copper grid that was subjected to a glow-discharge treatment prior to sample loading. A 2.0 μL aliquot of each EV sample was also spotted over the entire grid area. Excess solvent was removed by blotting with filter paper after ~30 s. The grids were stained for 30 s with 2% uranyl acetate, dried, and imaged on a JEOL JEM 1400plus transmission electron microscope equipped with a 4000 × 4000-pixel Gatan CCD camera. [84, 85]
Immunoelectron labeling with anti-TSG101 (Abcam, ab125011) was performed on keratinocyte-originated exosomes after fixing overnight in 4% paraformaldehyde diluted in 0.1M cacodylate buffer (pH 7.4). Fixed exosome preparations (20 μl) were applied to a carbon-Formvar coated 200 mesh nickel grids, and samples were allowed to stand for 30 min before wiping off excess liquid using Whatman filter paper. Grids were floated (sample side down) onto a 20 μl drop of 1M Ammonium Chloride for 30 min to quench aldehyde groups from the fixation step, followed by floating on drops of blocking buffer (0.4% BSA in PBS) for 2 h. Grids were rinsed 3 times (5 min each) using 1xPBS and were allowed to incubate with either blocking buffer only (negative control) or primary antibody (CD63) diluted with blocking buffer (1:100) for 1 h. Rinsing of the grids using deionized water (3 times for 5 min each) and 1xPBS followed the incubation step. Grids were floated on drops of 1.4 nm anti-rabbit nanogold (Nanoprobes, Inc.) diluted 1:1000 in blocking buffer for 1 h. Enhancing of grids using HQ Silver (gold enhancement reagent, Nanoprobes, Inc.) was then performed for 1 min, followed by rinsing in deionized ice-cold water.
As a final step, negative staining in 2% aqueous Uranyl Acetate was performed, and samples were wicked dry and allowed to air dry. TEM examination was performed using JOEL JEM 1400plus transmission electron microscope equipped with a 4000 × 4000-pixel Gatan CCD camera.
The morphology of exosomes and other EVs was also accessed by scanning electron microscopy (SEM). Briefly, the were fixed using 10% glutaraldehyde for 10 min at RT and drop cast onto the stubs containing glass coverslips mounted on the carbon tape. Samples were dried and placed in a vacuum chamber for at least 12 h before analysis. Imaging was done after gold sputter coating at a beam energy of 10 kV using a field emission scanning electron microscope (JEOL 7800F, JEOL Japan).[86]
Fluorescence correlation spectroscopy (FCS).
Qualitative evidence binding of fluorescently labeled antibody (TSG101-PE) with the isolated was measured by the change in diffusion using a two-channel fluorescence correlation spectroscopy system (Confocor 2, Zeiss) attached to an Axiovert200 M inverted microscope (Zeiss) as described previously.[30, 87] The system measures the characteristic diffusion time () of the fluorophore as determined by fitting fluorescent decay within a confocal volume to an autocorrelation curve using the Eq. (1), where N is particle concentration and Q is a factor relating to the ellipticity of the confocal volume:
| (1) |
Autocorrelation best-fit curves identify the change in particle diffusion with curves shifted to the left demonstrating faster diffusion (smaller ); exemplary best fit curves are shown, where the curves are fitted using a one-component fit identifying an average characteristic diffusion time for the fluorophores.
Super-resolution dSTORM Microscopy. `
Super-resolution fluorescent microscopy analyses of EVs and exosomes were performed with 100x oil immersion objective using Nanoimager S Mark II microscope from ONI (Oxford Nanoimaging, Oxford, UK) equipped with 405 nm/150 mW, 488 nm/1 W, 560 nm/500mW, 640 nm/1 W lasers. Fluorescence excited by 488, 560, and 640 lasers were recorded using bandpass filters 498–551nm, 576 – 620, and 665–705nm respectively. Primary antibodies against human Keratin 5 (clone: Poly19055; Biolegend, Cat # 905501) and keratin 14 (clone: Poly19053; Biolegend, Cat # 905301) were conjugated with Cy®3 and Atto-488 NHS Ester respectively using the Apex Antibody Labeling Kit (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s protocol. Isolated EVs from human wound fluid were incubated overnight at 4 °C with anti-Keratin 5-Cy3, anti-keratin 14 -Atto 488, and Alexa Fluor conjugated anti-CD9, anti-CD63, and anti-CD 81 (Oxford Nanoimaging, Oxford, UK) antibodies using ONI EV Profiler Kit (EV-MAN-1.0) as per manufacturer’s instruction. The samples were imaged using ONI BCubed dSTORM Imaging Buffer. Three-channel (647, 560, and 488) dSTORM data (2000 frames per channel) were acquired sequentially at 30 Hertz in total reflection fluorescence (TIRF) mode using 35% laser power for the 640 nm and 40% laser power for the 488 nm laser. Before each imaging session, beads slide calibration was performed to align fluorescent channels, achieving a channel mapping precision smaller than 12 nm. Single-molecule localization data was filtered using NimOS (Version 1.18.3, ONI, Oxford, UK) based on the point spread function shape, photon count, and localization precision to minimize background noise and remove low-precision localizations. Localization data were analyzed using algorithms developed by ONI via the collaborative discovery (CODI) online analysis platform (https://alto.codi.bio/, 3 October 2021) using the drift correction pipeline version 0.2.3. CODI was used for clustering analysis with min size and sample size of 15 defining a single EV during clustering. Further constraints on clusters were applied to the number of localizations per cluster from 16–400 localizations/cluster with a size range of a 10–200 nm radius. The positivity of each tetraspanin was calculated by counting the number of localizations up to 200nm from the center of the centroid for a cluster. Tetraspanin channel was counted positive if the number of localizations was more than 2.
Characterization of .
The characterization of keratinocyte-derived exosomes was performed in batches across multiple datasets using the Nanometrix software version 1.1b (Nanometrix Ltd, Oxford, UK). The 2-channel datasets were exported as localizations from the Nanoimager (ONI, Oxford, UK) and loaded into the Nanometrix software for analysis. Exosome identification, labelling and characterization were undertaken using the Nanometrix software’s inbuilt exosome batch multi-channel characterization pipeline that categorized the exosomes into three groups: KRT5-/Exo+, KRT5+/Exo-, and KRT5+/Exo+ exosomes, and measured their morphology, distribution, composition and colocalization. Additional filters were applied to discard exosomes smaller than 30nm, larger than 500nm, or comprising fewer than 20 localizations. Potential exosome labeling error was determined automatically based on the morphology and density of exosomes that have significantly different spatial properties to the majority of other exosomes within the same analysis batch.
Exosome antibody array.
Characterization of keratinocyte-originated exosome was performed using commercially available Exo-Check Exosome Ab Array kit (System Biosciences, Palo Alto, CA, EXORAY210B-8) as per manufacturer’s protocol. Protein concentration in keratinocyte originated exosome was determined by BCA assay. 50ug of protein was used for the array and blots were developed and imaged under Azure biosystems.
Flow cytometry.
Exosome marker HSP90 and potential cell organelle-derived contaminant prohibitin were evaluated by incubating exosome conjugated beads with either FITC anti-HSP90 (1:100, NB200–112PE, Novus Biologicals) or PE anti-prohibitin (Abcam ab2251651:200) antibodies for 90 min at room temperature as described previously. Alexa Fluor 488 fluorescence and PE fluorescence were determined using the FITC channel and PE channel respectively.[30, 83]
High-resolution automated electrophoresis of RNA.
The RNA isolated from was analyzed in Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, California).[30]
Zeta potential analysis.
The surface charge (ζ potential) measurement of was determined by Zetasizer (Nano-Z, Malvern Instruments Ltd., UK) as described previously.[30, 82–84] All samples were dispersed in double-distilled water and tested in volume-weighted size distribution mode.
Immunohistochemistry (IHC), Immunocytochemistry (ICC), and microscopy.
Immunohistochemistry on human tissues was performed as per the protocol described previously.71 Briefly, the cryosections of human tissues were blocked with 10% normal goat serum and incubated with the specific antibodies as indicated at 4°C overnight. Rabbit isotype control (Abcam, ab27478; 1:400) was used for the validation of specific antibodies. Signal was envisioned by following the incubation with fluorescence-tagged (Alexa 488-tagged α-rabbit, 1:200; Alexa 568-tagged α-rabbit, 1:200; Alexa 568-tagged α-rat, 1:200,) and counterstained with DAPI. Immunohistochemistry was performed on cryosections of human wound-edge skin sample using K14 (Covance, PRB-155P-100; 1:400), hnRNPA2B1(Sigma-Aldrich; HPA001666, 1:50), and TSG101 (Abcam, ab125011; 1:200). Immunocytochemistry was performed on monocyte-derived macrophages exposed to either non-diabetic or diabetic Keratinocyte originated exosomes (107) using F4/80 (Bio-Rad, MCA497R; 1:200), Arginase (Abcam; 203490, 1:100) and iNOS (Abcam; ab115819, 1:100). Imaging was done on Axio Scan (Z1, Zeiss, Germany) and super-resolution airyscan laser-scanning confocal system (CARL ZEISS confocal microscope LSM 888). Quantification of fluorescent intensity was examined using ImageJ software with colocalization plugin and Zen software (Zen blue 3.1) and ImageJ software with colocalization plugin.[89, 90]
Exosome uptake assay.
For the cellular uptake of keratinocyte-originated exosome by monocyte-derived macrophages, exosome concentration was measured by NTA and stained with ExoGlow™-Membrane EV Labeling Kit (System Biosciences, Palo Alto, CA, EXOGM600A-1). The Exoglow™-stained exosomes were added to the macrophages and live-cell imaging was performed using LSM 880 confocal microscope (Zeiss).
Cytokine profiling array.
Cytokine profiling from macrophages conditioned media was performed after exposing them for seven days either with or diaxosomes using the Proteome Profiler Human XL Cytokine Array Kit (R&D Biosystems, Minneapolis, MN, USA, ARY022B). Briefly, membranes were blocked for 1 h using array buffer and incubated overnight at 4°C with conditioned media combined with array buffer. Membranes were washed, incubated with antibody cocktail for 1 h at RT, washed, and incubated with streptavidin-HRP for 30 min. After treatment with a chemiluminescent reagent, membranes were imaged under Azure biosystems.
Enzyme-linked immunosorbent assay (ELISA).
The macrophages conditioned media was collected after exposing them for seven days either with or diaexosomes. The levels of IL-18 (R&D Biosystems, DY8936–05), CXCL5 (R&D Biosystems, DY254–05), TIM-3 (R&D Biosystems, DY2365), VEGF (R&D Biosystems, DY293B-05) were measured using the ELISA kits as per manufacturer’s instructions.
Monocytes derived macrophages chemotaxis assay.
Cytoselect cell migration assay was performed according to the manufacturer instructions (cell biolabs, #CBA-105). Briefly, in feeder tray, 150Ul of conditioned media from MDM cells treated with keratinocytes-originated exosomes were added and 40,000 MDM cells were seeded in 96 well plate membrane chamber in serum free RPMI media. Next day, migratory cells were dissociated from membrane with the help of cell detachment buffer to the lower chamber. Migratory cells were lysed and quantified using CyQuant fluorescent GR dye by noting down the fluorescence at 480 nm/520 nm.
Statistical analysis.
GraphPad Prism (GraphPad Software) v8.0 was used for statistical analyses. No statistical methods were used to predetermine the sample size. The ΔΔCt value was used for statistical analysis of all RT-qPCR data. Statistical analysis between multiple groups was performed using a one-way analysis of variance with the post-hoc Bonferroni multiple comparison test. Statistical analysis between two groups was performed using unpaired Student’s two-sided ttests. P < 0.05 was considered statistically significant. Significance levels and exact P values were indicated in all relevant figures. Data were checked for normality prior to analysis. Data for independent experiments were presented as means ± SEM unless otherwise stated.
Supplementary Material
Highlights.
The discarded negative pressure wound therapy sponge dressing are enriched in nanosized membrane-bound extracellular vesicles.
Immunomagnetic two-step separation method using KRT14 and tetraspanins antibodies can successfully isolate exosomes of keratinocyte-origin in chronic wound fluid of non-diabetic and diabetic patients.
Uptake of keratinocyte-originated exosomes by macrophages was low for diabetics versus non-diabetics leading to compromised cell-cell crosstalk between wound-edge keratinocytes and macrophages.
Keratinocyte-originated exosomes in diabetic patients caused sustained expression of inflammatory chemokines known to recruit macrophages
ACKNOWLEDGMENTS
We would like to acknowledge Dr. Pradeep Barak of Oxford Nanoimager for his critical input on the image acquisition of super-resolution microscopic images. We would also like to acknowledge Dr. Alexandre Kitching of Nanometrix for his help in data analysis. We would also like to acknowledge the Integrated Nanosystems Development Institute (INDI) for use of their JEOL 7800-f Field Emission Scanning Electron Microscope, which was awarded through NSF grant MRI-1229514. The 15 T Bruker SolariXR FT-ICR instrument was supported by NIH Award Number Grant S10 OD018507. This study primarily supported NIH grant DK129592 and the junior faculty startup pack from ICRME to SG.
ABBREVIATIONS
- 3D
Three Dimensional
- AMPs
Anti-Microbial Peptides
- ANOVA
Analysis of Variance
- ANXA5
Annexin A5
- AU
Arbitrary units
- BCA
Bicinchoninic acid assay
- CODI
Collaborative Discovery
- CXCL
C-X-C Motif Chemokine Ligand
- DAPI
4′,6-diamidino-2-phenylindole
- DiD
1,1′-dioctadecyl-3,3,3′,3′- tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate salt
- dSTORM
Direct Stochastic Optical Reconstruction Microscopy
- EpCAM
Epithelial Cellular Adhesion Molecule
- ESCRT-I
Endosomal Sorting Complexes Required for Transport
- EVs
Extracellular Vesicles
- FCS
Fluorescence Correlation Spectroscopy
- FITC
Fluorescein Isothiocyanate
- HbA1C
Hemoglobin A1C
Human Extracellular Vesicles from Keratinocytes
Human Keratinocyte-Originated Exosomes
- hnRNPA2B1
Heterogeneous Nuclear RibonucleoproteinA2B1
- IG-TEM
Immunogold Transmission Electron Microscopy
- IMS
Immunomagnetic Separation
- KRT
Keratin
- MCSF
Macrophage Colony-Stimulating Factor
- MDM
Monocyte-Derived Macrophages
- MVB
Multivesicular Bodies
- mϕ
Macrophages
- NPWT
Negative Pressure Wound Therapy
- NTA
Nanoparticle Tracking Analysis
- Ø
Diameter
- PCA
Principal Component Analysis
- PE
Phycoerythrin
- SEM
Scanning Electron Microscopy
- TEM
Transmission Electron Microscopy
- TIM3
T-cell Immunoglobulin Mucin-3
- TSG101
Tumor Susceptibility Gene 101 Protein
- VEGF
Vascular Endothelial Growth Factor
- VIP
Variable Importance on Projection
- ζ
Zeta potentials
- ρ
Density
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge.
Immunomagnetic separation method using anti-pan-keratin conjugated to superparamagnetic Dynabeads™ is not specific for isolating exosomes of keratinocyte origin (Fig. S1); Expression of KRT14 is specific for Keratinocytes (Fig. S2); Validation of super-resolution dSTORM immunofluorescence images (Fig. S3); Validation of KRT14 immunomagnetic separation method (Fig. S4); Validation of super-resolution dSTORM immunofluorescence images, size distribution of and loading plot of Raman spectra (Fig. S5); Zoomed out immunohistochemistry images of human non-diabetic and diabetic wound-edge epidermis and loading plot of Raman spectra (Fig. S6); Mycoplasma contamination detection and phase contrast images of mϕ exposed to and diaexosomes (Fig. S7); Z-stack image of exosome uptake assay by mϕ (Fig. S8). Immunofluorescence staining of mϕ and Human XL cytokine array of mϕ conditioned media exposed to either or diaexosomes (Fig. S9). Proteomics analysis of four different density fractions (Table S1). Live-cell confocal images showing uptake of by blood monocyte-derived macrophages using LSM 880 (mp4 movies S1). Live-cell confocal images showing uptake of diaexosomes by blood monocyte-derived macrophages using LSM 880 (mp4 movies S2).
Author’s Statement
All human studies were approved by The Ohio State University and Indiana University Institutional Review Board. Declaration of Helsinki protocols was followed, and patients gave their written informed consent.
Financial Interest Statement
The authors declare no competing financial interest.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Théry C, Ostrowski M, Segura E, Membrane vesicles as conveyors of immune responses, Nat Rev Immunol, 9 (2009) 581–593. [DOI] [PubMed] [Google Scholar]
- [2].Gallo A, Tandon M, Alevizos I, Illei GG, The majority of microRNAs detectable in serum and saliva is concentrated in exosomes, PLoS One, 7 (2012) e30679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Admyre C, Johansson SM, Qazi KR, Filén JJ, Lahesmaa R, Norman M, Neve EP, Scheynius A, Gabrielsson S, Exosomes with immune modulatory features are present in human breast milk, J Immunol, 179 (2007) 1969–1978. [DOI] [PubMed] [Google Scholar]
- [4].Street JM, Koritzinsky EH, Glispie DM, Star RA, Yuen PS, Urine Exosomes: An Emerging Trove of Biomarkers, Adv Clin Chem, 78 (2017) 103–122. [DOI] [PubMed] [Google Scholar]
- [5].Mathieu M, Martin-Jaular L, Lavieu G, Théry C, Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication, Nat Cell Biol, 21 (2019) 9–17. [DOI] [PubMed] [Google Scholar]
- [6].Skotland T, Sagini K, Sandvig K, Llorente A, An emerging focus on lipids in extracellular vesicles, Advanced Drug Delivery Reviews, 159 (2020) 308–321. [DOI] [PubMed] [Google Scholar]
- [7].Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sánchez-Madrid F, Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs, Nature Communications, 4 (2013) 2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Janas T, Janas MM, Sapoń K, Janas T, Mechanisms of RNA loading into exosomes, FEBS Letters, 589 (2015) 1391–1398. [DOI] [PubMed] [Google Scholar]
- [9].Zhang W, Jiang X, Bao J, Wang Y, Liu H, Tang L, Exosomes in Pathogen Infections: A Bridge to Deliver Molecules and Link Functions, Front Immunol, 9 (2018) 90–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Margolis L, Sadovsky Y, The biology of extracellular vesicles: The known unknowns, PLoS Biol, 17 (2019) e3000363-e3000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].O’Brien K, Breyne K, Ughetto S, Laurent LC, Breakefield XO, RNA delivery by extracellular vesicles in mammalian cells and its applications, Nature Reviews Molecular Cell Biology, 21 (2020) 585–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Han S, Gonzalo DH, Feely M, Rinaldi C, Belsare S, Zhai H, Kalra K, Gerber MH, Forsmark CE, Hughes SJ, Stroma-derived extracellular vesicles deliver tumor-suppressive miRNAs to pancreatic cancer cells, Oncotarget, 9 (2017) 5764–5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Luga V, Zhang L, Viloria-Petit AM, Ogunjimi AA, Inanlou MR, Chiu E, Buchanan M, Hosein AN, Basik M, Wrana JL, Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration, Cell, 151 (2012) 1542–1556. [DOI] [PubMed] [Google Scholar]
- [14].Webber JP, Spary LK, Sanders AJ, Chowdhury R, Jiang WG, Steadman R, Wymant J, Jones AT, Kynaston H, Mason MD, Tabi Z, Clayton A, Differentiation of tumour-promoting stromal myofibroblasts by cancer exosomes, Oncogene, 34 (2015) 290–302. [DOI] [PubMed] [Google Scholar]
- [15].Duan P, Tan J, Miao Y, Zhang Q, Potential role of exosomes in the pathophysiology, diagnosis, and treatment of hypoxic diseases, Am J Transl Res, 11 (2019) 1184–1201. [PMC free article] [PubMed] [Google Scholar]
- [16].Hofmann L, Ludwig S, Vahl JM, Brunner C, Hoffmann TK, Theodoraki MN, The Emerging Role of Exosomes in Diagnosis, Prognosis, and Therapy in Head and Neck Cancer, Int J Mol Sci, 21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, Ayre DC, Bach JM, Bachurski D, Baharvand H, Balaj L, Baldacchino S, Bauer NN, Baxter AA, Bebawy M, Beckham C, Bedina Zavec A, Benmoussa A, Berardi AC, Bergese P, Bielska E, Blenkiron C, Bobis-Wozowicz S, Boilard E, Boireau W, Bongiovanni A, Borràs FE, Bosch S, Boulanger CM, Breakefield X, Breglio AM, Brennan M, Brigstock DR, Brisson A, Broekman ML, Bromberg JF, Bryl-Górecka P, Buch S, Buck AH, Burger D, Busatto S, Buschmann D, Bussolati B, Buzás EI, Byrd JB, Camussi G, Carter DR, Caruso S, Chamley LW, Chang YT, Chen C, Chen S, Cheng L, Chin AR, Clayton A, Clerici SP, Cocks A, Cocucci E, Coffey RJ, Cordeiro-da-Silva A, Couch Y, Coumans FA, Coyle B, Crescitelli R, Criado MF, D’Souza-Schorey C, Das S, Datta Chaudhuri A, de Candia P, De Santana EF, De Wever O, Del Portillo HA, Demaret T, Deville S, Devitt A, Dhondt B, Di Vizio D, Dieterich LC, Dolo V, Dominguez Rubio AP, Dominici M, Dourado MR, Driedonks TA, Duarte FV, Duncan HM, Eichenberger RM, Ekström K, El Andaloussi S, Elie-Caille C, Erdbrügger U, Falcón-Pérez JM, Fatima F, Fish JE, Flores-Bellver M, Försönits A, Frelet-Barrand A, Fricke F, Fuhrmann G, Gabrielsson S, Gámez-Valero A, Gardiner C, Gärtner K, Gaudin R, Gho YS, Giebel B, Gilbert C, Gimona M, Giusti I, Goberdhan DC, Görgens A, Gorski SM, Greening DW, Gross JC, Gualerzi A, Gupta GN, Gustafson D, Handberg A, Haraszti RA, Harrison P, Hegyesi H, Hendrix A, Hill AF, Hochberg FH, Hoffmann KF, Holder B, Holthofer H, Hosseinkhani B, Hu G, Huang Y, Huber V, Hunt S, Ibrahim AG, Ikezu T, Inal JM, Isin M, Ivanova A, Jackson HK, Jacobsen S, Jay SM, Jayachandran M, Jenster G, Jiang L, Johnson SM, Jones JC, Jong A, Jovanovic-Talisman T, Jung S, Kalluri R, Kano SI, Kaur S, Kawamura Y, Keller ET, Khamari D, Khomyakova E, Khvorova A, Kierulf P, Kim KP, Kislinger T, Klingeborn M, Klinke DJ 2nd, Kornek M, Kosanović MM, Kovács Á F, Krämer-Albers EM, Krasemann S, Krause M, Kurochkin IV, Kusuma GD, Kuypers S, Laitinen S, Langevin SM, Languino LR, Lannigan J, Lässer C, Laurent LC, Lavieu G, LázaroIbáñez E, Le Lay S, Lee MS, Lee YXF, Lemos DS, Lenassi M, Leszczynska A, Li IT, Liao K, Libregts SF, Ligeti E, Lim R, Lim SK, Linē A, Linnemannstöns K, Llorente A, Lombard CA, Lorenowicz MJ, Lörincz Á M, Lötvall J, Lovett J, Lowry MC, Loyer X, Lu Q, Lukomska B, Lunavat TR, Maas SL, Malhi H, Marcilla A, Mariani J, Mariscal J, Martens-Uzunova ES, Martin-Jaular L, Martinez MC, Martins VR, Mathieu M, Mathivanan S, Maugeri M, McGinnis LK, McVey MJ, Meckes DG Jr., Meehan KL, Mertens I, Minciacchi VR, Möller A, Møller Jørgensen M, Morales-Kastresana A, Morhayim J, Mullier F, Muraca M, Musante L, Mussack V, Muth DC, Myburgh KH, Najrana T, Nawaz M, Nazarenko I, Nejsum P, Neri C, Neri T, Nieuwland R, Nimrichter L, Nolan JP, Nolte-’t Hoen EN, Noren Hooten N, O’Driscoll L, O’Grady T, O’Loghlen A, Ochiya T, Olivier M, Ortiz A, Ortiz LA, Osteikoetxea X, Østergaard O, Ostrowski M, Park J, Pegtel DM, Peinado H, Perut F, Pfaffl MW, Phinney DG, Pieters BC, Pink RC, Pisetsky DS, Pogge von Strandmann E, Polakovicova I, Poon IK, Powell BH, Prada I, Pulliam L, Quesenberry P, Radeghieri A, Raffai RL, Raimondo S, Rak J, Ramirez MI, Raposo G, Rayyan MS, Regev-Rudzki N, Ricklefs FL, Robbins PD, Roberts DD, Rodrigues SC, Rohde E, Rome S, Rouschop KM, Rughetti A, Russell AE, Saá P, Sahoo S, Salas-Huenuleo E, Sánchez C, Saugstad JA, Saul MJ, Schiffelers RM, Schneider R, Schøyen TH, Scott A, Shahaj E, Sharma S, Shatnyeva O, Shekari F, Shelke GV, Shetty AK, Shiba K, Siljander PR, Silva AM, Skowronek A, Snyder OL 2nd, Soares RP, Sódar BW, Soekmadji C, Sotillo J, Stahl PD, Stoorvogel W, Stott SL, Strasser EF, Swift S, Tahara H, Tewari M, Timms K, Tiwari S, Tixeira R, Tkach M, Toh WS, Tomasini R, Torrecilhas AC, Tosar JP, Toxavidis V, Urbanelli L, Vader P, van Balkom BW, van der Grein SG, Van Deun J, van Herwijnen MJ, Van Keuren-Jensen K, van Niel G, van Royen ME, van Wijnen AJ, Vasconcelos MH, Vechetti IJ Jr., Veit TD, Vella LJ, Velot É, Verweij FJ, Vestad B, Viñas JL, Visnovitz T, Vukman KV, Wahlgren J, Watson DC, Wauben MH, Weaver A, Webber JP, Weber V, Wehman AM, Weiss DJ, Welsh JA, Wendt S, Wheelock AM, Wiener Z, Witte L, Wolfram J, Xagorari A, Xander P, Xu J, Yan X, Yáñez-Mó M, Yin H, Yuana Y, Zappulli V, Zarubova J, Žėkas V, Zhang JY, Zhao Z, Zheng L, Zheutlin AR, Zickler AM, Zimmermann P, Zivkovic AM, Zocco D, Zuba-Surma EK, Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines, J Extracell Vesicles, 7 (2018) 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li X, Corbett AL, Taatizadeh E, Tasnim N, Little JP, Garnis C, Daugaard M, Guns E, Hoorfar M, Li ITS, Challenges and opportunities in exosome research-Perspectives from biology, engineering, and cancer therapy, APL Bioeng, 3 (2019) 011503–011503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ramirez MI, Amorim MG, Gadelha C, Milic I, Welsh JA, Freitas VM, Nawaz M, Akbar N, Couch Y, Makin L, Cooke F, Vettore AL, Batista PX, Freezor R, Pezuk JA, Rosa-Fernandes L, Carreira ACO, Devitt A, Jacobs L, Silva IT, Coakley G, Nunes DN, Carter D, Palmisano G, Dias-Neto E, Technical challenges of working with extracellular vesicles, Nanoscale, 10 (2018) 881–906. [DOI] [PubMed] [Google Scholar]
- [20].Willms E, Cabañas C, Mäger I, Wood MJA, Vader P, Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression, Front Immunol, 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Van Deun J, Mestdagh P, Agostinis P, Akay O, Anand S, Anckaert J, Martinez ZA, Baetens T, Beghein E, Bertier L, Berx G, Boere J, Boukouris S, Bremer M, Buschmann D, Byrd JB, Casert C, Cheng L, Cmoch A, Daveloose D, De Smedt E, Demirsoy S, Depoorter V, Dhondt B, Driedonks TA, Dudek A, Elsharawy A, Floris I, Foers AD, Gartner K, Garg AD, Geeurickx E, Gettemans J, Ghazavi F, Giebel B, Kormelink TG, Hancock G, Helsmoortel H, Hill AF, Hyenne V, Kalra H, Kim D, Kowal J, Kraemer S, Leidinger P, Leonelli C, Liang Y, Lippens L, Liu S, Lo Cicero A, Martin S, Mathivanan S, Mathiyalagan P, Matusek T, Milani G, Monguio-Tortajada M, Mus LM, Muth DC, Nemeth A, Nolte-’t Hoen EN, O’Driscoll L, Palmulli R, Pfaffl MW, Primdal-Bengtson B, Romano E, Rousseau Q, Sahoo S, Sampaio N, Samuel M, Scicluna B, Soen B, Steels A, Swinnen JV, Takatalo M, Thaminy S, Thery C, Tulkens J, Van Audenhove I, van der Grein S, Van Goethem A, van Herwijnen MJ, Van Niel G, Van Roy N, Van Vliet AR, Vandamme N, Vanhauwaert S, Vergauwen G, Verweij F, Wallaert A, Wauben M, Witwer KW, Zonneveld MI, De Wever O, Vandesompele J, Hendrix A, EV-TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research, Nat Methods, 14 (2017) 228–232. [DOI] [PubMed] [Google Scholar]
- [22].Sinha M, Sen CK, Singh K, Das A, Ghatak S, Rhea B, Blackstone B, Powell HM, Khanna S, Roy S, Direct conversion of injury-site myeloid cells to fibroblast-like cells of granulation tissue, Nat Commun, 9 (2018) 936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Montini E, Cesana D, Schmidt M, Sanvito F, Bartholomae CC, Ranzani M, Benedicenti F, Sergi LS, Ambrosi A, Ponzoni M, Doglioni C, Di Serio C, von Kalle C, Naldini L, The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy, J Clin Invest, 119 (2009) 964–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hu L, Wang J, Zhou X, Xiong Z, Zhao J, Yu R, Huang F, Zhang H, Chen L, Exosomes derived from human adipose mensenchymal stem cells accelerates cutaneous wound healing via optimizing the characteristics of fibroblasts, Scientific Reports, 6 (2016) 32993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Li B, Luan S, Chen J, Zhou Y, Wang T, Li Z, Fu Y, Zhai A, Bi C, The MSC-Derived Exosomal lncRNA H19 Promotes Wound Healing in Diabetic Foot Ulcers by Upregulating PTEN via MicroRNA-152–3p, Mol Ther Nucleic Acids, 19 (2020) 814–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang X, Jiao Y, Pan Y, Zhang L, Gong H, Qi Y, Wang M, Gong H, Shao M, Wang X, Jiang D, Fetal Dermal Mesenchymal Stem Cell-Derived Exosomes Accelerate Cutaneous Wound Healing by Activating Notch Signaling, Stem Cells International, 2019 (2019) 2402916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Liu W, Yu M, Xie D, Wang L, Ye C, Zhu Q, Liu F, Yang L, Melatonin-stimulated MSC-derived exosomes improve diabetic wound healing through regulating macrophage M1 and M2 polarization by targeting the PTEN/AKT pathway, Stem Cell Research & Therapy, 11 (2020) 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wu P, Zhang B, Shi H, Qian H, Xu W, MSC-exosome: A novel cell-free therapy for cutaneous regeneration, Cytotherapy, 20 (2018) 291–301. [DOI] [PubMed] [Google Scholar]
- [29].Rani S, Ritter T, The Exosome - A Naturally Secreted Nanoparticle and its Application to Wound Healing, Advanced Materials, 28 (2016) 5542–5552. [DOI] [PubMed] [Google Scholar]
- [30].Zhou X, Brown BA, Siegel AP, El Masry M, Zeng X, Song W, Das A, Khandelwal P, Clark A, Singh K, Guda PR, Gorain M, Timsina L, Xuan Y, Jacobson SC, Novotny MV, Roy S, Agarwal M, Lee RJ, Sen CK, Clemmer DE, Ghatak S, Exosome-Mediated Crosstalk between Keratinocytes and Macrophages in Cutaneous Wound Healing, ACS Nano, (2020). [DOI] [PMC free article] [PubMed]
- [31].Van Deun J, Mestdagh P, Sormunen R, Cocquyt V, Vermaelen K, Vandesompele J, Bracke M, De Wever O, Hendrix A, The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling, Journal of Extracellular Vesicles, 3 (2014) 24858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Théry C, Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes, Proceedings of the National Academy of Sciences, 113 (2016) E968–E977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zaver V, Kankanalu P, Negative Pressure Wound Therapy, StatPearls, StatPearls Publishing Copyright © 2022, StatPearls Publishing LLC., Treasure Island (FL), 2022. [PubMed] [Google Scholar]
- [34].Zhang J, Hu ZC, Chen D, Guo D, Zhu JY, Tang B, Effectiveness and safety of negative-pressure wound therapy for diabetic foot ulcers: a meta-analysis, Plast Reconstr Surg, 134 (2014) 141–151. [DOI] [PubMed] [Google Scholar]
- [35].Kubiak BD, Albert SP, Gatto LA, Snyder KP, Maier KG, Vieau CJ, Roy S, Nieman GF, Peritoneal negative pressure therapy prevents multiple organ injury in a chronic porcine sepsis and ischemia/reperfusion model, Shock, 34 (2010) 525–534. [DOI] [PubMed] [Google Scholar]
- [36].McDaniel JC, Roy S, Wilgus TA , Neutrophil activity in chronic venous leg ulcers--a target for therapy?, Wound Repair Regen, 21 (2013) 339–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ghatak PD, Schlanger R, Ganesh K, Lambert L, Gordillo GM, Martinsek P, Roy S, A Wireless Electroceutical Dressing Lowers Cost of Negative Pressure Wound Therapy, Adv Wound Care (New Rochelle), 4 (2015) 302–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Gemoll T, Rozanova S, Röder C, Hartwig S, Kalthoff H, Lehr S, ElSharawy A, Habermann JK, Protein Profiling of Serum Extracellular Vesicles Reveals Qualitative and Quantitative Differences After Differential Ultracentrifugation and ExoQuick(TM) Isolation, J Clin Med, 9 (2020) 1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Livshits MA, Khomyakova E, Evtushenko EG, Lazarev VN, Kulemin NA, Semina SE, Generozov EV, Govorun VM, Isolation of exosomes by differential centrifugation: Theoretical analysis of a commonly used protocol, Sci Rep, 5 (2015) 17319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chhoy P, Brown CW, Amante JJ, Mercurio AM, Protocol for the separation of extracellular vesicles by ultracentrifugation from in vitro cell culture models, STAR Protocols, 2 (2021) 100303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhang P, Yeo JC, Lim CT, Advances in Technologies for Purification and Enrichment of Extracellular Vesicles, SLAS Technol, 24 (2019) 477–488. [DOI] [PubMed] [Google Scholar]
- [42].Brennan K, Martin K, FitzGerald SP, O’Sullivan J, Wu Y, Blanco A, Richardson C, Mc Gee MM, A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum, Scientific Reports, 10 (2020) 1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhang Y, Liu Y, Liu H, Tang WH, Exosomes: biogenesis, biologic function and clinical potential, Cell Biosci, 9 (2019) 19–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, Imamichi T, Chang W, DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update), Nucleic Acids Res, (2022). [DOI] [PMC free article] [PubMed]
- [45].Miettinen M, Fetsch JF, Distribution of keratins in normal endothelial cells and a spectrum of vascular tumors: Implications in tumor diagnosis, Human Pathology, 31 (2000) 1062–1067. [DOI] [PubMed] [Google Scholar]
- [46].Battifora H, Expression of keratins by endothelial cells: atavism or anarchy?, Ultrastruct Pathol, 14 (1990) iii−v. [PubMed] [Google Scholar]
- [47].Katagata Y, Takeda H, Ishizawa T, Hozumi Y, Kondo S, Occurrence and comparison of the expressed keratins in cultured human fibroblasts, endothelial cells and their sarcomas, Journal of Dermatological Science, 30 (2002) 1–9. [DOI] [PubMed] [Google Scholar]
- [48].Merjava S, Neuwirth A, Mandys V, Jirsova K, Cytokeratins 8 and 18 in adult human corneal endothelium, Exp Eye Res, 89 (2009) 426–431. [DOI] [PubMed] [Google Scholar]
- [49].Guo Y, Redmond CJ, Leacock KA, Brovkina MV, Ji S, Jaskula-Ranga V, Coulombe PA, Keratin 14-dependent disulfides regulate epidermal homeostasis and barrier function via 14–3-3σ and YAP1, Elife, 9 (2020) e53165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Alam H, Sehgal L, Kundu ST, Dalal SN, Vaidya MM, Novel function of keratins 5 and 14 in proliferation and differentiation of stratified epithelial cells, Mol Biol Cell, 22 (2011) 4068–4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Feng X, Coulombe PA, Complementary roles of specific cysteines in keratin 14 toward the assembly, organization, and dynamics of intermediate filaments in skin keratinocytes, J Biol Chem, 290 (2015) 22507–22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Vassar R, Rosenberg M, Ross S, Tyner A, Fuchs E, Tissue-specific and differentiation-specific expression of a human K14 keratin gene in transgenic mice, Proc Natl Acad Sci U S A, 86 (1989) 1563–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Sümer C, Boz Er AB, Dinçer T, Keratin 14 is a novel interaction partner of keratinocyte differentiation regulator: receptor-interacting protein kinase 4, Turk J Biol, 43 (2019) 225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Fuchs E, Weber K, Intermediate filaments: structure, dynamics, function, and disease, Annu Rev Biochem, 63 (1994) 345–382. [DOI] [PubMed] [Google Scholar]
- [55].Andreu Z, Yáñez-Mó M, Tetraspanins in extracellular vesicle formation and function, Front Immunol, 5 (2014) 442-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kim DK, Lee J, Kim SR, Choi DS, Yoon YJ, Kim JH, Go G, Nhung D, Hong K, Jang SC, Kim SH, Park KS, Kim OY, Park HT, Seo JH, Aikawa E, BajKrzyworzeka M, van Balkom BW, Belting M, Blanc L, Bond V, Bongiovanni A, Borras FE, Buee L, Buzas EI, Cheng L, Clayton A, Cocucci E, Dela Cruz CS, Desiderio DM, Di Vizio D, Ekstrom K, Falcon-Perez JM, Gardiner C, Giebel B, Greening DW, Gross JC, Gupta D, Hendrix A, Hill AF, Hill MM, Nolte-’t Hoen E, Hwang DW, Inal J, Jagannadham MV, Jayachandran M, Jee YK, Jorgensen M, Kim KP, Kim YK, Kislinger T, Lasser C, Lee DS, Lee H, van Leeuwen J, Lener T, Liu ML, Lotvall J, Marcilla A, Mathivanan S, Moller A, Morhayim J, Mullier F, Nazarenko I, Nieuwland R, Nunes DN, Pang K, Park J, Patel T, Pocsfalvi G, Del Portillo H, Putz U, Ramirez MI, Rodrigues ML, Roh TY, Royo F, Sahoo S, Schiffelers R, Sharma S, Siljander P, Simpson RJ, Soekmadji C, Stahl P, Stensballe A, Stepien E, Tahara H, Trummer A, Valadi H, Vella LJ, Wai SN, Witwer K, Yanez-Mo M, Youn H, Zeidler R, Gho YS, EVpedia: a community web portal for extracellular vesicles research, Bioinformatics, 31 (2015) 933–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Keerthikumar S, Chisanga D, Ariyaratne D, Al Saffar H, Anand S, Zhao K, Samuel M, Pathan M, Jois M, Chilamkurti N, Gangoda L, Mathivanan S, ExoCarta: A Web-Based Compendium of Exosomal Cargo, J Mol Biol, 428 (2016) 688–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zhang H, Silva AC, Zhang W, Rutigliano H, Zhou A, Raman Spectroscopy characterization extracellular vesicles from bovine placenta and peripheral blood mononuclear cells, PLOS ONE, 15 (2020) e0235214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Park J, Hwang M, Choi B, Jeong H, Jung J.-h., Kim HK, Hong S, Park J.-h., Choi Y, Exosome Classification by Pattern Analysis of Surface-Enhanced Raman Spectroscopy Data for Lung Cancer Diagnosis, Analytical Chemistry, 89 (2017) 6695–6701. [DOI] [PubMed] [Google Scholar]
- [60].Guerrini L, Garcia-Rico E, O’Loghlen A, Giannini V, Alvarez-Puebla RA, Surface-Enhanced Raman Scattering (SERS) Spectroscopy for Sensing and Characterization of Exosomes in Cancer Diagnosis, Cancers (Basel), 13 (2021) 2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Luyet PP, Falguières T, Pons V, Pattnaik AK, Gruenberg J, The ESCRT-I subunit TSG101 controls endosome-to-cytosol release of viral RNA, Traffic, 9 (2008) 2279–2290. [DOI] [PubMed] [Google Scholar]
- [62].Ju Y, Bai H, Ren L, Zhang L, The Role of Exosome and the ESCRT Pathway on Enveloped Virus Infection, International journal of molecular sciences, 22 (2021) 9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Kumar B, Dutta D, Iqbal J, Ansari MA, Roy A, Chikoti L, Pisano G, Veettil MV, Chandran B, ESCRT-I Protein Tsg101 Plays a Role in the Post-macropinocytic Trafficking and Infection of Endothelial Cells by Kaposi’s Sarcoma-Associated Herpesvirus, PLOS Pathogens, 12 (2016) e1005960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Miller CJ, Kassem HS, Pepper SD, Hey Y, Ward TH, Margison GP, Mycoplasma infection significantly alters microarray gene expression profiles, Biotechniques, 35 (2003) 812–814. [DOI] [PubMed] [Google Scholar]
- [65].Feng N, Huang X, Jia Y, Mycoplasma contamination affects cell characteristics and decreases the sensitivity of BV2 microglia to LPS stimulation, Cytotechnology, 71 (2019) 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Khine YY, Callari M, Lu H, Stenzel MH, Direct Correlation Between Zeta Potential and Cellular Uptake of Poly(methacrylic acid) Post-Modified with Guanidinium Functionalities, Macromolecular Chemistry and Physics, 217 (2016) 2302–2309. [Google Scholar]
- [67].Jeon S, Clavadetscher J, Lee D-K, Chankeshwara SV, Bradley M, Cho W-S, Surface Charge-Dependent Cellular Uptake of Polystyrene Nanoparticles, Nanomaterials (Basel), 8 (2018) 1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kaplanski G, Interleukin-18: Biological properties and role in disease pathogenesis, Immunological Reviews, 281 (2018) 138–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Metzemaekers M, Vanheule V, Janssens R, Struyf S, Proost P, Overview of the Mechanisms that May Contribute to the Non-Redundant Activities of Interferon-Inducible CXC Chemokine Receptor 3 Ligands, Front Immunol, 8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Nouailles G, Dorhoi A, Koch M, Zerrahn J, Weiner J 3rd, Faé KC, Arrey F, Kuhlmann S, Bandermann S, Loewe D, Mollenkopf H-J, Vogelzang A, Meyer-Schwesinger C, Mittrücker H-W, McEwen G, Kaufmann SHE, CXCL5-secreting pulmonary epithelial cells drive destructive neutrophilic inflammation in tuberculosis, The Journal of Clinical Investigation, 124 (2014) 1268–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Leavy O, TIM3: dual role in immunity, Nature Reviews Immunology, 8 (2008) 4–4. [Google Scholar]
- [72].Wang J, Li C, Fu J, Wang X, Feng X, Pan X, Tim-3 regulates inflammatory cytokine expression and Th17 cell response induced by monocytes from patients with chronic hepatitis B, Scandinavian Journal of Immunology, 89 (2019) e12755. [DOI] [PubMed] [Google Scholar]
- [73].Dai YD, Dias P, Exosomes or Microvesicles, a Secreted Subcellular Organelle Contributing to Inflammation and Diabetes, Diabetes, 67 (2018) 2154–2156. [DOI] [PubMed] [Google Scholar]
- [74].Ashrafizadeh M, Kumar AP, Aref AR, Zarrabi A, Mostafavi E, Exosomes as Promising Nanostructures in Diabetes Mellitus: From Insulin Sensitivity to Ameliorating Diabetic Complications, Int J Nanomedicine, 17 (2022) 1229–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Li X, Shi S, Jing D, Li X, Zhang B, Bie Q, Signal transduction mechanism of exosomes in diabetic complications (Review), Exp Ther Med, 23 (2022) 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Kalluri R, LeBleu VS, The biology, function, and biomedical applications of exosomes, Science, 367 (2020) eaau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Yáñez-Mó M, Siljander PRM, Andreu Z, Zavec AB, Borràs FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, Colás E, Cordeiro-da Silva A, Fais S, Falcon-Perez JM, Ghobrial IM, Giebel B, Gimona M, Graner M, Gursel I, Gursel M, Heegaard NHH, Hendrix A, Kierulf P, Kokubun K, Kosanovic M, Kralj-Iglic V, Krämer-Albers E-M, Laitinen S, Lässer C, Lener T, Ligeti E, Linē A, Lipps G, Llorente A, Lötvall J, MančekKeber M, Marcilla A, Mittelbrunn M, Nazarenko I, Nolte-’t Hoen ENM, Nyman TA, O’Driscoll L, Olivan M, Oliveira C, Pállinger É, Del Portillo HA, Reventós J, Rigau M, Rohde E, Sammar M, Sánchez-Madrid F, Santarém N, Schallmoser K, Ostenfeld MS, Stoorvogel W, Stukelj R, Van der Grein SG, Vasconcelos MH, Wauben MHM, De Wever O, Biological properties of extracellular vesicles and their physiological functions, Journal of extracellular vesicles, 4 (2015) 27066–27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Dai J, Su Y, Zhong S, Cong L, Liu B, Yang J, Tao Y, He Z, Chen C, Jiang Y, Exosomes: key players in cancer and potential therapeutic strategy, Signal Transduction and Targeted Therapy, 5 (2020) 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Das A, Ganesh K, Khanna S, Sen CK, Roy S, Engulfment of apoptotic cells by macrophages: a role of microRNA-21 in the resolution of wound inflammation, J Immunol, 192 (2014) 1120–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Ganesh K, Das A, Dickerson R, Khanna S, Parinandi NL, Gordillo GM, Sen CK, Roy S, Prostaglandin E(2) induces oncostatin M expression in human chronic wound macrophages through Axl receptor tyrosine kinase pathway, J Immunol, 189 (2012) 25632573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Roy S, Dickerson R, Khanna S, Collard E, Gnyawali U, Gordillo GM, Sen CK, Particulate beta-glucan induces TNF-alpha production in wound macrophages via a redox-sensitive NF-kappabeta-dependent pathway, Wound Repair Regen, 19 (2011) 411–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Ghatak S, Li J, Chan YC, Gnyawali SC, Steen E, Yung BC, Khanna S, Roy S, Lee RJ, Sen CK, AntihypoxamiR functionalized gramicidin lipid nanoparticles rescue against ischemic memory improving cutaneous wound healing, Nanomedicine, 12 (2016) 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Li J, Ghatak S, El Masry MS, Das A, Liu Y, Roy S, Lee RJ, Sen CK, Topical Lyophilized Targeted Lipid Nanoparticles in the Restoration of Skin Barrier Function following Burn Wound, Mol Ther, 26 (2018) 2178–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Ghatak S, Khona DK, Sen A, Huang K, Jagdale G, Singh K, Gopalakrishnan V, Cornetta KG, Roy S, Khanna S, Baker LA, Sen CK, Electroceutical fabric lowers zeta potential and eradicates coronavirus infectivity upon contact, Sci Rep, 11 (2021) 21723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Brown BA, Guda PR, Zeng X, Anthony A, Couse A, Barnes LF, Sharon EM, Trinidad JC, Sen CK, Jarrold MF, Ghatak S, Clemmer DE, Analysis of Keratinocytic Exosomes from Diabetic and Nondiabetic Mice by Charge Detection Mass Spectrometry, Anal Chem, (2022). [DOI] [PMC free article] [PubMed]
- [86].Deng B, Ghatak S, Sarkar S, Singh K, Das Ghatak P, Mathew-Steiner SS, Roy S, Khanna S, Wozniak DJ, McComb DW, Sen CK, Novel Bacterial Diversity and Fragmented eDNA Identified in Hyperbiofilm-Forming Pseudomonas aeruginosa Rugose Small Colony Variant, iScience, 23 (2020) 100827–100827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Garg S, Tang J, Rühe J, Naumann C, Actin-induced perturbation of PS lipid–cholesterol interaction: A possible mechanism of cytoskeleton-based regulation of membrane organization, Journal of structural biology, 168 (2009) 11–20. [DOI] [PubMed] [Google Scholar]
- [88].Rhoades E, Ramlall TF, Webb WW, Eliezer D, Quantification of α-Synuclein Binding to Lipid Vesicles Using Fluorescence Correlation Spectroscopy, Biophysical Journal, 90 (2006) 4692–4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, Fiji: an open-source platform for biological-image analysis, Nature Methods, 9 (2012) 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Bolte S, Cordelieres FP, A guided tour into subcellular colocalization analysis in light microscopy, J Microsc, 224 (2006) 213–232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.