Abstract
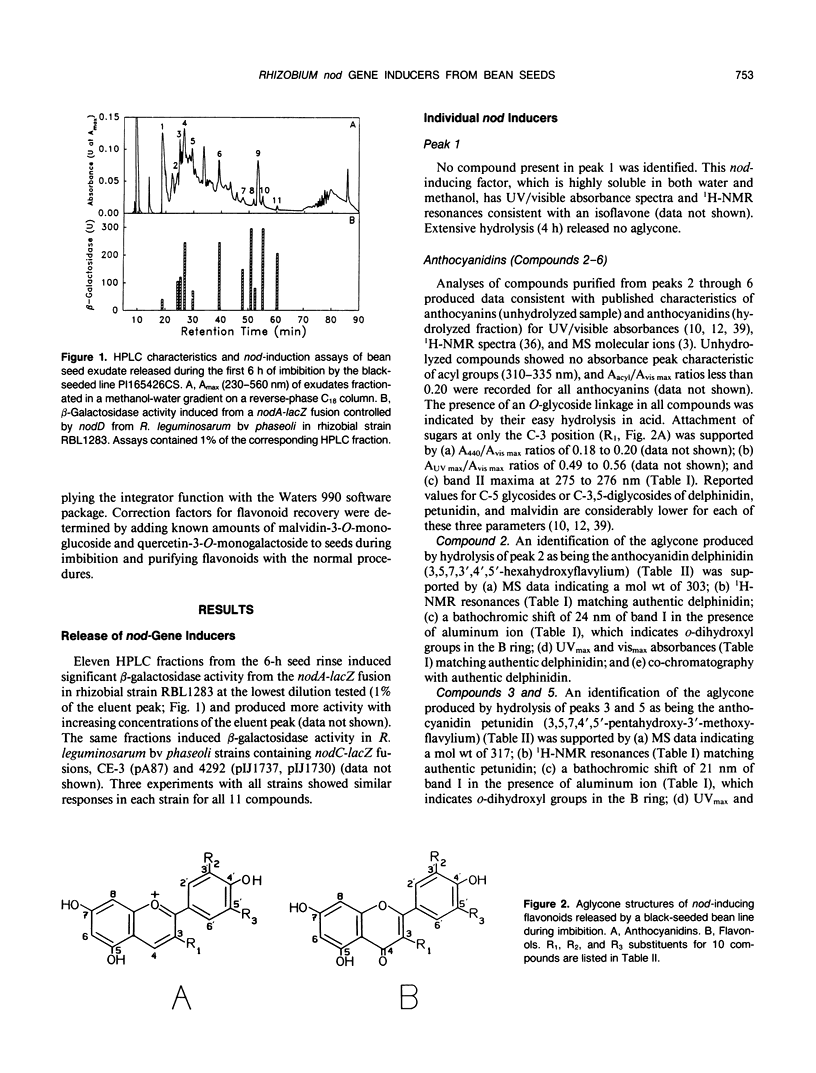
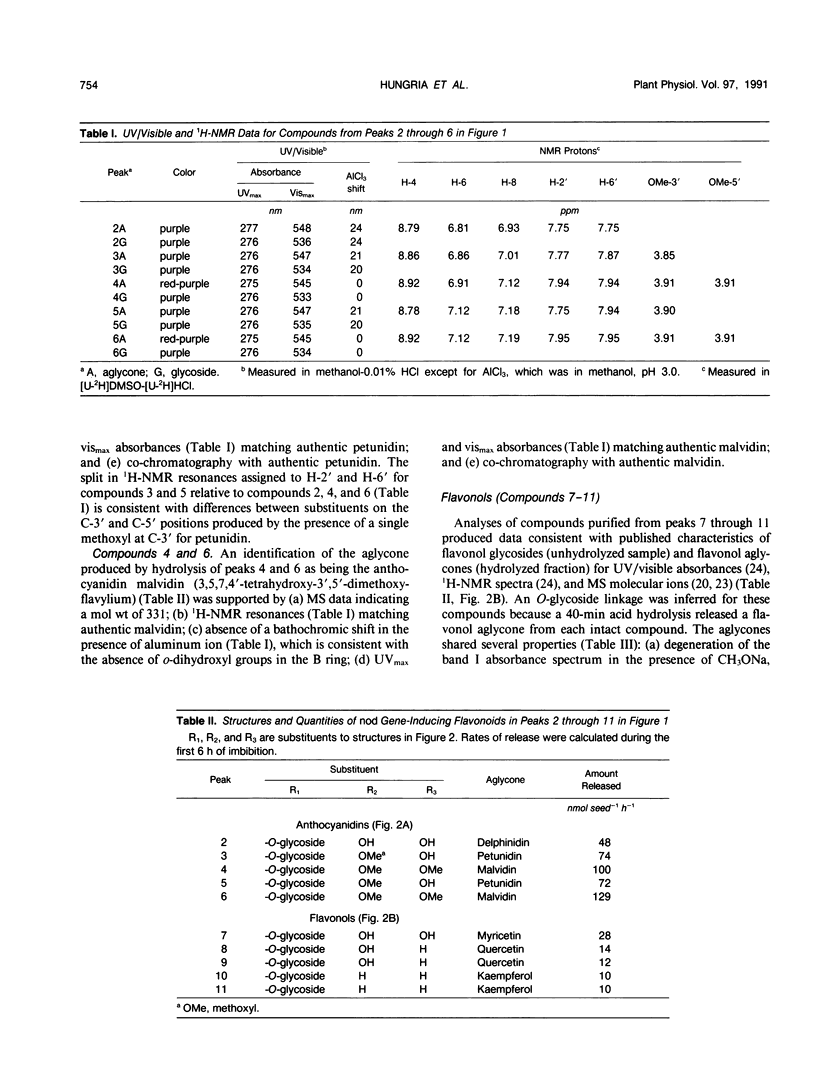
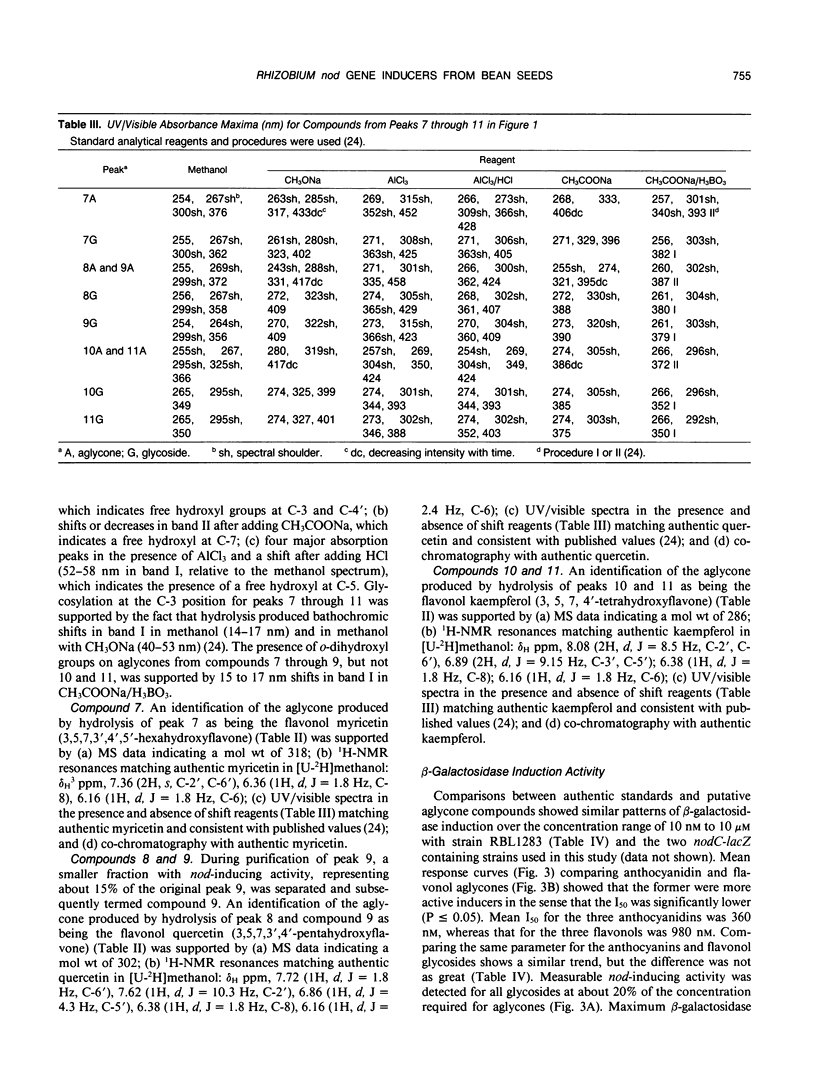
Eleven compounds released from germinating seeds of a black-seeded bean (Phaseolus vulgaris L., cv PI165426CS) induce transcription of nod genes in Rhizobium leguminosarum biovar phaseoli. Aglycones from 10 of those compounds were identified by spectroscopic methods (ultraviolet/visible, proton nuclear magnetic resonance, and mass spectroscopy), and their biological activities were demonstrated by induction of β-galactosidase activity in R. leguminosarum strains containing nodA-lacZ or nodC-lacZ fusions controlled by R. leguminosarum biovar phaseoli nodD genes. By making comparisons with authentic standards, the chemical structures for aglycones from the 10 molecules were confirmed as being anthocyanidins (delphinidin, petunidin, and malvidin) and flavonols (myricetin, quercetin, and kaempferol). All anthocyanidins and flavonols had 3-O-glycosylation and free hydroxyl groups at the 4′, 5, and 7 positions. Hydrolysis experiments showed that the mean concentration required for half-maximum nod gene induction (I50) by the 10 glycosides was about half that of the corresponding aglycones. The mean I50 value for the three anthocyanidins (360 nanomolar) was less (P ≤ 0.05) than that of the three flavonol aglycones (980 nanomolar). Each seed released approximately 2500 nanomoles of anthocyanidin and 450 nanomoles of flavonol nod gene inducers in conjugated forms during the first 6 hours of imbibition. Based on amounts and activities of the compounds released, anthocyanins contributed approximately 10-fold more total nod-inducing activity than flavonol glycosides. These anthocyanidins from bean seeds represent the first nod-inducing compounds identified from that group of flavonoids.
Full text
PDF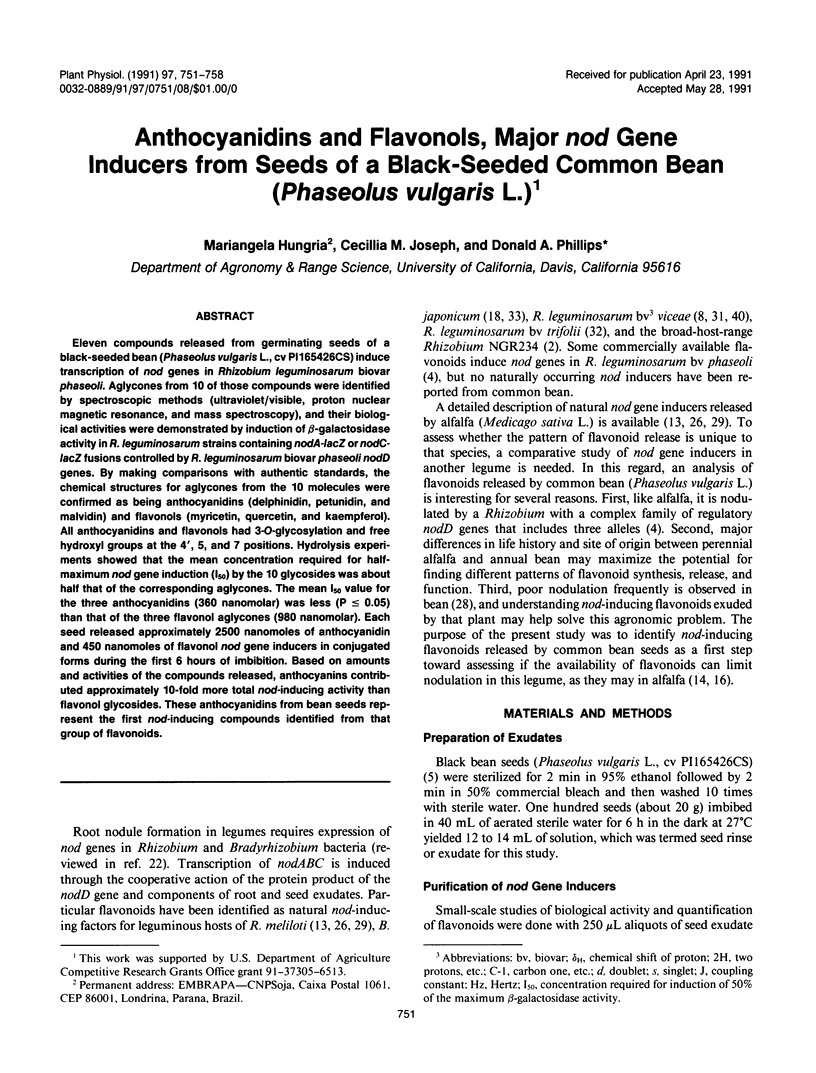



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bassam B. J., Djordjevic M. A., Redmond J. W., Batley M., Rolfe B. G. Identification of a nodD-dependent locus in the Rhizobium strain NGR234 activated by phenolic factors secreted by soybeans and other legumes. Mol Plant Microbe Interact. 1988 Apr;1(4):161–168. doi: 10.1094/mpmi-1-161. [DOI] [PubMed] [Google Scholar]
- Davis E. O., Johnston A. W. Regulatory functions of the three nodD genes of Rhizobium leguminosarum biovar phaseoli. Mol Microbiol. 1990 Jun;4(6):933–941. doi: 10.1111/j.1365-2958.1990.tb00666.x. [DOI] [PubMed] [Google Scholar]
- Djordjevic M. A., Redmond J. W., Batley M., Rolfe B. G. Clovers secrete specific phenolic compounds which either stimulate or repress nod gene expression in Rhizobium trifolii. EMBO J. 1987 May;6(5):1173–1179. doi: 10.1002/j.1460-2075.1987.tb02351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig U. A., Maxwell C. A., Joseph C. M., Phillips D. A. Chrysoeriol and Luteolin Released from Alfalfa Seeds Induce nod Genes in Rhizobium meliloti. Plant Physiol. 1990 Jan;92(1):116–122. doi: 10.1104/pp.92.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapulnik Y., Joseph C. M., Phillips D. A. Flavone limitations to root nodulation and symbiotic nitrogen fixation in alfalfa. Plant Physiol. 1987 Aug;84(4):1193–1196. doi: 10.1104/pp.84.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosslak R. M., Bookland R., Barkei J., Paaren H. E., Appelbaum E. R. Induction of Bradyrhizobium japonicum common nod genes by isoflavones isolated from Glycine max. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7428–7432. doi: 10.1073/pnas.84.21.7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosslak R. M., Joshi R. S., Bowen B. A., Paaren H. E., Appelbaum E. R. Strain-Specific Inhibition of nod Gene Induction in Bradyrhizobium japonicum by Flavonoid Compounds. Appl Environ Microbiol. 1990 May;56(5):1333–1341. doi: 10.1128/aem.56.5.1333-1341.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S. R. Rhizobium-legume nodulation: life together in the underground. Cell. 1989 Jan 27;56(2):203–214. doi: 10.1016/0092-8674(89)90893-3. [DOI] [PubMed] [Google Scholar]
- Maxwell C. A., Hartwig U. A., Joseph C. M., Phillips D. A. A Chalcone and Two Related Flavonoids Released from Alfalfa Roots Induce nod Genes of Rhizobium meliloti. Plant Physiol. 1989 Nov;91(3):842–847. doi: 10.1104/pp.91.3.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters N. K., Frost J. W., Long S. R. A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science. 1986 Aug 29;233(4767):977–980. doi: 10.1126/science.3738520. [DOI] [PubMed] [Google Scholar]
- Peters N. K., Long S. R. Alfalfa Root Exudates and Compounds which Promote or Inhibit Induction of Rhizobium meliloti Nodulation Genes. Plant Physiol. 1988 Oct;88(2):396–400. doi: 10.1104/pp.88.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recourt K., Schripsema J., Kijne J. W., van Brussel A. A., Lugtenberg B. J. Inoculation of Vicia sativa subsp. nigra roots with Rhizobium leguminosarum biovar viciae results in release of nod gene activating flavanones and chalcones. Plant Mol Biol. 1991 May;16(5):841–852. doi: 10.1007/BF00015076. [DOI] [PubMed] [Google Scholar]
- Sadowsky M. J., Olson E. R., Foster V. E., Kosslak R. M., Verma D. P. Two host-inducible genes of Rhizobium fredii and characterization of the inducing compound. J Bacteriol. 1988 Jan;170(1):171–178. doi: 10.1128/jb.170.1.171-178.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaat S. A., Schripsema J., Wijffelman C. A., van Brussel A. A., Lugtenberg B. J. Analysis of the major inducers of the Rhizobium nodA promoter from Vicia sativa root exudate and their activity with different nodD genes. Plant Mol Biol. 1989 Aug;13(2):175–188. doi: 10.1007/BF00016136. [DOI] [PubMed] [Google Scholar]


