Abstract
Background
The clinical and microbial factors associated with Klebsiella pneumoniae bloodstream infections (BSIs) are not well characterized. Prior studies have focused on highly resistant or hypervirulent isolates, limiting our understanding of K. pneumoniae strains that commonly cause BSI. We performed a record review and whole-genome sequencing to investigate the clinical characteristics, bacterial diversity, determinants of antimicrobial resistance, and risk factors for in-hospital death in a cohort of patients with K. pneumoniae BSI.
Methods
We identified 562 patients at Massachusetts General Hospital with K. pneumoniae BSIs between 2016 and 2022. We collected data on comorbid conditions, infection source, clinical outcomes, and antibiotic resistance and performed whole-genome sequencing on 108 sequential BSI isolates from 2021 to 2022.
Results
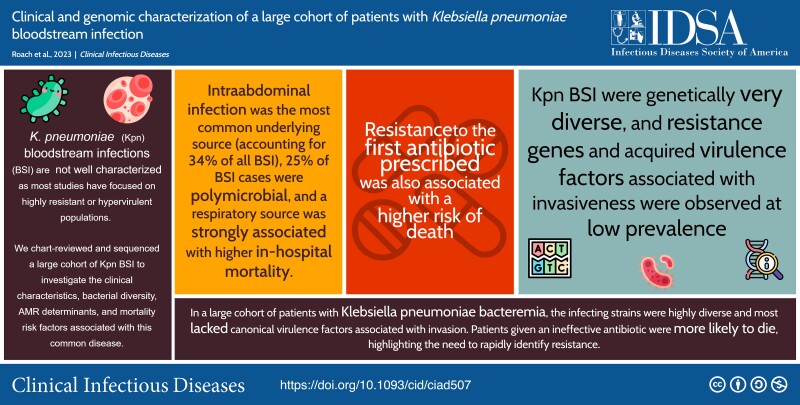
Intra-abdominal infection was the most common source of infection accounting for 34% of all BSIs. A respiratory tract source accounted for 6% of BSIs but was associated with a higher in-hospital mortality rate (adjusted odds ratio, 5.4 [95% confidence interval, 2.2–12.8]; P < .001 for comparison with other sources). Resistance to the first antibiotic prescribed was also associated with a higher risk of death (adjusted odds ratio, 5.2 [95% confidence interval, 2.2–12.4]; P < .001). BSI isolates were genetically diverse, and no clusters of epidemiologically and genetically linked cases were observed. Virulence factors associated with invasiveness were observed at a low prevalence, although an unexpected association between O-antigen type and the source of infection was found.
Conclusions
These observations demonstrate the versatility of K. pneumoniae as an opportunistic pathogen and highlight the need for new approaches for surveillance and the rapid identification of patients with invasive antimicrobial-resistant K. pneumoniae infection.
Keywords: Klebsiella pneumoniae, whole-genome sequencing, bacteremia, virulence factors, mortality modeling
In a cohort of patients with Klebsiella pneumonia bacteremia, most infections were caused by strains that lacked canonical features associated with invasion. Patients given an ineffective first antibiotic were more likely to die, highlighting the need to rapidly identify resistance.
Graphical Abstract
Graphical Abstract.
This graphical abstract is also available at Tidbit: https://tidbitapp.io/tidbits/clinical-and-genomic-characterization-of-a-large-cohort-of-patients-with-klebsiella-pneumoniae-bloodstream-infection/update
Klebsiella pneumoniae is a leading cause of healthcare-associated infections [1] and neonatal sepsis [2]. K. pneumoniae colonizes the gut and respiratory tract of humans and animals, persists in environmental reservoirs, and is permissive of horizontal gene transfer [3, 4]. These characteristics have contributed to the emergence of K. pneumoniae with high levels of antimicrobial resistance (AMR) [5]. In 2019, an estimated 600 000 deaths were attributable to AMR K. pneumoniae globally [6].
K. pneumoniae infections are associated with impaired host defense, but infections also occur in nonimmunocompromised individuals, and bacterial factors play a role in determining host range [7, 8]. Hypervirulent strains have thick capsules, express multiple siderophores, and are associated with invasive disease following community acquisition in healthy hosts [9]. Classic K. pneumoniae, in contrast, has variable capsule production, may not express siderophores, and is opportunistic [8, 10].
Few studies have systematically evaluated the clinical and microbiologic factors associated with K. pneumoniae bloodstream infections (BSIs). Most research has focused on BSI in highly selected K. pneumoniae populations, such as those displaying carbapenem resistance or hypervirulence, but less attention has been devoted to understanding K. pneumoniae BSI in general clinical scenarios [11, 12]. As K. pneumoniae displays different infection characteristics in distinct populations [3, 6], a better understanding is needed of risk factors for acquisition of and death from K. pneumoniae BSI.
In the current study, we evaluated a cohort of patients with K. pneumoniae BSI at a tertiary care hospital in Boston, Massachusetts in the United States. We extracted data on all patients with K. pneumoniae BSI from 2016 to 2022 and coupled this with whole-genome sequencing (WGS) on isolates collected over the last year of the study. Here, we report the clinical characteristics of patients with K. pneumoniae BSIs, define risk factors for in-hospital death, and perform a genomic survey of the diversity and virulence factors of the bacterial population.
METHODS
Patient Population
The study was conducted at Massachusetts General Hospital in Boston, Massachusetts, a 1000-bed tertiary care hospital with >11 000 admissions to the internal medicine service annually [13]. Patients who had K. pneumoniae isolated in ≥1 blood culture from 1 March 2016 to 31 May 2022 were included; patients ranged in age from 2 months to 96 years. We reviewed records retrospectively from March 2016 to June 2021 and prospectively from July 2021 to May 2022. Isolates from all K. pneumoniae BSIs identified during the prospective period were subcultured for sequencing and hypermucoidity testing. The study was approved by the MassGeneral Brigham Institutional Review Board.
Record Review
We extracted clinical and microbiologic data from the medical record; this procedure is detailed in the Supplementary Methods. K. pneumoniae BSI was defined as isolation of the bacterium in ≥1 blood culture bottle during the study period. Infectious diseases clinicians (D. J. R., R. C. L., and J. B. H.) reviewed each case to ensure that the primary infection sites met the National Healthcare Safety Network definitions [14]. A primary catheter-associated infection was documented if the catheter was present for >48 hours before the positive culture and no other site of infection was identified. In cases where no site met the above criteria, the source was listed as unidentified. Hospital-acquired infection was defined as a positive blood culture collected >48 hours after admission. Hardware-associated infections were classified when source hardware was present for >48 hours before the BSI.
Microbiology
K. pneumoniae isolated from blood cultures in the Massachusetts General Hospital Clinical Microbiology Laboratory underwent identification using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF) (bioMérieux V3.2 database) and antimicrobial susceptibility testing per Clinical and Laboratory Standards Institute standards. Multidrug resistance (MDR) was defined as resistance to ceftriaxone and ≥2 non–β-lactam drug classes [15]. Isolates with intermediate susceptibility were categorized as resistant. Hypermucoidity testing was performed using methods described by Short et al [7]. In brief, cultures were grown in lysogeny broth, a 1:100 dilution of this was subcultured and normalized to an optical density (OD) of 1 and centrifuged at 1000g for 5 minutes, and mucoidity was expressed as a ratio of the OD at 600 nm (OD600) of the supernatant to the OD600 of the original culture, with the top 15% of ratios considered hypermucoid.
Genome Sequencing
WGS was performed on the first K. pneumoniae bloodstream isolate collected from patients between July 2021 and May 2022. Sequencing was performed using 150-base paired-end reads on an Illumina platform with an average of 10 million reads per isolate. Reads were aligned to the reference genome NTUH-K2044 [16] using bwa software v0.7.17 [17]. Pilon v1.24 was used for single-nucleotide polymorphism (SNP) calling [18]. Core SNPs were used to create a phylogenetic tree using RAxML v8.2.12 [19]. Sequence types (STs), AMR genes, virulence factors, and K and O types were identified using Kleborate v2.2.0 [20], and fim and mrk genes were identified using the basic local alignment search tool (BLAST) v2.9.0+ [21]. We calculated the Shannon diversity index (SDI) of STs using the R package “vegan.” All sequences were uploaded to the Sequence Read Archive (BioProject PRJNA978102).
Statistical Analysis
The association between in-hospital death and patient and bacterial characteristics was assessed using χ2 tests for categorical variables and t or Wilcoxon tests for continuous variables. Covariates included age, sex, comorbid conditions (diabetes, liver disease, chronic kidney disease, cancer, solid tumor, and hematologic cancer), Charlson comorbidity index, immunosuppressed status, neutropenic status, underlying bacteremia source (abdominal, urinary tract, central catheter, respiratory tract, or unidentified), intensive care unit (ICU) admission at the time of culture, resistance to particular antibiotics (ceftriaxone, cefepime, piperacillin-tazobactam, aminoglycoside, and fluoroquinolones), time to first effective antibiotic, hospital-acquired infection, whether an incorrect antibiotic was prescribed, and whether the infectious diseases service was consulted. Bivariate associations were first assessed; subsequently, adjusted odds ratios (ORs) and 95% confidence intervals (CIs) were obtained from a multivariable logistic regression model to evaluate associations with in-hospital death, with adjustment for covariates. All analyses were performed with R software, v4.2.1 [22], or SAS v9.4 software (SAS Institute), and differences were considered significant at P < .05 (2-sided) .
RESULTS
A total of 562 unique patients meeting criteria were identified. Of these, 454 patients (81%) were included in the retrospective review, and 108 (19%) were identified prospectively. No patients were excluded.
Patient Characteristics
Table 1 shows the cohort characteristics. K. pneumoniae BSI occurred more often in older, male patients. Polymicrobial infection occurred in 25% of K. pneumoniae BSI (Supplementary Figure 1) and was present across sources (Supplementary Table 1). K. pneumoniae BSI was associated with severe illness: 207 patients (37%) were admitted to the intensive care unit, and 81 (14%) died before discharge. The most common comorbid conditions were diabetes and cancer. Of 178 patients with cancer, 104 (58%) had solid organ cancer, while 74 (42%) had hematologic cancer. Thirty-nine patients (6.9%) had recurrent K. pneumoniae BSI. Ten recurrences were in the prospective cohort; of these, 6 reinfections were due to the same strain, and 4 due to different strains.
Table 1.
Demographic Characteristics and Comorbid Conditions in Patients With Klebsiella pneumoniae Bloodstream Infections
| Characteristic or Condition | Patients, No. (%)a | ||
|---|---|---|---|
| Total Cohort (N = 562) | Prospective Cohort (n = 108) | Retrospective Cohort (n = 454) | |
| Male sex | 335 (59.6) | 68 (63) | 267 (58.8) |
| Age, mean (SD), y | 62.1 (16.8) | 62.9 (19.7) | 61.9 (16.0) |
| BMI, mean (SD)b | 26.9 (7.3) | 26.7 (7.2) | 26.9 (7.3) |
| Polymicrobial bacteremiac | 140 (24.9) | 29 (26.9) | 111 (24.4) |
| Immunosuppressed | 284 (50.5) | 51 (47.2) | 233 (51.3) |
| CCI, mean (SD) | 5.37 (2.9) | 5.21 (3.15) | 5.41 (2.9) |
| Comorbid conditionsd | |||
| Cancer | 178 (31.7) | 32 (29.6) | 146 (32.2) |
| Diabetes | 182 (32.4) | 32 (29.6) | 150 (33.0) |
| Coronary artery disease | 109 (19.4) | 24 (22.2) | 85 (18.7) |
| COPD or asthma | 90 (16.0) | 17 (15.7) | 73 (16.1) |
| Liver disease | 94 (16.7) | 16 (14.8) | 78 (17.2) |
| Chronic kidney disease | 145 (25.8) | 31 (28.7) | 114 (25.1) |
| ICU admission | 207 (36.8) | 39 (36.1) | 168 (37.0) |
| Hospital death | 81 (14.4) | 11 (10.2) | 70 (15.4) |
Abbreviations: BMI, body mass index; CCI, Charlson comorbidity index; COPD, chronic obstructive lung disease; ICU, intensive care unit, SD, standard deviation.
aData represent no. (%) of patients unless otherwise specified.
bBMI calculated as weight in kilograms divided by height in meters squared.
cSee Supplementary Figure 1 for further details.
dThe 6 most common comorbid conditions are listed.
Source of Infection
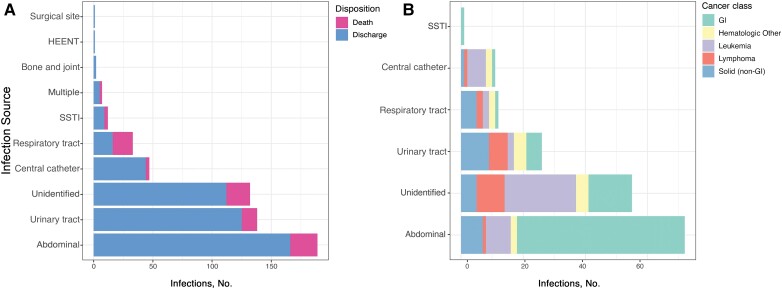
We identified a source in 430 patients (77%) (Table 2). Intra-abdominal infections were most common, followed by urinary tract, central venous catheter, and respiratory tract infections (Figure 1A ). One hundred sixty-one infections (29%) were healthcare associated; in those patients, the median time from admission to positive blood culture was 12 days (interquartile range [IQR], 6–22 days). However, 434 patients (77%) were hospitalized in the preceding year, suggesting high exposure to hospital environments. One hundred eighty-five BSIs (33%) were hardware associated. An unidentified source was most common in patients with hematologic cancers, in whom gut translocation was frequently suspected (Figure 1B ).
Table 2.
Source of Klebsiella pneumoniae Bloodstream Infection
| BSI Type or Outcome | Patients With BSI by Infection Source, No. % | |||||||
|---|---|---|---|---|---|---|---|---|
| Overall (N = 562) | Abdominal (n = 189) | Urinary Tract (n = 138) | Unidentified (n = 132) | Central Catheter (n = 47) | Respiratory Tract (n = 33) | SSTI (n = 12) | Multiple Sources (n = 7) | |
| Hospital acquired | 161 (29) | 39 (21) | 27 (20) | 50 (38) | 21 (45) | 18 (55) | 2 (17) | 3 (43) |
| MDR | 87 (16) | 21 (11) | 28 (20) | 15 (11) | 8 (17) | 10 (30) | 2 (17) | 2 (29) |
| ESBL phenotype | 112 (20) | 29 (15) | 32 (23) | 22 (17) | 10 (21) | 12 (36) | 4 (33) | 2 (29) |
| Hardware | 185 (33) | 86 (46) | 42 (30) | 0 (0) | 47 (100) | 7 (21) | 0 (0) | 3 (43) |
| ICU admission | 207 (37) | 56 (30) | 39 (28) | 54 (41) | 17 (36) | 28 (85) | 7 (58) | 4 (57) |
| Hospital death | 81 (14) | 23 (12) | 13 (9) | 20 (15) | 3 (6) | 17 (52) | 3 (25) | 2 (29) |
Abbreviations: BSI, bloodstream infection; ESBL, extended-spectrum β-lactamase; ICU, intensive care unit; MDR, multidrug resistance; SSTI, skin and soft-tissue infection.
Figure 1.
Outcome stratified by infection sources and distribution of cancer class. A, Number of each primary infection source resulting in bacteremia in the population, delineated by outcome (discharge vs in-hospital death). B, Primary infection sources in patients with cancer according to cancer type; “hematologic other” indicates hematologic cancer other than leukemia. Abbreviations: GI, gastrointestinal; HEENT, head, ears, eyes, nose, and throat; SSTI, skin and soft-tissue infection.
Antibiotic Administration
We determined the time to delivery of the “first antibiotic” and the “first effective antibiotic” after each initial positive culture was obtained, limiting this inquiry to antibiotics with gram-negative activity. The first effective antibiotic was defined as the first antibiotic given after the culture was obtained to which the isolate was susceptible. Seventeen different first antibiotics were used by clinicians; in 8 cases, no antibiotic was given because patients elected for comfort measures only. Cefepime was the most frequently used first antibiotic (47.9%) followed by piperacillin-tazobactam (20.3%) and ceftriaxone (12.2%) (Supplementary Table 2).
The median time to delivery of the first effective antibiotic was 1.9 hours (IQR, 0.48–12.15 hours). In 195 patients, there was a >4-hour delay from when cultures were drawn to when an effective antibiotic was given. In 40 (7%) of the 554 cases with administered antibiotics, the isolate was resistant to the first antibiotic given, and a prolonged time to delivery of the first effective antibiotic was observed (median, 30.8 hours; IQR, 16.9–42.8 hours). Of these 40 patients, only 9 had prior cultures (from any site) growing gram-negative organisms with resistance to the given antibiotic.
Bacterial Characteristics
Antimicrobial Resistance
Supplementary Table 3 shows resistance patterns of the 562 isolates; 112 (20%) were resistant to ceftriaxone, and 89 (16%) were MDR. Cefepime resistance was detected in 43 isolates (8%), and carbapenem resistance in 18 (3%). Healthcare-associated infections were more resistant to all antibiotic classes.
Genomic Diversity
We assembled 108 complete genomes from the prospective collection. Ninety-four of these were identified through WGS as K. pneumoniae, while in 14 instances there was discordance between MALDI-TOF and WGS species typing, with 9 identified as Klebsiella quasipneumoniae and 5 as Klebsiella variicola. This rate of discordant classification has been observed in other WGS studies of K. pneumoniae [23, 24].
The isolates were diverse (Figure 2). Among the 94 confirmed K. pneumoniae isolates, 70 STs were represented. Of the global MDR STs, only ST307 (4 isolates), ST14 (3 isolates) and ST20 (4 isolates) were present in our population [25, 26]. We calculated the ST diversity of the K. pneumoniae isolates using the SDI and compared it to findings of another study [27] including isolates from multiple anatomic sources. Even though our cohort was restricted to bloodstream isolates, the K. pneumoniae BSI isolates from our cohort were more diverse (SDI, 4.14 vs 3.53 for all K. pneumoniae; 4.07 vs 4.08 for carbapenem-sensitive K. pneumoniae). There were 2 pairs of isolates (KPN0005 and KPN0021; KPN0019 and KPN0090), which both had 38 pairwise SNPs between them. However, we found no epidemiologic links between the patients, suggesting that there were no transmission events in this cohort.
Figure 2.
Phylogenetic tree of the 94 Klebsiella pneumoniae bloodstream infection isolates, a midpoint-rooted tree based on core single-nucleotide polymorphism aligned to reference strain NTUH-K2044. Column 1 shows the sequence types (STs) with ≥3 isolates represented; column 2, ceftriaxone resistance pattern; and column 3, the infection source. Branches are colored black or red for community-acquired or hospital-acquired infections, respectively. Abbreviations: AST, antimicrobial susceptibility testing; SSTI, skin and soft-tissue infection.
Genetic Determinants of Resistance
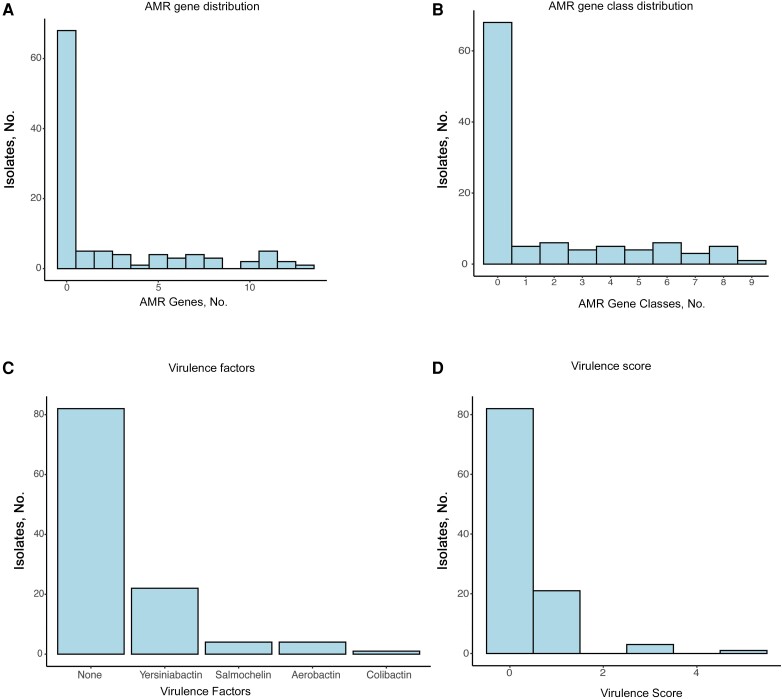
Sixty-eight (63%) of 108 sequenced isolates had no resistance genes other than the chromosomal sulf-hydryl variable common to all K. pneumoniae, and the remainder had 1–13 AMR genes spanning 9 drug classes (Figure 3A and 3B ). Of 108 sequenced isolates, 25 (23%) were resistant to ceftriaxone (Supplementary Table 4). blaCTX-M-15 was present in 16 (64%) of the 25 ceftriaxone-resistant isolates. Genes encoding carbapenemases (blaKPC-2 [2 isolates], blaKPC-3, blaOXA-48, and blaNDM-5) were identified in 5 of 7 carbapenem-resistant isolates, while blaCTX-M-15 and blaTEM-1 genes with concomitant porin mutations were present in the other 2.
Figure 3.
Acquired determinants of antimicrobial resistance (AMR) and virulence factors within the prospective cohort. A, Histogram of the number of AMR genes present across isolates, which demonstrates that most isolates lacked resistance genes. B, Histogram of the number of antibiotic classes present across isolates. C, Histogram of the individual virulence factors across the population, which shows low prevalence of all factors other than yersiniabactin. D, Histogram of virulence scores across the population.
Virulence
We assessed the number of acquired virulence factors and the overall “virulence score” for each isolate. This metric, developed by Lam et al [20], incorporates the number and type of virulence genes present with well-supported clinical risks (Figure 3C and 3D ). One isolate (KPN0084) belonging to ST23, a hypervirulent ST associated with East Asia [28], had the highest possible score of 5 and was hypermucoid. The infected patient was a previously healthy man originally from East Asia admitted with liver abscesses, a presentation consistent with the hypervirulent genotype. Eighty-two isolates (77%) had a virulence score of 0, while 3 isolates (2.8%) had a virulence score of 3 (aerobactin only) and 21 (20%) had a virulence score of 1 (yersiniabactin only) (Figure 3). The 4 isolates with virulence scores ≥3 also carried the rmpADC/rmpA_2 genes associated with hypermucoidity. Three of these isolates (KPN0084, KPN0029, and KPN0035) had a hypermucoid phenotype [7]. We assessed the prevalence of the fim and mrk fimbrial operons within our population, and both were present in >90% of the isolates (Supplementary Table 5).
Serotype
Capsular polysaccharide (K) and lipopolysaccharide (O) antigens are used to characterize K. pneumoniae serologically, and different serotypes demonstrate differential capacity in evading host defenses [29]. We identified the serotypes of the prospective cohort using Kleborate, which are listed in Supplementary Table 6. Among the 94 isolates confirmed as K. pneumoniae, 56 capsule-encoding loci (KL) were identified, and 2 strains had unidentified KL. The most common loci, KL102 and KL2 accounted for 5 (5%) and 6 (6%) of confirmed K. pneumoniae BSIs, respectively. Eight known O types were identified, and 7 isolates had unknown O types based on WGS. The most common O types were O1 and O3b, which accounted for 35 (37%) and 19 (20%) of the isolates, respectively. As shown in Supplementary Table 7, a respiratory tract source was associated with higher odds of O1 infection (OR 5.9 [95% CI: 1.1–31.1]), while a gastrointestinal source was associated with higher odds of O3b infection (3.8 [1.3–10.7]) and lower odds of O1 infection (0.36 [.14–.97]).
Risk Factors for In-Hospital Death
In an unadjusted analysis increasing age, ICU admission at time of culture, higher Charlson comorbidity index, cancer, AMR, and a respiratory tract source were associated with increased mortality rate (Table 3). After adjustment for confounding in the multivariable model, independent predictors of in-hospital death were ICU admission, hematologic cancer (aOR 4.48 [95% CI: 2.13–9.41]), a respiratory tract source (aOR 5.35 [2.23–12.79]), and resistance of the infection strain to the first antibiotic given (aOR5.22 [2.21–12.37]). Resistance to ceftriaxone and higher-order β-lactams was associated with increased odds of death, but this phenotype was tightly correlated with incorrect first antibiotic and was not included in the final multivariable model.
Table 3.
Odds Ratios Obtained With Unadjusted and Adjusted Logistic Regression Assessing the Relationship of Variables to In-Hospital Death
| Variable | Unadjusted Regression Model | Adjusted Regression Model | ||
|---|---|---|---|---|
| OR (95% CI) | P Value | OR (95% CI) | P Value | |
| Age (in years) | 1.0 (1.0–1.0) | .04 | 1.01 (.99–1.04) | .3 |
| Male sex | 0.89 (.55–1.4) | .63 | 0.57 (.33–1.00) | .05 |
| Diabetes | 1.3 (.80–2.2) | .29 | … | … |
| Liver disease | 1.5 (.82–2.7) | .19 | … | … |
| Chronic kidney disease | 1.1 (.6–1.8) | .85 | … | … |
| Cancer | 1.7 (1.0–2.7) | .05 | … | … |
| Solid tumor | 0.89 (.47–1.7) | .72 | 0.91 (.40–2.08) | .83 |
| Hematologic cancer | 2.5 (1.4–4.6) | <.01 | 4.48 (2.13–9.41) | <.001 |
| Immunosuppression | 1.4 (.83–2.2) | .23 | … | … |
| ANC <500/µL | 2.4 (1.2–4.9) | .02 | … | … |
| Charlson comorbidity index | 1.1 (1.0–1.2) | .04 | 1.14 (.99–1.30) | .06 |
| Infection source | ||||
| Abdominal | 0.8 (.47–1.3) | .39 | … | … |
| Urinary tract | 0.58 (.3–1.1) | .09 | … | … |
| Central catheter | 0.42 (.13–1.4) | .15 | … | … |
| Respiratory tract | 8.1 (3.9–16.9) | <.001 | 5.35 (2.23–12.79) | <.001 |
| Unidentified | 0.92 (.52–1.7) | .79 | … | … |
| Antibiotic resistant | ||||
| Ceftriaxone | 2.1 (1.2–3.5) | .01 | … | … |
| Cefepime | 3.8 (1.9–7.7) | <.001 | … | … |
| Piperacillin-tazobactam | 2.9 (1.5–5.5) | .00 | … | … |
| Aminoglycoside | 1.9 (.93–3.9) | .08 | … | … |
| Fluoroquinolone | 1.4 (.76–2.4) | .29 | … | … |
| MDR | 1.7 (.94–3.1) | .08 | … | … |
| Delay of ≥4 h to antibiotic treatment | 1.5 (.9–2.4) | .12 | … | … |
| Incorrect 1st antibiotic | 3.8 (1.9–7.7) | <.001 | 5.22 (2.21–12.37) | <.001 |
| ID consultation | 0.83 (.51–1.3) | .44 | … | … |
| Hospital-acquired infection | 2.8 (1.7–4.5) | <.001 | … | … |
| ICU admission | 4.72 (2.86–7.79) | <.001 | 5.06 (2.82–9.07) | <.001 |
Abbreviations: ANC, absolute neutrophil count; CI, confidence interval; ICU, intensive care unit; ID, infectious diseases; MDR, multidrug resistance; OR, odds ratio.
In the prospective cohort, we evaluated the association between in-hospital death and 4 K. pneumoniae virulence factors (yersiniabactin, colibactin, salmochelin, and aerobactin), as well as the association with a virulence score ≥1, using Fisher's exact test (Supplementary Table 8). Salmochelin was the only factor that reached statistical significance (OR, 10 [95% CI: 1.3–79.8]), but it was present in isolates from only 3 patients, 2 of whom died.
DISCUSSION
K. pneumoniae BSIs were opportunistic in our cohort, with most cases occurring in individuals with hospital exposure, immunocompromised status, and indwelling hardware. Biliary tract infection was the single most common source, a finding that aligns with a population-based survey in Canada from 2000 to 2007 [11]. Patients with hepatobiliary hardware may consequently be ideal candidates for studies of vaccines to prevent invasive K. pneumoniae infections [30].
We found that K. pneumoniae BSI arises from a genetically heterogeneous population of bacteria, with an unexpectedly low prevalence of acquired virulence determinants. This differs from prior molecular epidemiologic studies that focused on K. pneumoniae with specific phenotypes, including carbapenem-resistant or hypervirulent isolates, which found they were dominated by specific lineages [31, 9]. In our large, unselected cohort we found that K. pneumoniae BSI was not driven by a restricted population of bacteria with increased potential for invasion and dissemination, but rather by diverse strains with potential to cause invasive disease in compromised hosts. A 2021 study in the United Kingdom also found that bloodstream isolates were diverse and that canonical virulence genes were observed at low frequency [12]. Similarly, a study that included both BSI and non-BSI clinical K. pneumoniae showed extensive diversity and a low prevalence of acquired virulence genes [23]. Taken together, these studies provide evidence that while a convergence of resistance and increasing virulence may drive the expansion of selected lineages [32], K. pneumoniae BSI in highly resourced hospital settings arises from a heterogenous population of opportunistic pathogens.
The diversity observed in our study suggests either that community acquisition is the dominant driver of K. pneumoniae infection or that there is a large reservoir of K. pneumoniae strains in the healthcare environment. This has implications for the design of epidemiologic studies; mapping transmission events will require deep probing of environmental and human reservoirs to identify risk factors for the transmission of closely related strains, thereby lending support to evolving strategies, such as institutional wastewater sampling [33, 34]. This finding is also consistent with the observation that many K. pneumoniae infections are caused by colonizing strains, as opposed to outbreaks of highly resistant, transmissible, or hypervirulent strains, and it adds support for implementing bundled decolonization protocols in certain high-risk populations [35, 24, 36].
The diversity of the K. pneumoniae strains also extends to their serotype. While the KL2 locus has been associated with invasiveness and represents the most common capsule type, this accounted for a small fraction of our characterized isolates. Our observation that certain O types were associated with distinct anatomic sites of infection was unexpected and not previously reported. If confirmed, these associations suggest that O-polysaccharide–specific biologic properties may affect the anatomic niche and shape the outcome of host-pathogen interactions in invasive K. pneumoniae.
Our study informs the approach to antibiotic therapy when K. pneumoniae BSI is suspected or identified. First, when K. pneumoniae BSI is confirmed, concomitant bacteremia with other enteric pathogens is common. Almost a quarter of K. pneumoniae BSIs involved additional bloodstream organisms. This suggests that broad antibiotic coverage should be maintained until concomitant infection is excluded. In addition, the excess mortality rate in patients given the wrong first antibiotic underscores the need for approaches to rapidly identify patients at risk for resistant organisms [37, 38].
Our study has limitations. First, we only present data on patients with K. pneumoniae BSI and are unable to estimate population-based incidence rates. Second, our study was conducted in a tertiary care hospital, which may not be representative of all medical settings. Third, we had bacterial sequencing data only from the prospective cohort; however, this patient population was well-matched to the retrospective cohort, and we expect it to be representative. Fourth, we used short-read sequencing, which limited our ability to investigate the role of plasmid transmission among isolates [27]. Finally, given the discordance in species typing we identified between MALDI-TOF and WGS, some proportion of the retrospective cohort may be Klebsiella species other than K. pneumoniae.
In conclusion, the current study demonstrates that K. pneumoniae BSI is a life-threatening infection caused by diverse strains. The typical genotypic and phenotypic characteristics associated with hypervirulent K. pneumoniae strains were observed at an unexpectedly low frequency, and the convergence of increasing AMR and virulence factors reported in other studies was not seen in this cohort. Nonetheless, association between AMR, the incorrect first choice of antibiotic, and in-hospital death was profound, pointing to a need to prevent K. pneumoniae BSI and to rapidly identify patients who are at risk of AMR K. pneumoniae infection.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
David J Roach, The Broad Institute of MIT and Harvard, Boston, Massachusetts, USA; Division of Infectious Diseases, Brigham and Women's Hospital, Boston, Massachusetts, USA; Division of Infectious Diseases, Massachusetts General Hospital, Boston, Massachusetts, USA.
Sushmita Sridhar, The Broad Institute of MIT and Harvard, Boston, Massachusetts, USA; Division of Infectious Diseases, Massachusetts General Hospital, Boston, Massachusetts, USA.
Elizabeth Oliver, Division of Infectious Diseases, Massachusetts General Hospital, Boston, Massachusetts, USA.
Sowmya R Rao, Department of Global Health, Boston University, Boston, Massachusetts, USA.
Damien M Slater, Division of Infectious Diseases, Massachusetts General Hospital, Boston, Massachusetts, USA.
Wontae Hwang, Division of Infectious Diseases, Massachusetts General Hospital, Boston, Massachusetts, USA.
Kian Hutt Vater, Division of Infectious Diseases, Massachusetts General Hospital, Boston, Massachusetts, USA.
Anupama Dinesh, Division of Infectious Diseases, Massachusetts General Hospital, Boston, Massachusetts, USA.
Firdausi Qadri, Dhaka Hospital, International Centre for Diarrheal Disease Research, Bangladesh.
Mohammod J Chisti, Dhaka Hospital, International Centre for Diarrheal Disease Research, Bangladesh.
Virginia M Pierce, Department of Pathology, University of Michigan Medical School, Ann Arbor, Michigan, USA.
Sarah E Turbett, Division of Infectious Diseases, Massachusetts General Hospital, Boston, Massachusetts, USA; Department of Pathology, Massachusetts General Hospital, Boston, Massachusetts, USA.
Roby P Bhattacharyya, The Broad Institute of MIT and Harvard, Boston, Massachusetts, USA; Division of Infectious Diseases, Massachusetts General Hospital, Boston, Massachusetts, USA.
Colin J Worby, The Broad Institute of MIT and Harvard, Boston, Massachusetts, USA.
Ashlee M Earl, The Broad Institute of MIT and Harvard, Boston, Massachusetts, USA.
Regina C LaRocque, Division of Infectious Diseases, Massachusetts General Hospital, Boston, Massachusetts, USA.
Jason B Harris, Division of Infectious Diseases, Massachusetts General Hospital, Boston, Massachusetts, USA; Department of Pediatrics, Harvard Medical School, Boston, Massachusetts, USA.
Notes
Acknowledgments. The authors thank the Massachusetts General Hospital Clinical Microbiology Laboratory for sample collection.
Financial support. This work was supported by the National Institutes of Health (training grant T32AI007061-44 to D. J. R.), Massachusetts General Hospital (Center for Global Health Research Development Award to S. S. and institutional support to J. B. H.), and the National Institute of Allergy and Infectious Diseases (grant U19AI110818 to C. J. W. and A. M. E.).
Data sharing. Whole-genome sequencing data can be found at the Sequence Read Archive (BioProject PRJNA978102).
References
- 1. Magill SS, O’Leary E, Janelle SJ, et al. Changes in prevalence of health care–associated infections in U.S. hospitals. N Engl J Med 2018; 379:1732–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sands K, Carvalho MJ, Portal E, et al. Characterization of antimicrobial-resistant gram-negative bacteria that cause neonatal sepsis in seven low- and middle-income countries. Nat Microbiol 2021; 6:512–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wyres KL, Lam MMC, Holt KE. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol 2020; 18:344–59. [DOI] [PubMed] [Google Scholar]
- 4. Holt KE, Wertheim H, Zadoks RN, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci 2015; 112:E3574–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev 2017; 41:252–75. [DOI] [PubMed] [Google Scholar]
- 6. Murray CJ, Ikuta KS, Sharara F, et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 2022; 399:629–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Short FL, Di Sario G, Reichmann NT, Kleanthous C, Parkhill J, Taylor PW. Genomic profiling reveals distinct routes to complement resistance in Klebsiella pneumoniae. Infect Immun 2020; 88:e00043-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev 2019; 32:e00001-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shankar C, Jacob JJ, Vasudevan K, et al. Emergence of multidrug resistant hypervirulent ST23 Klebsiella pneumoniae: multidrug resistant plasmid acquisition drives evolution. Front Cell Infect Microbiol 2020; 10:575289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lin ZW, Zheng JX, Bai B, et al. Characteristics of hypervirulent Klebsiella pneumoniae: does low expression of rmpA contribute to the absence of hypervirulence? Front Microbiol 2020; 11:436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meatherall BL, Gregson D, Ross T, Pitout JDD, Laupland KB. Incidence, risk factors, and outcomes of Klebsiella pneumoniae bacteremia. Am J Med 2009; 122:866–73. [DOI] [PubMed] [Google Scholar]
- 12. Lipworth S, Vihta KD, Chau K, et al. Ten-year longitudinal molecular epidemiology study of Escherichia coli and Klebsiella species bloodstream infections in Oxfordshire, UK. Genome Med 2021; 13:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Massachusetts General Hospital. General internal medicine treatments & services. Available at: https://www.massgeneral.org/medicine/internal-medicine/treatments-and-services. Accessed 4 August 2023.
- 14. 2021 National Health Safety Network patient safety component Manual. Published online 2021:428.
- 15. Salipante SJ, Roach DJ, Kitzman JO, et al. Large-scale genomic sequencing of extraintestinal pathogenic Escherichia coli strains. Genome Res 2015; 25:119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu KM, Li LH, Yan JJ, et al. Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J Bacteriol 2009; 191:4492–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009; 25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Walker BJ, Abeel T, Shea T, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 2014; 9:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014; 30:1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun 2021; 12:4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sayers EW, Bolton EE, Brister JR, et al. Database resources of the national center for biotechnology information. Nucleic Acids Res 2021; 50(D1):D20–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. R: The R Project for Statistical Computing. Available at: https://www.r-project.org/. Accessed 19 September 2022.
- 23. Gorrie CL, Mirčeta M, Wick RR, et al. Genomic dissection of Klebsiella pneumoniae infections in hospital patients reveals insights into an opportunistic pathogen. Nat Commun 2022; 13:3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gorrie CL, Mirčeta M, Wick RR, et al. Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin Infect Dis 2017; 65:208–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wyres KL, Holt KE. Klebsiella pneumoniae population genomics and antimicrobial-resistant clones. Trends Microbiol 2016; 24:944–56. [DOI] [PubMed] [Google Scholar]
- 26. Wyres KL, Hawkey J, Hetland MAK, et al. Emergence and rapid global dissemination of CTX-M-15-associated Klebsiella pneumoniae strain ST307. J Antimicrob Chemother 2019; 74:577–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Salamzade R, Manson AL, Walker BJ, et al. Inter-species geographic signatures for tracing horizontal gene transfer and long-term persistence of carbapenem resistance. Genome Med 2022; 14:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lam MMC, Wyres KL, Duchêne S, et al. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat Commun 2018; 9:2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gonzalez-Ferrer S, Peñaloza HF, Budnick JA, et al. Finding order in the chaos: outstanding questions in Klebsiella pneumoniae pathogenesis. Infect Immun 2021; 89:e00693-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Assoni L, Girardello R, Converso TR, Darrieux M. Current stage in the development of Klebsiella pneumoniae vaccines. Infect Dis Ther 2021; 10:2157–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bowers JR, Kitchel B, Driebe EM, et al. Genomic analysis of the emergence and rapid global dissemination of the clonal group 258 Klebsiella pneumoniae pandemic. PLoS One 2015; 10:e0133727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li P, Liang Q, Liu W, et al. Convergence of carbapenem resistance and hypervirulence in a highly-transmissible ST11 clone of K. pneumoniae: an epidemiological, genomic and functional study. Virulence 2021; 12:377–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Munk P, Brinch C, Møller FD, et al. Genomic analysis of sewage from 101 countries reveals global landscape of antimicrobial resistance. Nat Commun 2022; 13:7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chau KK, Barker L, Budgell EP, et al. Systematic review of wastewater surveillance of antimicrobial resistance in human populations. Environ Int 2022; 162:107171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Willems RPJ, van Dijk K, Vehreschild MJGT, et al. Incidence of infection with multidrug-resistant gram-negative bacteria and vancomycin-resistant enterococci in carriers: a systematic review and meta-regression analysis. Lancet Infect Dis 2023; 23:719–31. [DOI] [PubMed] [Google Scholar]
- 36. Hayden MK, Lin MY, Lolans K, et al. Prevention of colonization and infection by Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae in long-term acute-care hospitals. Clin Infect Dis 2015; 60:1153–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fowler VG, Jezek A, Spivak ES, Talkington K; Infectious Diseases Society of America and The Pew Charitable Trusts . Urgent, comprehensive federal action needed to stem mortality and Medicare costs associated with antimicrobial resistance. Clin Infect Dis 2022; 74:1107–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mo Y. Rapid diagnostics for antibiotic resistance: urgent need for strong clinical evidence. Clin Infect Dis 2022; 75:2076–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.