Abstract
Microglia are the resident immune cells in the brain that play a key role in driving neuroinflammation, a hallmark of neurodegenerative disorders. Inducible microglia-like cells have been developed as an in vitro platform for molecular and therapeutic hypothesis generation and testing. However, there has been no systematic assessment of similarity of these cells to primary human microglia along with their responsiveness to external cues expected of primary cells in the brain. In this study, we performed transcriptional characterization of commercially available human inducible pluripotent stem cell (iPSC)-derived microglia-like (iMGL) cells by bulk and single cell RNA sequencing to assess their similarity with primary human microglia. To evaluate their stimulation responsiveness, iMGL cells were treated with Liver X Receptor (LXR) pathway agonists and their transcriptional responses characterized by bulk and single cell RNA sequencing. Bulk transcriptome analyses demonstrate that iMGL cells have a similar overall expression profile to freshly isolated human primary microglia and express many key microglial transcription factors and functional and disease-associated genes. Notably, at the single-cell level, iMGL cells exhibit distinct transcriptional subpopulations, representing both homeostatic and activated states present in normal and diseased primary microglia. Treatment of iMGL cells with LXR pathway agonists induces robust transcriptional changes in lipid metabolism and cell cycle at the bulk level. At the single cell level, we observe heterogeneity in responses between cell subpopulations in homeostatic and activated states and deconvolute bulk expression changes into their corresponding single cell states. In summary, our results demonstrate that iMGL cells exhibit a complex transcriptional profile and responsiveness, reminiscent of in vivo microglia, and thus represent a promising model system for therapeutic development in neurodegeneration.
Subject terms: Neuroimmunology, Genomics, Neurodegeneration, Microglia
Introduction
Microglia are the primary resident immune cells in the brain. Derived from a myeloid lineage, microglia fulfill critical roles in immune surveillance and phagocytosis of cells and debris caused by injury, disease, and aging1. Additionally, microglia play important roles in neuronal homeostasis by regulating synaptogenesis and synaptic pruning2. Dysregulation of microglial functions are heavily implicated in neurodegenerative disorders including Alzheimer’s disease (AD)3,4 and Parkinson’s disease (PD)5. Thus, studying the role of microglia in neurodegenerative disorders is essential for developing effective therapies.
Currently, it is untenable to purify sufficient amounts of ex vivo human microglia from brain tissue to perform multifaceted experiments. Additionally, microglia isolated from brain tissue undergo rapid transcriptomic and phenotypic changes when transferred to in vitro conditions6. These challenges have facilitated the development of protocols to differentiate iPSCs into iMGL cells7–12. These protocols transition iPSCs through hematopoietic precursor cells (HPCs), erythro-myeloid progenitors (EMPs), and finally into iMGL cells in culture13. Alternatively, HPCs can be transplanted into early postnatal mouse brain where they develop into microglia14–16. These xenotransplant microglia (xMG) more closely resemble the transcriptomic profile of ex vivo microglia than iMGL cells, however they are difficult to produce at large scale.
The goal of this study is to assess commercially available iMGL cells, from Fujifilm Cellular Dynamics, Inc. (Commercial iMGL cells), as a platform for functional studies and target discovery in neurodegeneration. To characterize transcriptional and cellular heterogeneity of Commercial iMGL cells in comparison to primary human microglia, we performed single-cell and bulk RNA sequencing of iMGLs cells and compared results with a number of publicly available transcriptomic data. Then, to assess transcriptional responsiveness of different iMGL cell subpopulations to known neuroprotective agents, we treated Commercial iMGL cells with two LXR pathway agonists17–19.
Methods
Cell culture
Cell culture was performed as described previously12 with modifications as described below. iPSC-derived microglia were obtained from FujiFilm Cellular Dynamics, Inc. (clone01279.107, lot 105093 and lot 105887). Cells were thawed at 37 °C. Each vial was rinsed with 1 mL of culture medium (Table 1) and spun at 1000×g for 10 min. Cells were transferred to 8 mL of culture medium and plated at a density of 5 × 104 cells/cm2 in a plate format based on the cell input requirements for a given experiment. Specifically, 6-well format for single-cell and 48-well format for bulk RNA-seq experiments. Cells were grown between 0 and 4 days in culture based on the recommended 3 day recovery period from cryopreservation (FujiFilm Cellular Dynamics, Inc.).
Table 1.
Composition of culture medium.
| Culture media component | Concentration | Volume (mL) per 100 mL of media | Vendor |
|---|---|---|---|
| Dulbecco’s Modified Eagle Medium (DMEM)/F-12 HEPES, no phenol red | 93.3 | Thermo Fisher Scientific | |
| N-2 Supplement | 100× | 0.5 | Thermo Fisher Scientific |
| B-27™ Supplement | 50× | 1 | Thermo Fisher Scientific |
| 10% BSA in DPBS | 0.5 | MilliporeSigma | |
| 1-Thioglycerol | 11.5 M | 0.004 | MilliporeSigma |
| Ascorbic acid | 20 mg/mL | 0.25 | MilliporeSigma |
| Penicillin–streptomycin | 1 | Thermo Fisher Scientific | |
| GlutaMAX® Supplement | 1 | Thermo Fisher Scientific | |
| MEM Non-essential Amino Acids | 100× | 1 | Thermo Fisher Scientific |
| Insulin-Transferrin-Selenium | 100× | 1 | Thermo Fisher Scientific |
| Human Insulin Solution | 0.05 | MilliporeSigma | |
| Recombinant Human M-CSF Protein (rhM-CSF) | 100 μg/mL | 0.025 | PeproTech |
| Recombinant Human TGF beta 1 protein (rhTGFb1) | 100 μg/mL | 0.05 | R&D Systems |
| Recombinant Human IL-34 (rhIL-34) | 100 μg/mL | 0.1 | PeproTech |
| Recombinant human Fractalkine (rhFractalkine) | 100 μg/mL | 0.1 | PeproTech |
| Recombinant human CD200 (rhCD200) | 100 μg/mL | 0.1 | Acro Biosystems |
Immunostaining
Cells were plated on a black glass bottom 96-well plate at a density of ~ 30,000 cells/cm2. On Day 8, cells were washed with 1× phosphate buffered solution (PBS) and fixed with 4% paraformaldehyde in PBS for 15 min at room temperature. Permeabilization of the cells was done with 1× PBS containing 0.25% Triton™X-100. Following 1 h blocking in 1× PBS with 5% goat serum and donkey serum at room temperature, primary rabbit anti-IBA1 (Abcam plc, Cat. No. ab178846; 1:1000), rabbit anti-TREM2 (Cell Signaling, Cat. No. D8I4C; 1:200), and rabbit anti-P2Y12 (Thermo Fisher Scientific Inc, Cat. No. 4H5L19; 1:100) antibodies were added in the blocking solution and incubated at 4 °C overnight. The next day, cells were washed 3 times with PBS for 5 min and stained with donkey anti-rabbit Alexa Fluor® conjugated secondary antibody (Thermo Fisher Scientific Inc., Cat. No. A-21206) at 1:500 for 1 h at room temperature in the dark. After secondary antibody staining, cells were washed 3 times with PBS and imaged on a Zeiss® LSM 710 microscope. Images were segmented and quantified using CellProfiler™.
RNA isolation, cDNA synthesis, and bulk RNA sequencing library preparation
For bulk RNA sequencing experiments, medium was removed and cells were lysed directly on plate in RNA lysis buffer and processed using the MagMAX® mirVana Total RNA Isolation Kit (Thermo Fisher Scientific Inc., Cat. No. A27828) according to the manufacturer’s instructions. cDNA was generated from total RNA using SuperScript® IV Reverse Transcriptase (Thermo Fisher Scientific Inc., Cat No. 18090050). Bulk RNA-seq library preparation was performed with 500 ng of total RNA using the NEBNext® Poly(A) mRNA Magnetic Isolation Module (New England Biolabs, Inc., Cat. No. E7490) and NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina® (New England Biolabs, Inc., Cat. No. E7420). All libraries were dual-indexed using 12 cycles of PCR amplification using NEBNext® Multiplex Oligos for Illumina® Dual Index Primer Set (New England Biolabs, Inc., Cat. No. E7600). Library quality control was performed by measuring concentration with Qubit® dsDNA HS Assay Kit (Thermo Fisher Scientific Inc., Cat. No. Q32854) and fragment size distribution with the Agilent® High Sensitivity DNA Kit (Agilent Technologies, Inc., Cat. No. 5067-4626). Libraries were sequenced as paired-end 150 × 150 bp on a NovaSeq® 6000 System (Illumina, Inc.).
Single cell RNA-sequencing library preparation
Single cell experiments were performed in parallel with the bulk RNA-seq experiments described above. Cells were first washed with PBS, then trypsinized for 10 min at 37 °C using TrypLE®Express Enzyme (Thermo Fisher Scientific Inc., Cat. No. 12604013). Trypsinization was halted by the addition of equal volume of warm medium, cells were spun at 1000×g for 10 min and resuspended in 0.04% PBS/bovine serum albumin (BSA). Trypan blue was used to assess cell number and viability (> 85% in all cases, typically > 90%) using a Cellometer® automated cell counter (Nexcelcom Bioscience, LLC). Gel bead-in-emulsion (GEM) encapsulation and single cell indexing reactions were performed using a Chromium™ Controller instrument (10× Genomics, Inc., Cat. No. 1000202, version 3.1 chemistry). Single-cell 3’ RNA-seq libraries were prepared using the Chromium™ Next GEM Single Cell 3’ GEM, Library and Gel Bead Kit (10× Genomics, Inc., Cat. No. 1000121), according to the manufacturer’s instructions, specifically targeting 2–3000 cells per replicate and 13 rounds of PCR amplification. Technical replicates were processed independently from trypsinization through sequencing (cells thawed together then plated in independent wells). Libraries were sequenced on a NovaSeq® 6000 System, with paired end 150 × 150 bp sequencing.
LXR agonist treatment
Cells were plated at 150,000 cells per well on 24-well plates in culture medium. After 24 h at 37 °C, medium was replaced with fresh medium. At day 3, cells were treated with either dimethyl sulfoxide (DMSO) or different doses (30 nM or 100 nM) of T0901317 (MedChemExpress LLC, Cat. No. HY-10626) and GW3965 (30 nM or 300 nM) (MedChem Express LLC, Cat. No. HY-10627A) and incubated for 24 h. After 24 h, medium was discarded, and cells were directly lysed on the plate for RNA extraction using the MagMax® mirVana Total RNA Isolation Kit. Bulk and single cell RNA sequencing were performed as described above.
Time course bulk RNA-seq processing
FastQ files were downloaded from 5 external datasets of microglia related samples6,14,15,20,21 (Accession numbers—Gosselin: dbGaP phs001373.v2.p2, Olah: Synapse syn11468526, Galatro: GEO GSE99074, Hasselmann: GEO GSE133432, Svoboda: GEO GSE139194). Samples were removed with less than 10,000,000 reads. FastQ files were processed through a uniform RNA-sequencing pipeline. Briefly, reads were aligned to the hg38 genome with STAR22 and RSEM was used23 to quantify the expression of genes in transcripts per million (TPM) using Gencode annotation version 2424. Samples were removed with less than 60% alignment rate after mapping. For external datasets downloaded from literature sources, paired-end samples were removed with greater than 50% duplicate read fraction after mapping. Ribosomal and mitochondrial genes were removed, those that start with “RP” or “MT”. Gene expression values were transformed into log2 (TPM + 0.01) to stabilize the variance. This dataset was used for the assessment of progenitor and monocyte markers. For principal components analyses (PCA) and the assessment of key transcription factors (TFs), marker genes, and disease genes, lowly expressed genes with a median TPM < 1 were removed, leaving a dataset of 12,392 genes. ComBat25 was used to correct for batch effects using the dataset source as the correction factor.
Principal components analysis
The set of all expressed genes was used as features to run PCA. Before running PCA, the expression levels of each gene were scaled across samples by subtracting the mean value and dividing by the standard deviation. PCA was run using the prcomp function in R. For the quantitative comparison in Fig. 1D, Euclidean distances were calculated between all pairs of samples using their loadings on PC1 and PC2 with the pdist function in R.
Figure 1.
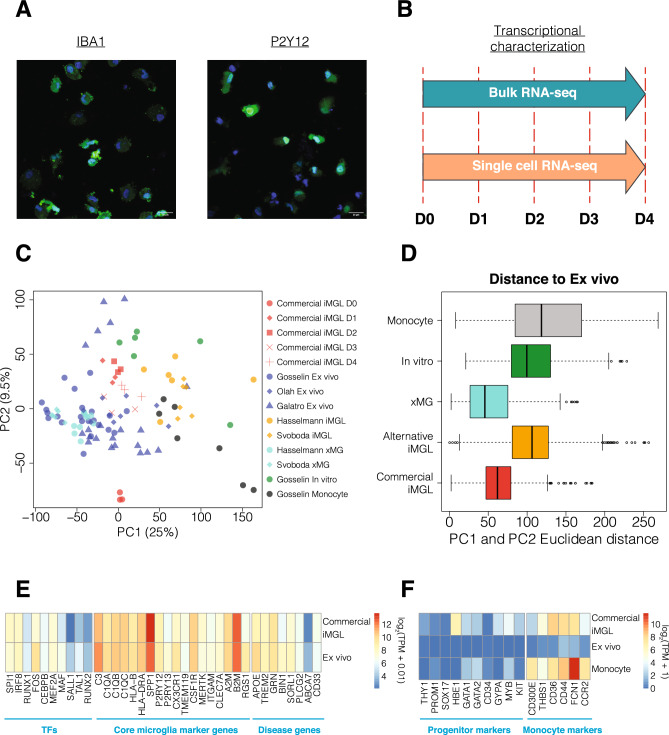
Gene expression comparison between Commercial iMGL cells and microglia-related datasets. (A) Representative immunofluorescence images of Commercial iMGL cells stained with IBA1 or P2Y12 (green) and DAPI (blue). The scale bar represents 20 μm. (B) This study performed bulk and single cell transcriptional profiling of Commercial iMGL cells between 0 and 4 days in culture. (C) PCA clustering using top 2 PCs for Commercial iMGL cell samples and microglia comparator datasets. Variance explained by each principal component is shown in parenthesis. (D) Euclidean distances for all pairs of samples between ex vivo microglia and each of the other microglia groups. Euclidean distances were calculated using sample loadings on PC1 and PC2. (E) Comparison of gene expression levels for key microglia TFs, core marker genes, and disease risk genes between Commercial iMGL cells at Days 1–4 and ex vivo microglia. Expression levels displayed are the median values across samples. (F) Comparison of gene expression levels for HPC and EMP progenitor and monocyte markers between Commercial iMGL cells at Days 1–4, ex vivo microglia, and monocytes. Expression levels displayed are the median values across samples.
Statistical comparison of cell sources using PCA
For the statistical analysis in Supplementary Fig. 3, PCA was run separately across all pairs of cell sources. For each independent pair, meaningful principal components (PCs) were identified which explained a greater proportion of variance than random noise. One hundred permutated datasets were created by randomly shuffling sample labels for each gene and the variance explained by the top PC in each permutated dataset was calculated. A threshold for variance explained by meaningful PCs was defined as 2 standard deviations above the mean value for the top component across the 100 permuted datasets. Sample loadings on the meaningful PCs were used to quantitatively access separability for the pair of datasets using 2 methods: SigClust26 and Silhouette27. The SigClust score was calculated using the sigclust package in R. The Silhouette score was calculated using the cluster package in R. One hundred permuted datasets were generated by shuffling the sample labels for the meaningful PCs and re-running the 2 methods. For each method, an empirical p-value was calculated to assess the significance of dataset separability by identifying the proportion of permutated datasets with a larger separability score then the actual score. In the case of Sigclust, this was the proportion of permutated datasets with a lower score, because lower SigClust scores correspond to greater separability. For Silhouette, this was the proportion of permutated datasets with a higher score, because higher Silhouette scores correspond to greater separability.
Time course single cell RNA-seq processing
FastQ files for each single cell RNA-seq sample were processed using Cell Ranger (10× Genomics, Inc.) to generate count matrices of genes per cell. Count matrices were processed for downstream analyses using Seurat version 3.228. Low quality cells were removed, potentially empty droplets or doublets by imposing the following filtering criteria for each cell: number of genes detected > 1500, number of reads per cell < 100,000, and mitochondrial RNA (mtRNA) fraction < 20%. We also filtered any genes that were detected in less than 3 individual cells. This left us with a dataset of 18,426 genes across 31,984 cells collectively. Gene expression values were log-transformed using the NormalizeData function and the top 2000 variable genes were identified using the FindVariableFeatures function. Expression levels for each gene were scaled and centered using the ScaleData function. Louvain clustering was performed with the top 20 principal components using the FindNeighbors and FindClusters functions with a resolution parameter of 0.4. For visualization, the variance across the top 20 principal components was reduced into 2 Uniform Manifold Approximation Projection (UMAP) dimensions using the RunUMAP function. For cells in each cluster, preferentially expressed marker genes were identified using the FindAllMarkers function with the following criteria: adjusted Wilcox p-value < 0.01, detected percentage > 40%, and log2(fold-change) > 0.6. GO enrichment analyses was performed for the marker gene sets using the clusterProfiler package in R29.
Single cell RNA-seq integration analysis
Single cell RNA-seq integration was performed on the time course dataset with two ex vivo microglia datasets30,31 using Seurat version 4.332. The cell by gene count matrices and cell cluster identities were downloaded for each dataset (Olah: https://github.com/vilasmenon/Microglia_Olah_et_al_2020, Sankowski: https://github.com/rsankowski/sankowski-et-al-microglia) and loaded into separate Seurat objects. Prior to integration, for each dataset, gene expression values were normalized, and the top 2000 variable genes were identified as described above. Data integration was performed using the functions SelectIntegrationFeatures and FindIntegrationAnchors. For visualization, the variance across the top 30 principal components was reduced into 2 UMAP dimensions as described above.
LXR agonist treatment bulk RNA-seq processing
FastQ files were processed as described above. Differential gene expression analysis was run using the DESeq2 package in R33. The DESeq function was used to perform median of ratios normalization of the count data. Four contrasts were run: GW3965 30 nM vs DMSO, GW3965 300 nM vs DMSO, T0901317 30 nM vs DMSO, and T0901317 100 nM vs DMSO. To identify DEGs, the following criteria were used: absolute value of log2 fold change > 0.6, adjusted p-value < 0.05, and the maximum TPM value across samples > 1. The enrichR package in R34 was used to run pathway enrichment analysis. For each contrast, both up- and downregulated DEGs were mixed and run against public databases including KEGG_2019_Human, GO_Biological_Process_2018 and Reactome_2016. The significance of the enrichment and overlapping genes were generated by the enrichr function for each pathway. Significantly enriched pathways were identified with an adjusted p-value < 0.05.
LXR agonist treatment single cell RNA-seq processing
FastQ files were processed as described above. After filtering of low quality cells, we started with a dataset of 19,517 genes in 43,312 cells, collectively. Data normalization and clustering were performed as described above, expect we used a resolution parameter of 0.2 for Louvain clustering. For each of the 4 contrasts, DEGs were identified in each cluster using the FindAllMarkers function with the following criteria: absolute value of log2 fold change > 0.6, adjusted Wilcox p-value < 0.05, and percentage of detected cells > 10%. Pathway enrichments were identified as described above.
Results
Comparison of Commercial iMGL cell bulk transcriptome data with publicly available primary microglia-related datasets
We verified expression of microglia markers IBA1, P2Y12, and TREM2 on Commercial iMGL cells by immunostaining (Fig. 1A and Supplementary Fig. 1) and quantified > 99% of cells as positive for each marker (“Methods”). Bulk RNA sequencing was performed on cells grown between 0 and 4 days in culture (Fig. 1B and “Methods”). As comparator datasets, we collected published bulk RNA sequencing data from five studies6,14,15,20,21, encompassing three human primary microglia datasets (ex vivo), two alternative iMGL cell datasets, two xMG datasets, one in-vitro cultured human primary microglia dataset, and one human ex vivo monocyte dataset (Supplementary Table 1). For all RNA sequencing samples, we started with FastQ files, processed them using a uniform pipeline, and performed batch correction to minimize sources of technical variation between datasets (“Methods” and Supplementary Fig. 2). We noticed that Commercial iMGL samples from day 3 have higher duplicate read ratios than samples from the other days (Supplementary Table 1), indicative of technical differences in these samples during library preparation.
To assess genome-wide patterns in gene expression, we conducted principal components analysis (PCA) with all expressed genes. We found that xMG and Commercial iMGL cell samples are more closely related to ex vivo microglia than alternative iMGL cells, in vitro cultured microglia, and monocytes by visual (Fig. 1C) and quantitative (Fig. 1D) comparisons using the top two principal components (PCs). We also examined two different similarity metrics, SigClust and Silhouette index, with all PCs above noise, which has been determined by permutation tests (“Methods”). The overall similarity was observed in both metrics used (Supplementary Fig. 3). These results demonstrate that the overall expression profile is similar between ex vivo microglia and Commercial iMGL cells.
Next, we compared the expression of key microglia TFs35, core marker genes associated with microglia functions36, and AD risk genes37 between the datasets. We found the expression of most TFs, marker genes, and disease genes to be similar between ex vivo microglia and Commercial iMGL cells (Fig. 1E and Supplementary Fig. 4). However, there are differences including lower expression of SALL1 and higher expression of SPP1 in Commercial iMGL cells. Loss of SALL1 expression and increase in SPP1 expression have been associated with an increase in inflammation and phagocytic activity in mouse microglia38,39, suggesting that Commercial iMGL cells might have an elevated level of microglial activation. We also assessed the expression of iPSC, HPC, and EMP marker genes as well as monocyte enriched marker genes21,40 (Fig. 1F and Supplementary Fig. 4). We found moderate expression of monocyte markers as well as multiple progenitor markers including HBE1, GATA1, GATA2, GYPA, MYB, and KIT, suggesting the presence of a progenitor subpopulation within the Commercial iMGL cell culture. Taken together, these results demonstrate that Commercial iMGL cells largely recapitulate the basal transcriptional profile of ex vivo microglia.
Single cell transcriptomics identifies multiple subpopulations of cells within Commercial iMGL cells
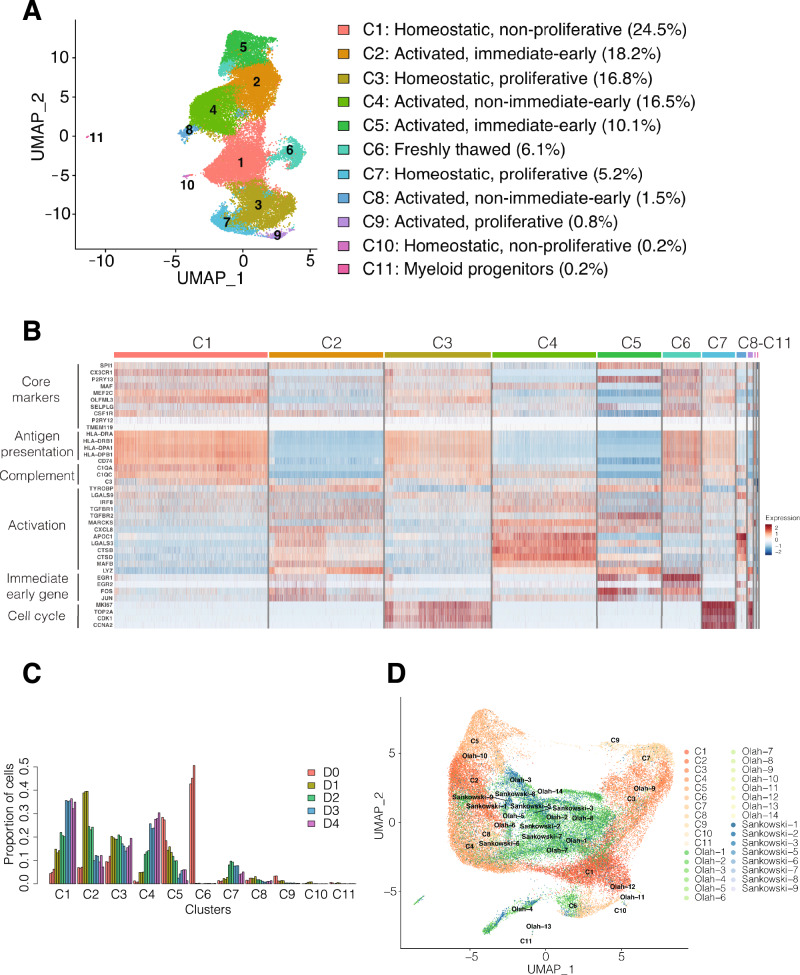
Microglia exhibit a variety of transcriptional states in response to the local environment31,35. These states are indicative of individual microglia performing different roles including surveillance and neuronal homeostasis (homeostatic microglia) as well as inflammatory response and phagocytosis (activated microglia). To characterize the heterogeneity of Commercial iMGL cells in culture, we performed single-cell RNA sequencing on cells grown between 0 and 4 days in culture (Fig. 1B and “Methods”). After removing low quality cells (Supplementary Fig. 5), we generated a dataset of 31,984 cells. Using de novo clustering, we identified 11 clusters of cells within Commercial iMGL cell cultures (Fig. 2A and Supplementary Data 1). To assign functional states for each cluster, we assessed the expression patterns of genes, from a single cell study of human primary microglia, that mark important aspects of microglial biology including antigen presentation, complement pathway, immune activation, and cell cycle31 (Fig. 2B and Supplementary Fig. 6). Additionally, we generated gene ontology enrichments of differentially expressed cluster marker genes (“Methods”, Supplementary Table 2, and Supplementary Figs. 7, 8). We identified 4 clusters, C1, C3, C7, and C10, indicative of homeostatic microglia with high expression of genes involved in antigen presentation and complement pathways. We identified 5 clusters, C2, C4, C5, C8, and C9, indicative of activated microglia with high expression of immune related genes. Among activated microglia, 2 clusters, C2 and C5, have high expression of immediate-early genes which are rapidly induced upon stimulation41. Among homeostatic and activated microglia, we identified 3 clusters of proliferating cells, C3, C7, and C9 with high expression of cell cycle genes. In concordance with bulk expression patterns (Fig. 1F), we identified 1 cluster, C11, corresponding to myeloid progenitors (Supplementary Fig. 9). Reassuringly, the proportion of total cells within C11 is less than one percent (Fig. 2C), demonstrating the low prevalence of incompletely differentiated cells. Of note, AD risk genes were differentially expressed across clusters (Supplementary Fig. 10), supporting the roles of multiple microglia functions and pathways in the etiology of AD42.
Figure 2.
Single cell RNA-seq profiling of Commercial iMGL cell cultures. (A) Identification of 11 cellular clusters in Commercial iMGL cell cultures. Functional annotations were assigned based on the expression of microglia functional marker genes as described in the “Methods” and shown in (B). (B) Gene expression heatmap of selected microglia functional marker genes in the time course single cell RNA-seq dataset. Heatmap with a more comprehensive list of marker genes is shown in Supplementary Fig. 6. (C) Proportion of cells in each cluster in the time course single cell RNA-seq dataset. (D) Integration of Commercial iMGL single cell RNA-seq dataset with two published ex vivo microglia single cell RNA-seq datasets (Olah and Sankowski).
We directly compared the single cell transcriptomes of Commercial iMGL cells with two ex vivo microglia single cell RNA-seq datasets30,31 (“Methods”). The two ex vivo datasets integrate tightly together, with the Commercial iMGL cells located around the periphery (Fig. 2D). On the UMAP projection, functionally similar ex vivo and Commercial iMGL cell clusters have some intermixing (Fig. 2D and Supplementary Fig. 11). Activated Commercial iMGL clusters C2 and C8 are overlapping with activated ex vivo clusters Olah-5, Olah-6, Sankowski-1, and Sankowski-9. Proliferative Commercial iMGL clusters C3 and C7 are overlapping with proliferative ex vivo cluster Olah-9. These results demonstrate that Commercial iMGL cell clusters recapitulate the diverse cellular states of ex vivo microglia. The Commercial iMGL cell cluster transcriptional profiles closely resemble, but are not equivalent to, those of ex vivo microglia.
The proportion of cells within each cluster was dynamic over the 4-day time course (Fig. 2C). One cluster, C6, with high expression of homeostatic and activated marker genes (Fig. 2B), was only present at D0 and indicative of freshly thawed cells. The proportion of homeostatic microglia in cluster 1, those not expressing proliferating markers, increased over time in culture from 15% of cells at day 1 to 35% of cells at day 4. In contrast, the proportion of proliferative homeostatic microglia in cluster C3 was relatively stable between 1 and 4 days in culture. The proportion of activated microglia in cluster C4, those not expressing immediate-early genes, increased over time in culture from 5% at day 1 to 30% at day 4, while the proportion of activated microglia expressing immediate-early genes in cluster C2 decreased over time in culture from 40% at day 1 to 10% at day 4. Overall, the proportion of microglia subpopulations changes substantially over time in culture, approaching steady state between days 3 and 4.
Robust bulk transcriptional response of Commercial iMGL cells to LXR pathway agonists
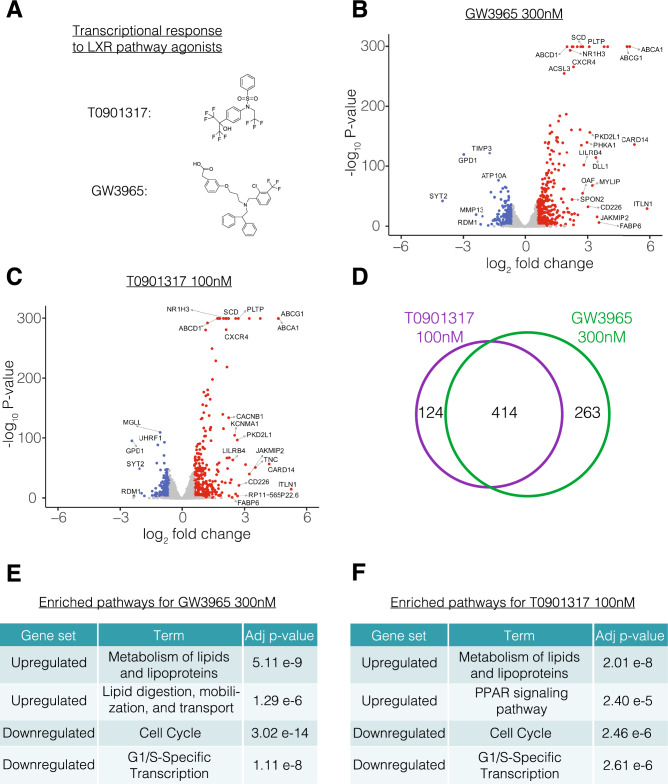
To assess the transcriptional responsiveness of Commercial iMGL cells, we selected two structurally distinct LXR pathway agonists: T0901317 and GW3965 (Fig. 3A)43,44. LXRs are lipid responsive TFs that form heterodimers with retinoid X receptors (RXRs) to regulate expression of key cholesterol homeostatic genes involved in AD45. In microglia, LXR agonists repress microglial activation by inhibiting nitric oxide production and nuclear factor-kappa B activity46,47. LXR agonists are potential therapeutic targets for neurodegenerative disorders and have previously been shown to reduce amyloid plaque burden, ameliorate neuroinflammation, and improve memory in preclinical AD models17–19.
Figure 3.
Bulk transcriptional responses of Commercial iMGL cells to LXR agonist treatment. (A) Chemical structures of LXR pathway agonists GW3965 and T0901317. (B) Volcano plot showing relationship between expression fold change and significance for differential gene expression between GW3965 300 nM and DMSO treated cells. Genes with significant upregulation and downregulation in GW3965 300 nM are colored red and blue, respectively. (C) Volcano plot showing relationship between expression fold change and significance for differential gene expression between T0901317 100 nM and DMSO treated cells. Genes with significant upregulation and downregulation in T0901317 100 nM are colored red and blue, respectively. (D) Overlap in DEGs between GW3965 300 nM and T0901317 100 nM treatments. (E) Selected pathway enrichments for DEGs with GW3965 300 nM treatment. (F) Selected pathway enrichments for DEGs with T0901317 100 nM treatment.
We treated Commercial iMGL cells with each LXR agonist starting on day 3 of in vitro culture and performed bulk RNA sequencing to assess their transcriptional responses 24 h later (“Methods” and Supplementary Table 3). We selected two doses of each compound to evaluate the consistency of responses across a dose curve which is an important aspect of developing a transcriptomic bioassay48,49. We found a marked increase in expression of the established LXR pathway target genes50 ABCA1, ABCG1, and APOE (Supplementary Fig. 12). We systematically identified differentially expressed genes (DEGs) for each treatment (“Methods”). We identified 296, 677, 227, and 538 DEGs for GW3965 30 nM, GW3965 300 nM, T0901317 30 nM, and T0901317 100 nM, respectively (Supplementary Table 4). In all four stimulus conditions, ABCA1 and ABCG1 were among the most highly upregulated genes, as expected with stimulation of the LXR pathway (Fig. 3B,C and Supplementary Fig. 13). Additionally, for both compounds, the expression level of DEGs was dose-dependent, and almost all the DEGs identified at the lower dose were also identified at the higher dose (Supplementary Fig. 14). Comparing between the two compounds, we found highly overlapping lists of DEGs (Fig. 3D) and high concordance in gene fold changes (Supplementary Fig. 15), suggesting that both compounds are modulating the same cellular pathways. We ran gene ontology and pathway enrichments for the DEGs identified in each of the 4 stimulus conditions (Supplementary Tables 5–8). We found similar pathway enrichments in both compounds with upregulated genes enriched in lipid metabolism processes and downregulated genes enriched in cell cycle, extracellular matrix, and inflammatory processes (Fig. 3E,F). Overall, Commercial iMGL cells exhibited robust bulk transcriptional responses to LXR agonists.
Differential transcriptional responses to LXR pathway agonists across Commercial iMGL cell subpopulations
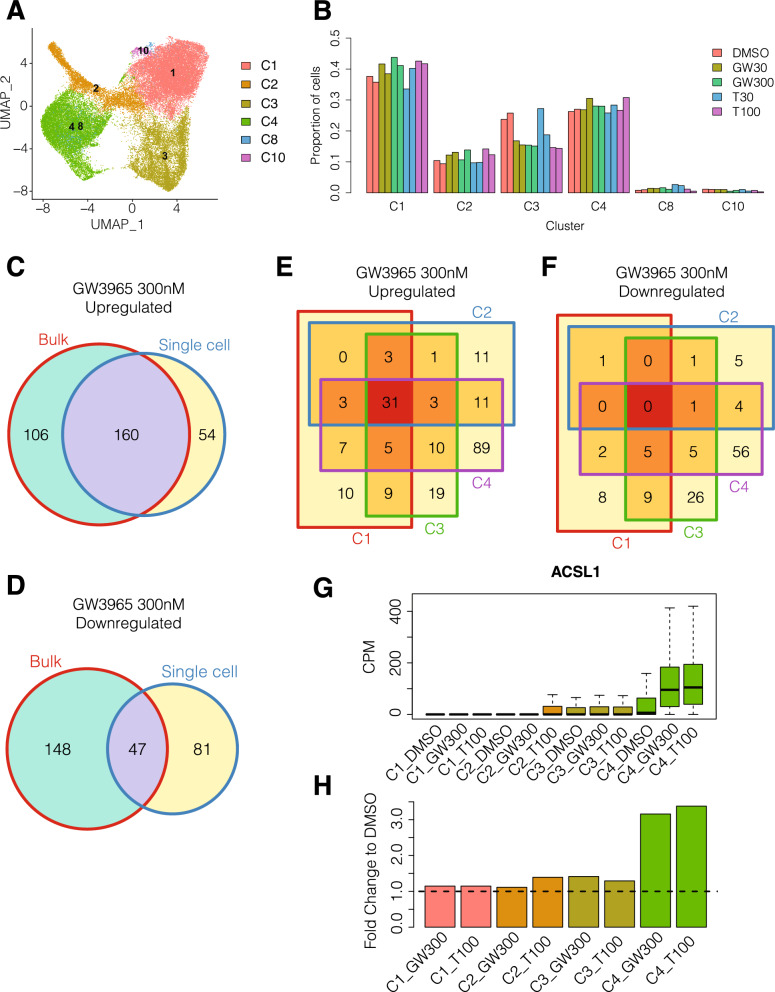
To assess changes in subpopulation abundances and cell-to-cell heterogeneity in transcriptional responses of Commercial iMGL cells to LXR pathway agonists, we performed single cell RNA sequencing (“Methods”). After removing low quality cells (Supplementary Fig. 16), we generated a dataset of 43,312 cells. Using Louvain clustering, we identified 6 clusters of cells (Fig. 4A and Supplementary Data 2) and matched their identities to the previously identified Commercial iMGL cell subpopulations (Fig. 2A) using a set of microglia functional marker genes31 (Supplementary Fig. 17). In general, the subpopulation abundances are unchanged between untreated and LXR agonist treated samples (Fig. 4B). There is a slight reduction in the proportion of proliferative homeostatic cells in cluster C3 which is consistent with the finding that downregulated genes in bulk RNA-seq are enriched within the cell cycle pathway (Fig. 3E,F).
Figure 4.
Single cell transcriptional responses of Commercial iMGL cells to LXR agonist treatment. (A) Identification of 6 single cell clusters in LXR agonist treated Commercial iMGL cells. Cluster names were matched to the time course single cell RNA-seq dataset (Fig. 2A) using microglia functional marker genes (Supplementary Fig. 17) as described in the “Methods”. (B) Proportion of cells belonging to each single cell cluster in the LXR agonist treatment single cell RNA-seq dataset. Treatments are abbreviated: DMSO, GW3965 30 nM (GW30), GW3965 300 nM (GW300), T0901317 30 nM (T30), and T0901317 100 nM (T100). (C) Overlap in upregulated DEGs between bulk RNA-seq and the union of single cell RNA-seq DEGs in clusters C1, C2, C3, C4, C8, and C10 with GW3965 300 nM treatment. Bulk DEGs were filtered to genes tested in at least one single cell contrast. (D) Overlap in downregulated DEGs between bulk RNA-seq and the union of single cell RNA-seq DEGs in clusters C1, C2, C3, C4, C8, and C10 with GW3965 300 nM treatment. Bulk DEGs were filtered to genes tested in at least one single cell contrast. (E) Overlap in upregulated DEGs for single cell clusters C1, C2, C3, and C4 with GW3965 300 nM treatment. (F) Overlap in downregulated DEGs for single cell clusters C1, C2, C3, and C4 with GW3965 300 nM treatment. (G) Expression of ACSL1 gene in single cells assigned to clusters C1, C2, C3, and C4 from DMSO, GW3965 300 nM (GW300), and T0901317 100 nM (T100) treatments. (H) Fold change of ACSL1 gene compared to DMSO in single cells assigned to clusters C1, C2, C3, and C4 from GW3965 300 nM (GW300) and T0901317 100 nM (T100) treatments.
We identified LXR agonist induced DEGs for each cluster (“Methods”, Supplementary Fig. 18, and Supplementary Table 9). First, we compared the DEG responses between high and low doses for each treatment within the same cluster. In general, fold change differences correlate with dose for each treatment (Supplementary Fig. 19). We noticed that clusters C1, C3, and C4 show higher correlations between doses than cluster C2, C8, and C10, for both treatments, suggesting that Commercial iMGL cell subpopulations have differing sensitivities to LXR agonists. Comparing between the two compounds, we found highly overlapping lists of DEGs within each cluster (Supplementary Fig. 20), and similar gene ontology and pathway enrichments (Supplementary Table 10).
Next, we compared the responses between bulk and single cell RNA-seq to evaluate the performance of the two technologies in identifying LXR agonist induced DEGs. In general, the gene expression changes detected by bulk and single cell analysis are quite similar (cor = 0.7–0.81). However, there are many genes that exhibit differences in fold-changes between the two technologies (Supplementary Fig. 21). The number of DEGs is greater in bulk as compared to single cell, however there is substantial overlap between the two methods (Fig. 4C,D and Supplementary Fig. 22). We found that DEGs detected by bulk only exhibit lower expression levels in the single cell RNA-seq dataset when compared to DEGs detected by both methods (Supplementary Fig. 23). This finding is consistent with previous reports showing that identification of lowly expressed DEGs by single cell analysis is challenging51.
Finally, we compared the DEG responses for the high dose of each compound between the four major clusters C1, C2, C3, and C4. Between clusters, the gene expression responses were positively correlated, however the fold change extents were different (Supplementary Fig. 24). We found that the transcriptional response in cluster 1 was more comparable to cluster 3 (cor = 0.74–0.77) than cluster 2 and cluster 4 (cor = 0.52–0.64), which support our previous characterization of C1/C3 and C2/C4 as representing homeostatic and activated microglia, respectively. We compared the set of DEGs across different clusters in each treatment and found substantial differences in DEGs across clusters, with a particular emphasis on a large number that are specific to cluster C4 after GW3965 300 nM (Fig. 4E,F) and T0901317 100 nM treatments (Supplementary Fig. 25). One example of a gene with differential transcriptional responses across clusters is ACSL1. The expression of ACSL1 is upregulated in cluster C4 (FC = 3.16–3.38), in response to both compounds, whereas its expression is unchanged in clusters C1, C2 and C3 (FC = 1.11–1.42) (Fig. 4G,H). ACSL1 is significantly upregulated in bulk RNA-seq (FC = 2.6–2.75) (Supplementary Table 4), and the single cell dataset demonstrates that the bulk RNA-seq change in ACSL1 is predominantly driven by activated microglia cells in cluster C4. Similarly, we sorted the full list of bulk DEGs into their corresponding single cell clusters (Supplementary Table 11). These findings demonstrate that different subpopulations of iMGL cells exhibit heterogeneous transcriptional responses to LXR agonists.
Discussion
Microglia play important roles in the pathogenesis of neurodegenerative disorders. A substantial increase in neuroinflammation mediated by microglial activation and proliferation is a hallmark of late stage neurodegenerative disorders including AD, PD, and frontotemporal dementia52. Two of the most prominent genetic risk factors for late onset AD, APOE and TREM2, are highly expressed in microglia and are important regulators of cholesterol metabolism and transport in the brain53. Therefore, in vitro models, such as iMGL cells, are essential tools for preclinical development. Multiple studies have shown that iMGL cells have transcriptional profiles that are highly similar to ex vivo microglia12,15,54–58. In this study, we find that iMGL cells, from Fujifilm Cellular Dynamics, Inc., are an attractive commercial option.
Currently, multiple therapeutics targeting APOE and TREM2 in microglia are in development for AD59–61, including LXR pathway agonists62. This is the first study to perform a comprehensive bulk and single cell transcriptional characterization of LXR agonists on iMGL cells. At the bulk level, we find an upregulation of lipid metabolism pathways, consistent with previous findings that positive effects of LXR agonists on AD pathology in preclinical models are largely due to the induction of ABCA1 expression, which promotes cholesterol clearance and APOE lipidation17,53. Interestingly, at the single cell level, we identified heterogeneous responses across subpopulations of iMGL cells including a prominent response in activated microglia. We found a downregulation of inflammatory response pathways, consistent with a previous report that treatment of mouse microglia-derived cell line BV2 with GW3965 causes an attenuation of neuroinflammation63.
Although Commercial iMGL cells show promising potential, they do not completely recapitulate all aspects of microglial biology. The transcriptional factor SALL1, an important determinant of microglia identity64, is not expressed in Commercial iMGL cells (Fig. 1E and Supplementary Fig. 4). However, xenotransplant microglia exhibit expression of SALL1, indicating that iMGL cells have the potential to turn on the SALL1-mediated transcriptional program (Supplementary Fig. 4). Moving forward, optimization of differentiation protocols and culture media components as well as further methods development, including direct cell conversion54, will continue to make iMGL cells an attractive model system for therapeutic development in neurodegeneration.
Conclusions
In this study, we performed comprehensive bulk and single cell transcriptional characterization of Commercial iMGL cells. We find that Commercial iMGL cells closely resemble ex vivo microglia at the overall transcriptome level and express most, but not all, key microglia marker genes. We identified 11 subpopulations of cells representing homeostatic, activated, and proliferating states, replicating the heterogeneity of ex vivo microglia30,31. Commercial iMGL cells stabilize after 3 days in culture and we treated them with two distinct LXR pathway agonists to assess their transcriptional responsiveness. At the bulk level, Commercial iMGL cells respond by upregulation of lipid metabolism pathways and downregulation of cell cycle pathways. At the single cell level, the transcriptional responses differ between homeostatic and activated microglia. Overall, our results demonstrate that Commercial iMGL cells exhibit a basal transcriptional profile, cellular heterogeneity, and transcriptional plasticity that is comparable to in vivo microglia.
Supplementary Information
Acknowledgements
The results published here are in part based on data obtained from the AD Knowledge Portal (https://adknowledgeportal.org). The results published here are in part based on data obtained from dbGaP (PI: Glass, Institute: NINDS).
Abbreviations
- iPSC
Induced pluripotent stem cells
- iMGL cells
IPSC-derived microglia-like cells
- LXR
Liver X receptor
- xMG
Xenotransplant microglia
- PCA
Principal components analysis
- DEG
Differentially expressed gene
- DMSO
Dimethyl sulfoxide
- HPC
Hematopoietic precursor cells
- EMP
Erythro-myeloid progenitors
- AD
Alzheimer’s disease
- PD
Parkinson’s disease
- TF
Transcription factor
Author contributions
G.R. performed computational analyses and wrote the manuscript draft. Y.Y., B.A., B.M., J.W., and A.A.A. performed experiments. G.G., J.H., and Y.L. performed computational analyses. D.H., L.C.B., S.J.E., I.G., A.S., A.A.S., R.T.F., Y.L., and D.B. supervised the experiments and computational analyses. All authors aided in the interpretation of results; read, reviewed, and approved the manuscript for publication.
Funding
This research was funded by CAMP4 Therapeutics Corporation and Biogen Inc.
Data availability
Raw and processed data from bulk and single cell RNA sequencing experiments have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GSE226081.
Competing interests
G.R., Y.Y., B.A., B.M., J.W., G.G., J.H., A.A.A., A.A.S., Y.L., and D.B. are current employees of, and hold equity interest in, CAMP4 Therapeutics Corporation. D.H. and S.J.E. are current employees of Biogen Inc. L.C.B. is a stockholder and former employee of Biogen Inc. I.G., A.S., and R.T.F. are former employees of CAMP4 Therapeutics Corporation.
Footnotes
The original online version of this Article was revised: Iris Grossman was omitted from the author list in the original version of this Article. The Author Contributions section now reads: “G.R. performed computational analyses and wrote the manuscript draft. Y.Y., B.A., B.M., J.W., and A.A.A. performed experiments. G.G., J.H., and Y.L. performed computational analyses. D.H., L.C.B., S.J.E., I.G., A.S., A.A.S., R.T.F., Y.L., and D.B. supervised the experiments and computational analyses. All authors aided in the interpretation of results; read, reviewed, and approved the manuscript for publication.” Competing interests now reads: “G.R., Y.Y., B.A., B.M., J.W., G.G., J.H., A.A.A., A.A.S., Y.L., and D.B. are current employees of, and hold equity interest in, CAMP4 Therapeutics Corporation. D.H. and S.J.E. are current employees of Biogen Inc. L.C.B. is a stockholder and former employee of Biogen Inc. I.G., A.S., and R.T.F. are former employees of CAMP4 Therapeutics Corporation.”
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
4/23/2024
A Correction to this paper has been published: 10.1038/s41598-024-60234-z
Contributor Information
Gokul Ramaswami, Email: gokul@camp4tx.com.
David Bumcrot, Email: bumcrot@camp4tx.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-52311-0.
References
- 1.Salter MW, Stevens B. Microglia emerge as central players in brain disease. Nat. Med. 2017;23:1018–1027. doi: 10.1038/nm.4397. [DOI] [PubMed] [Google Scholar]
- 2.Schafer DP, Stevens B. Microglia function in central nervous system development and plasticity. Cold Spring Harb. Perspect. Biol. 2015;7:a020545. doi: 10.1101/cshperspect.a020545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nott A, Holtman IR, Coufal NG, Schlachetzki JCM, Yu M, Hu R, et al. Brain cell type-specific enhancer–promoter interactome maps and disease-risk association. Science. 2019;366:1134–1139. doi: 10.1126/science.aay0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leng F, Edison P. Neuroinflammation and microglial activation in Alzheimer disease: Where do we go from here? Nat. Rev. Neurol. 2021;17:157–172. doi: 10.1038/s41582-020-00435-y. [DOI] [PubMed] [Google Scholar]
- 5.Kam T-I, Hinkle JT, Dawson TM, Dawson VL. Microglia and astrocyte dysfunction in Parkinson’s disease. Neurobiol. Dis. 2020;144:105028. doi: 10.1016/j.nbd.2020.105028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gosselin D, Skola D, Coufal NG, Holtman IR, Schlachetzki JCM, Sajti E, et al. An environment-dependent transcriptional network specifies human microglia identity. Science. 2017;356:eaal3222. doi: 10.1126/science.aal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pandya H, Shen MJ, Ichikawa DM, Sedlock AB, Choi Y, Johnson KR, et al. Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nat. Neurosci. 2017;20:753–759. doi: 10.1038/nn.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haenseler W, Sansom SN, Buchrieser J, Newey SE, Moore CS, Nicholls FJ, et al. A highly efficient human pluripotent stem cell microglia model displays a neuronal-co-culture-specific expression profile and inflammatory response. Stem Cell Rep. 2017;8:1727–1742. doi: 10.1016/j.stemcr.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douvaras P, Sun B, Wang M, Kruglikov I, Lallos G, Zimmer M, et al. Directed differentiation of human pluripotent stem cells to microglia. Stem Cell Rep. 2017;8:1516–1524. doi: 10.1016/j.stemcr.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muffat J, Li Y, Yuan B, Mitalipova M, Omer A, Corcoran S, et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat. Med. 2016;22:1358–1367. doi: 10.1038/nm.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takata K, Kozaki T, Lee CZW, Thion MS, Otsuka M, Lim S, et al. Induced-pluripotent-stem-cell-derived primitive macrophages provide a platform for modeling tissue-resident macrophage differentiation and function. Immunity. 2017;47:183–198.e6. doi: 10.1016/j.immuni.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 12.Abud EM, Ramirez RN, Martinez ES, Healy LM, Nguyen CHH, Newman SA, et al. iPSC-derived human microglia-like cells to study neurological diseases. Neuron. 2017;94:278–293.e9. doi: 10.1016/j.neuron.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasselmann J, Blurton-Jones M. Human iPSC-derived microglia: A growing toolset to study the brain’s innate immune cells. Glia. 2020;68:721–739. doi: 10.1002/glia.23781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasselmann J, Coburn MA, England W, Figueroa Velez DX, Kiani Shabestari S, Tu CH, et al. Development of a chimeric model to study and manipulate human microglia in vivo. Neuron. 2019;103:1016–1033.e10. doi: 10.1016/j.neuron.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Svoboda DS, Barrasa MI, Shu J, Rietjens R, Zhang S, Mitalipova M, et al. Human iPSC-derived microglia assume a primary microglia-like state after transplantation into the neonatal mouse brain. Proc. Natl. Acad. Sci. 2019;116:25293–25303. doi: 10.1073/pnas.1913541116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mancuso R, Van Den Daele J, Fattorelli N, Wolfs L, Balusu S, Burton O, et al. Stem-cell-derived human microglia transplanted in mouse brain to study human disease. Nat. Neurosci. 2019;22:2111–2116. doi: 10.1038/s41593-019-0525-x. [DOI] [PubMed] [Google Scholar]
- 17.Carter AY, Letronne F, Fitz NF, Mounier A, Wolfe CM, Nam KN, et al. Liver X receptor agonist treatment significantly affects phenotype and transcriptome of APOE3 and APOE4 Abca1 haplo-deficient mice. PLoS One. 2017;12:e0172161. doi: 10.1371/journal.pone.0172161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui X, Chopp M, Zacharek A, Cui Y, Roberts C, Chen J. The neurorestorative benefit of GW3965 treatment of stroke in mice. Stroke. 2013;44:153–161. doi: 10.1161/STROKEAHA.112.677682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savage JC, Jay T, Goduni E, Quigley C, Mariani MM, Malm T, et al. Nuclear receptors license phagocytosis by Trem2+ myeloid cells in mouse models of Alzheimer’s disease. J. Neurosci. 2015;35:6532–6543. doi: 10.1523/JNEUROSCI.4586-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olah M, Patrick E, Villani A-C, Xu J, White CC, Ryan KJ, et al. A transcriptomic atlas of aged human microglia. Nat. Commun. 2018;9:539. doi: 10.1038/s41467-018-02926-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galatro TF, Holtman IR, Lerario AM, Vainchtein ID, Brouwer N, Sola PR, et al. Transcriptomic analysis of purified human cortical microglia reveals age-associated changes. Nat. Neurosci. 2017;20:1162–1171. doi: 10.1038/nn.4597. [DOI] [PubMed] [Google Scholar]
- 22.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Hayes DN, Nobel A, Marron JS. Statistical significance of clustering for high-dimension, low-sample size data. J. Am. Stat. Assoc. 2008;103:1281–1293. doi: 10.1198/016214508000000454. [DOI] [Google Scholar]
- 27.Rousseeuw PJ. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987;20:53–65. doi: 10.1016/0377-0427(87)90125-7. [DOI] [Google Scholar]
- 28.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation. 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olah M, Menon V, Habib N, Taga MF, Ma Y, Yung CJ, et al. Single cell RNA sequencing of human microglia uncovers a subset associated with Alzheimer’s disease. Nat. Commun. 2020;11:6129. doi: 10.1038/s41467-020-19737-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sankowski R, Böttcher C, Masuda T, Geirsdottir L, Sagar, Sindram E, et al. Mapping microglia states in the human brain through the integration of high-dimensional techniques. Nat. Neurosci. 2019;22:2098–2110. doi: 10.1038/s41593-019-0532-y. [DOI] [PubMed] [Google Scholar]
- 32.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holtman IR, Skola D, Glass CK. Transcriptional control of microglia phenotypes in health and disease. J. Clin. Investig. 2017;127:3220–3229. doi: 10.1172/JCI90604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masuda T, Sankowski R, Staszewski O, Prinz M. Microglia heterogeneity in the single-cell era. Cell Rep. 2020;30:1271–1281. doi: 10.1016/j.celrep.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 37.Bellenguez, C., Küçükali, F., Jansen, I., Andrade, V., Moreno-Grau, S., Amin, N. et al. New insights on the genetic etiology of Alzheimer’s and related dementia (2020).
- 38.Fixsen BR, Han CZ, Zhou Y, Spann NJ, Saisan P, Shen Z, et al. SALL1 enforces microglia-specific DNA binding and function of SMADs to establish microglia identity. Nat. Immunol. 2023;24:1188–1199. doi: 10.1038/s41590-023-01528-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Schepper S, Ge JZ, Crowley G, Ferreira LSS, Garceau D, Toomey CE, et al. Perivascular cells induce microglial phagocytic states and synaptic engulfment via SPP1 in mouse models of Alzheimer’s disease. Nat. Neurosci. 2023;26:406–415. doi: 10.1038/s41593-023-01257-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mizutani M, Pino PA, Saederup N, Charo IF, Ransohoff RM, Cardona AE. The fractalkine receptor but not CCR2 is present on microglia from embryonic development throughout adulthood. J. Immunol. 2012;188:29–36. doi: 10.4049/jimmunol.1100421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Howe CL, Mayoral S, Rodriguez M. Activated microglia stimulate transcriptional changes in primary oligodendrocytes via IL-1β. Neurobiol. Dis. 2006;23:731–739. doi: 10.1016/j.nbd.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 42.Hemonnot A-L, Hua J, Ulmann L, Hirbec H. Microglia in Alzheimer disease: Well-known targets and new opportunities. Front. Aging Neurosci. 2019;11:233. doi: 10.3389/fnagi.2019.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collins JL, Fivush AM, Watson MA, Galardi CM, Lewis MC, Moore LB, et al. Identification of a nonsteroidal liver X receptor agonist through parallel array synthesis of tertiary amines. J. Med. Chem. 2002;45:1963–1966. doi: 10.1021/jm0255116. [DOI] [PubMed] [Google Scholar]
- 44.Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, Li L, et al. Role of LXRs in control of lipogenesis. Genes Dev. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Courtney R, Landreth GE. LXR regulation of brain cholesterol: From development to disease. Trends Endocrinol. Metab. 2016;27:404–414. doi: 10.1016/j.tem.2016.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Secor McVoy JR, Oughli HA, Oh U. Liver X receptor-dependent inhibition of microglial nitric oxide synthase 2. J. Neuroinflamm. 2015;12:27. doi: 10.1186/s12974-015-0247-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang-Gandhi CX, Drew PD. Liver X receptor and retinoid X receptor agonists inhibit inflammatory responses of microglia and astrocytes. J. Neuroimmunol. 2007;183:50–59. doi: 10.1016/j.jneuroim.2006.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ji R-R, de Silva H, Jin Y, Bruccoleri RE, Cao J, He A, et al. Transcriptional profiling of the dose response: A more powerful approach for characterizing drug activities. PLoS Comput. Biol. 2009;5:e1000512. doi: 10.1371/journal.pcbi.1000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, Ho DJ, Henault M, Yang C, Neri M, Ge R, et al. DRUG-seq provides unbiased biological activity readouts for neuroscience drug discovery. ACS Chem. Biol. 2022;17:1401–1414. doi: 10.1021/acschembio.1c00920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang B, Tontonoz P. Liver X receptors in lipid signalling and membrane homeostasis. Nat. Rev. Endocrinol. 2018;14:452–463. doi: 10.1038/s41574-018-0037-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mou T, Deng W, Gu F, Pawitan Y, Vu TN. Reproducibility of methods to detect differentially expressed genes from single-cell RNA sequencing. Front. Genet. 2020;10:1331. doi: 10.3389/fgene.2019.01331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bachiller S, Jiménez-Ferrer I, Paulus A, Yang Y, Swanberg M, Deierborg T, et al. Microglia in neurological diseases: A road map to brain-disease dependent-inflammatory response. Front. Cell. Neurosci. 2018;12:488. doi: 10.3389/fncel.2018.00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nugent AA, Lin K, van Lengerich B, Lianoglou S, Przybyla L, Davis SS, et al. TREM2 regulates microglial cholesterol metabolism upon chronic phagocytic challenge. Neuron. 2020;105:837–854.e9. doi: 10.1016/j.neuron.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 54.Chen S-W, Hung Y-S, Fuh J-L, Chen N-J, Chu Y-S, Chen S-C, et al. Efficient conversion of human induced pluripotent stem cells into microglia by defined transcription factors. Stem Cell Rep. 2021;16:1363–1380. doi: 10.1016/j.stemcr.2021.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dräger NM, Sattler SM, Huang CT-L, Teter OM, Leng K, Hashemi SH, et al. A CRISPRi/a platform in human iPSC-derived microglia uncovers regulators of disease states. Nat. Neurosci. 2022;25:1149–1162. doi: 10.1038/s41593-022-01131-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McQuade A, Coburn M, Tu CH, Hasselmann J, Davtyan H, Blurton-Jones M. Development and validation of a simplified method to generate human microglia from pluripotent stem cells. Mol. Neurodegener. 2018;13:67. doi: 10.1186/s13024-018-0297-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Banerjee A, Lu Y, Do K, Mize T, Wu X, Chen X, et al. Validation of induced microglia-like cells (iMG cells) for future studies of brain diseases. Front. Cell. Neurosci. 2021;15:629279. doi: 10.3389/fncel.2021.629279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu R, Li X, Boreland AJ, Posyton A, Kwan K, Hart RP, et al. Human iPSC-derived mature microglia retain their identity and functionally integrate in the chimeric mouse brain. Nat. Commun. 2020;11:1577. doi: 10.1038/s41467-020-15411-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Williams T, Borchelt DR, Chakrabarty P. Therapeutic approaches targeting Apolipoprotein E function in Alzheimer’s disease. Mol. Neurodegener. 2020;15:8. doi: 10.1186/s13024-020-0358-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang S, Mustafa M, Yuede CM, Salazar SV, Kong P, Long H, et al. Anti-human TREM2 induces microglia proliferation and reduces pathology in an Alzheimer’s disease model. J. Exp. Med. 2020;217:e20200785. doi: 10.1084/jem.20200785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schlepckow K, Monroe KM, Kleinberger G, Cantuti-Castelvetri L, Parhizkar S, Xia D, et al. Enhancing protective microglial activities with a dual function TREM2 antibody to the stalk region. EMBO Mol. Med. 2020;12:e11227. doi: 10.15252/emmm.201911227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fitz NF, Nam KN, Koldamova R, Lefterov I. Therapeutic targeting of nuclear receptors, liver X and retinoid X receptors, for Alzheimer’s disease. Br. J. Pharmacol. 2019;176:3599–3610. doi: 10.1111/bph.14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zelcer N, Khanlou N, Clare R, Jiang Q, Reed-Geaghan EG, Landreth GE, et al. Attenuation of neuroinflammation and Alzheimer’s disease pathology by liver x receptors. Proc. Natl. Acad. Sci. 2007;104:10601–10606. doi: 10.1073/pnas.0701096104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buttgereit A, Lelios I, Yu X, Vrohlings M, Krakoski NR, Gautier EL, et al. Sall1 is a transcriptional regulator defining microglia identity and function. Nat. Immunol. 2016;17:1397–1406. doi: 10.1038/ni.3585. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed data from bulk and single cell RNA sequencing experiments have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GSE226081.