Abstract
Increasing evidence suggests that key cancer-causing driver genes continue to exert a sustained influence on the tumor microenvironment (TME), highlighting the importance of immunotherapeutic targeting of gene mutations in governing tumor progression. TP53 is a prominent tumor suppressor that encodes the p53 protein, which controls the initiation and progression of different tumor types. Wild-type p53 maintains cell homeostasis and genomic instability through complex pathways, and mutant p53 (Mut p53) promotes tumor occurrence and development by regulating the TME. To date, it has been wildly considered that TP53 is able to mediate tumor immune escape. Herein, we summarized the relationship between TP53 gene and tumors, discussed the mechanism of Mut p53 mediated tumor immune escape, and summarized the progress of applying p53 protein in immunotherapy. This study will provide a basic basis for further exploration of therapeutic strategies targeting p53 protein.
Keywords: TP53, Tumors, Immune escape, Immunotherapy
Introduction
Tumors are complex diseases involving multiple factors and stages. Immune escape plays a key role in tumorigenesis and tumor progression. The complex interaction between the immune system and tumor cells determines the tumor state through a process called “tumor immune editing,” which includes three phases: clearance, homeostasis and escape [1, 2]. In the immune clearance phase, the immune system recognizes, monitors, and removes most malignant cells; however, a small number of malignant cells survive and enter the homeostasis phase. At this stage, although continuous pressure from the adaptive immune system prevents tumor cell growth and expansion, tumor cells with genetic instability form immunogenically reduced tumor subclones under pressure, Finally, the tumor cells evade the antitumor immune mechanism and enter the immune escape stage [3, 4].The tumor microenvironment (TME) is a dynamic and complex environment where tumor cells arise, and its composition varies by tumor type.[5]. During the “tumor immune editing” process, each cellular and non-cellular component of the TME uniquely regulates tumor immune escape. Therefore, the goal of tumor immunotherapy is to counteract this immune escape by both maintaining the tumor-immune cycle in the TME and reactivating the anti-tumor immune response [6].
Specific genetic alterations in different types of cancer can influence tumor growth and metastasis in vivo by regulating the TME. The human melanoma model with progressive genome editing constructed by Regev et al. [7] revealed that the combination of tumor gene mutations can not only impact the cellular composition of the TME but also re-edit the cellular state of the individual cell types that constitute it, thus affecting tumorigenesis and progression. TP53 mutations are prevalent in tumor progression. TP53 encodes the p53 protein, which is a prominent core tumor suppressor [8]. As a transcription factor, p53 regulates the transcription of target genes by directly binding to p53 DNA-binding elements in their promoter regions [9]. wild-type p53 (WT p53) regulates cell apoptosis, cellular senescence, cell cycle arrest, DNA damage repair, metabolic adaptation, and other cellular stress responses to exert tumor-suppressive effects [10]. However, TP53 missense mutations could disrupt the structural domain of the p53 protein, thereby impairing the expression or function of WT p53 and depriving it of its tumor-suppressive activity [11]. Tumor-derived mutant p53 (Mut p53) proteins contain missense, frameshift, truncation and deletion mutations; of which, approximately 74% are missense mutations mostly occurring within the p53 DNA-binding domain (DBD) [12]. Mut p53 has traditionally been classified as a “conformational” or “DNA contact” mutation. The former mainly interferes with folding of the core domain of p53 and is unable to bind to DNA or activate target genes. The latter directly mutates the amino acid residues that bind to DNA [13]. Both types of Mut p53 are unable to transactivate the target genes of WT p53 and therefore cannot mediate tumor-suppressive processes.
In addition to the loss of the tumor suppressor function of WT p53, Mut p53 promotes tumor progression through a gain-of-function (GOF) mechanism. To date, various Mut p53 GOF activities have been reported, including the promotion of tumor cell proliferation and metastasis, genomic instability, metabolic reprogramming, cell dryness, tumor microenvironment remodeling, immunosuppression and cancer treatment drug resistance [14]. Mut p53 usually exhibits different GOF activities through different molecular mechanisms. Further, Mut p53 forms complexes with the transcription factor NF-Y and cofactor p300 and transcribes and activates target genes of NF-Y, such as cyclin A, cyclin B1 and cyclin-dependent kinase 1 (CDK1), to promote cancer cell proliferation [15]. Specifically, most studies on the molecular mechanism of Mut p53 promoting metastasis have been found in rectal and pancreatic cancer cells. Mut p53R248W stabilizes proteins through a heat shock protein 90 chaperone mechanism in pancreatic cancer and selectively binds to phosphorylated STAT3 in pancreatic cancer cells to form the Mut p53-pSTAT3 complex, which promotes tumor cell migration [16]. This evidence paves way for novel avenues for using Hsp90 inhibitors in patients with Mutp53 mutations. Notably, the metabolic changes in glucose, lipids and nucleotides are not only markers of tumor cells but are also key factors in tumor development [17]. Innate immune cells promote insulin resistance by secreting cytokines to regulate metabolism, reflecting the complex role between metabolism and immunity [18]. The Warburg effect is defined as a faster glucose uptake and lactic acid accumulation in tumor cells than in normal cells under aerobic conditions [19]. Mut p53 promotes the translocation of glucose transporter 1 to the cell membrane by activating RhoA, thereby promoting the Warburg effect in cancer cells [20]. Glycolysis and metabolism reshape the tumor microenvironment, and both computational and experimental analyses have shown that glycolysis increases PD-L1 expression in tumor [21]. Glycolytic lactic acid production can promote tumor cells and tumor-associated macrophages (TAMs) to secrete a series of factors supporting angiogenesis, whereas glucose deprivation and extracellular acidosis significantly inhibit the antitumor function of macrophages, CD4 + T cells, CD8 + T cells, and dendritic cells (DCs). However, they have little effect on immunosuppressive cells such as myeloid-derived suppressor cells (MDSCs) and regulatory T cells (Tregs), and cancer-associated fibroblasts (CAF) and tumor cells can promote glycolysis [22]. Therefore, tumors carrying Mut p53 protein may promote the transition from metabolism to glycolysis through the abovementioned mechanisms, leading to tumor progression. In addition, Mut p53 can increase the expression and activity of manganese superoxide dismutase, a key antioxidant detoxification enzyme, in melanoma cells through SIRT3-mediated deacetylation, which helps regulate the level of reactive oxygen species (ROS) and prevent its cytotoxicity [23]. Damages to the glycolysis pathway in tumor cells leads to the increase in ROS level, which downregulates c-FLIP, the key inhibitor of tumor necrosis factor-α-induced cell death and enhances CTLs-mediated bystander killing [24]. Therefore, the above results indicate a potential molecular mechanism of the Mut p53-induced glycolysis pathway in promoting tumor immune escape. Mevalonate pathway is important for lipid metabolism. Its metabolic intermediate mevalonate-5-phosphate (M5p) promotes the stability of Mut p53 by inhibiting the proteasomal degradation of Mut p53 mediated by the ubiquitin ligase CHIP, whereas Mut p53 binds to SREBP2 and promotes the mevalonate pathway to increase M5p levels, thus forming a positive feedback loop [25, 26]. In addition, Mut p53 binds to the ETS2 site in the target gene promoter to activate the expression of several nucleotide metabolic genes, such as dCK, TK1, and GMPS and promote nucleotide synthesis [27]. The activation of GMPS can mediate the production of inflammatory cytokines, INF-α, and tumor necrosis factor (TNF), thus activate STAT and NF-κB pathways in cancer cells, stimulate their growth, and increase chemotherapy resistance [28]. Therefore, to a large extent, Mut p53 achieves its GOF activity through metabolic reprogramming, thereby promoting tumor immune escape. Further studying the mechanism through which Mutp53 promotes tumor immune escape is crucial to identify potential antitumor therapeutic targets.
Recent studies have suggested that WT p53 protein plays an important role in the immune clearance phase of tumors. The Mut p53 can disrupt this function, thus affecting the tumor immune microenvironment [29]. Major histocompatibility complex-I (MHC-I)-mediated antigen presentation plays a key role in the antitumor adaptive immune response. WT p53 upregulates the expression of Tap1 and the aminotransferase Erap1, which are peptide transport proteins essential for MHC-I transport to the cell surface [30]. The expression of MHC-I on the cell surface decreased under TP53 mutation conditions. In addition, Ghosh et al. [31] found that Mut p53 inhibits the function of the cGAS-STING-TBK1-IRF3 pathway, a cytoplasmic DNA-sensing mechanism in the natural immune response, by binding to TANK-binding kinase-1 (TBK1) and reducing the infiltration of lymphocytes, such as natural killer (NK) cells and CD8+ T cells, in the TME, thereby contributing to tumor immune escape. Therefore, Mut p53 may promote immune escape by inhibiting the antitumor immune response to form an immunosuppressive TME.
A review of related studies revealed that Mut p53 forms a tumor-promoting immunosuppressive microenvironment by damaging its function, reducing its number, and inhibiting the recruitment of immune cells to the TME [32–34]. In addition, Mut p53 has been reported to regulate the function of tumor stromal cells, other than immune cells, to promote tumor immune escape [35–37]. This review aimed to describe how Mut p53 regulates the immune landscape in the TME to create an ecological niche for tumor immune escape and summarize the current status of research on immunotherapy related to targeting the p53 pathway.
Mut p53 mediates immune escape by regulating the immune cell component in TME
Mut p53-mediated tumor immune escape is dominated by tumor-induced immunosuppressive effects, resulting in an immunosuppressive TME [38]. The immune ecological niche of the TME is regulated by cytotoxic T lymphocytes (CTLs), NK cells, MDSCs, TAMs and Tregs. The abundance and composition of each immune cell type are different in different tumor types. Mut p53 can regulate the TME by affecting single-cell functions, such as reducing CTL and NK cell infiltration and inhibiting their antitumor activity while recruiting large numbers of immunosuppressive cells (MDSCs, TAMs and Tregs) into the TME, ultimately creating a pro-tumor immune ecological niche by controlling cytokine secretion.
Mut p53 attenuates CTL cell-mediated antitumor immune response
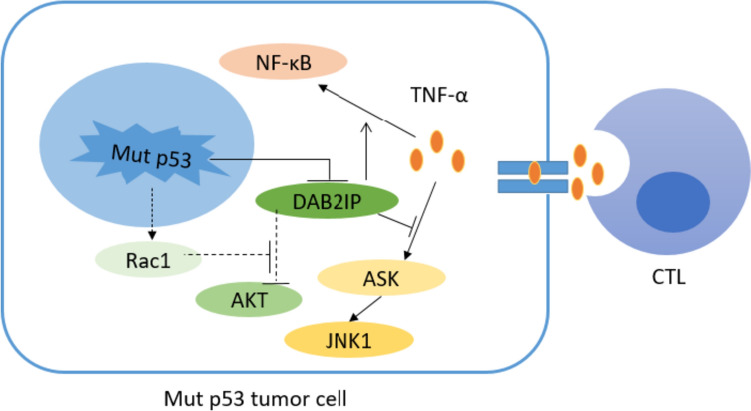
CTLs are important effector cells involved in killing tumor cells. Recent studies have suggested that Mut p53 promotes tumor immune escape by disrupting CTL function or reducing the number of CTLs in the TME. Activated CTLs kill target tumor cells mainly through two pathways [39–41]: first, CTL release perforin (PFN) to transport secreted granzymes A and B (GzmA and GzmB) into tumor cells after forming pores in the tumor cell membrane, triggering an enzyme chain reaction that leads to apoptosis of tumor cells. Second, the death factor Fas ligand (FasL) on the surface of CTL binds to the Fas receptor (FasR) on the surface of tumor cells and activates cystatin 8 to initiate apoptosis. Studies have shown that GzmB can induce the accumulation of WT p53 in the mitochondria of target tumor cells and interact with the anti-apoptotic protein B-cell lymphoma-2 (BCL-2), thereby antagonizing the inhibitory effect of BCL-2 on the pro-apoptotic factors Bcl-2 Associated X-protein (BAX) and truncated Bid (tBid), contributing to a large release of BAX and tBid, thereby facilitating the release of cytochrome C from mitochondria and triggering the apoptosis of tumor cells [42]. Blocking the WT p53/BCL-2 interaction significantly reduced CTL/NK-mediated cytotoxicity of WT p53 target cells. Hence, further research is needed to determine whether Mut p53 is related to the resistance of tumor cells to PFN/GzmB-and CTL/NK-mediated cell death. Additionally, WT p53 positively regulates TNF-related apoptosis-inducing ligand-induced apoptotic pathways by directly upregulating FasR and death receptors 4 and 5 [43]. Mut p53 was also found to reprogram TNF-α signal transduction in tumor cells by binding tumor suppressor DAB2IP and downregulate its expression, that is, it promoted the activation of NF-κB and inhibited the activation of TNF-α on ASK1/JNK and finally promoted the survival of tumor cells and resisted the tumor killing effect mediated by CTL [44]. Mutp53 also activates Rac1, promotes AKT activation, and promotes the survival of tumor cells [45]. However, the mechanism through which AKT activation promotes the survival of tumor cells remains unclear and requires further investigation; therefore, Mut p53 may diminish the killing effect of CTL on tumor cells and promote tumor immune escape (Fig. 1).
Fig. 1.
Schematic diagram of Mut p53 blocking anti-tumor immune response. Mut p53 down-regulates the expression of Tap1 and Erap1, which are necessary for MHC-I to be transported to the cell surface. The expression of MHC-I on the cell surface decreases and MHC-antigen complex decreases, which can not be recognized by T cells and finally blocks the anti-tumor adaptive immune response. On the other hand, Mut p53 inhibits TBK1 phosphorylation by binding with TBK1, thus inhibiting the function of cGAS-STING-TBK1-IRF3 pathway, which is a cytoplasmic DNA sensing mechanism in natural immune response, and reducing the infiltration of NK cells, CD8+T cells and other lymphocytes in TME, leading to tumor immune escape
It was also found that the number of bone marrow-infiltrating CTLs and T helper (Th) cells was reduced in patients with acute myeloid leukemia with Mut p53, whereas the number of Treg cells, which promote immune escape, was increased [46]. Mechanistically, Mut p53 reduced the number of CTLs and Th cells by downregulating the expression of the costimulatory molecule OX40 and increased the infiltration of Treg cells by upregulating the expression of the costimulatory molecule ICOS [47, 48]. In summary, Mut p53 may influence the immunosuppressive microenvironment by regulating immune cell function and numbers. Moreover, the reactivation of WT p53 function may be a novel approach to optimize CTL-mediated tumor killing.
Mut p53 inhibits normal NK cell function
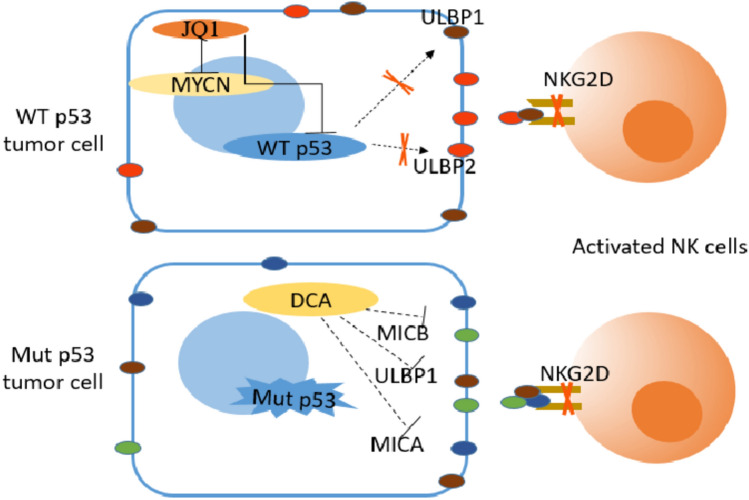
Activation of NK cells by effector molecules, such as interleukin (IL)-2, IL-12, IL-15 and IL-18. Downregulation of NK cell-inhibitory receptors (NK-IRs) or upregulation of NK cell-activating receptors (NK-ARs) can trigger the release of TNF-α and interferon (IFN)-γ from NK cells, which in turn exert antibody-dependent cell-mediated cytotoxic effects to kill tumor cells [49, 50]. NK cell-mediated tumor cell recognition and lysis is mainly dependent on the expression of NK-ARs NKG2D and DNAM-1. WT p53 upregulates the expression levels of ULBP1 and ULBP2, members of the ligand of the NKG2D (NKG2DL) family, on the surface of tumor cells and enhances the NK cell-mediated antitumor immune response [51]. Similarly, in oncogenic MYCN-amplified neuroblastoma, JQ1, a BET-bromo structural domain inhibitor of MYCN, downregulates c-MYC and p53 levels, which in turn downregulate NKG2DL expression and render neuroblastoma cells resistant to NK cell-mediated killing [52]. In addition, the glycolysis inhibitor dichloroacetate downregulates the expression of NKG2DL family members MICA, MICB, and ULBP-1 in tumor cells lacking WT p53 or expressing Mut p53, thereby mediating immune escape [53]. The above evidence suggests that Mut p53 inhibits the tumor-killing effect of NK cells, mainly by regulating the expression of stress ligands on the surface of tumor cells (Fig. 2). Therefore, increasing the number of NK cells, enhancing NK cell activity, and promoting NK cell infiltration have become major strategies for immunotherapy.
Fig. 2.
Schematic diagram of Mut p53 participating in regulating CTL cell-mediated anti-tumor immunity. Mut p53 reprogrammed the signal transduction of TNF-α in tumor cells by binding with tumor suppressor DAB2IP and down-regulating its expression, that is, it promoted the activation of NF-κB and inhibited the activation of TNF-α on ASK1/JNK at the same time, finally promoted the survival of tumor cells and resisted the tumor killing effect mediated by CTL. Mut p53 can also inhibit DAB2IP-AKT interaction by activating Rac1, promote AKT activation and lead to tumor cell survival
Mut p53 regulates the cell proliferation and differentiation of TAMs
As the most widely distributed type of immune cells in the TME, TAMs are mainly derived from circulating monocytes, which would differentiate into TAMs after being recruited to the TME and are stimulated by various cytokines [54]. TAMs are a highly plastic class of mixed phenotype cells, and consistent with the specific differentiation of activated macrophages into M1 and M2 types, TAMs are also polarized into M1-like and M2-like TAMs. M1-like TAMs are primarily activated by IFN-γ, transforming growth factor-α (TGF-α) or granulocyte–macrophage [55]. They are activated by IFN-γ, TGF-α or granulocyte–macrophage colony stimulating factor (GM-CSF); express CD68, CD80 and CD86; and secrete IL-1β, IL-6 and chemokine CXCL9, which exert antitumor effects. M2-like TAMs are primarily activated by IL-10 and TGF-β, expressing CD163, CD204 and CD206, secrete IL-10, TNF, CCL17, among others, which exerting pro-tumor effects [56].
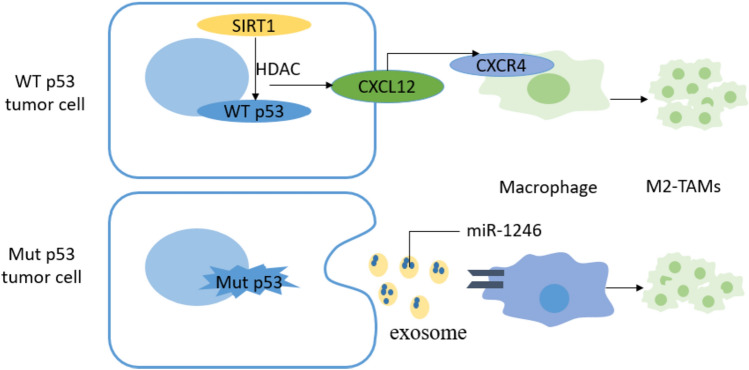
In a mouse model of colorectal cancer, alterations in p53 function affect the proliferation of TAMs and regulate macrophage polarization. First, p53 affects the recruitment of TAMs in primary colorectal cancer cells. The intrinsic mechanism is that p53 inhibits colony-stimulating factor 1 receptor (CSF1R) expression by inducing miR-34a, which further inhibits the first step of the STAT3-mediated tumor cell metastatic cascade epithelial–mesenchymal transformation and promotes tumor cell migration [57]. In contrast, the high expression of CSF1R caused by TP53 mutation is considered to affect both the differentiation and proliferation of TAMs. The overexpression of NAD-dependent deacetylase sirtuin 1 (SIRT1) in colorectal cancer promoted the expression of CXCL12 and CXCR4 on the surface of tumor cells and macrophages, respectively, facilitated the recruitment of M2-like TAMs and inhibited the cell proliferation and activity of CD8 + T cells, thus promoting colorectal cancer progression [58]. Simultaneously, Mut p53 colorectal cancer cells selectively release miR-1246-rich exosomes, and uptake of these exosomes by macrophages triggers miR-1246-dependent self-reprogramming, resulting in a pro-tumor state phenotype [59]. Using a set of 16 different genetically engineered mouse models of breast cancer, Wellensten et al. [60] revealed that p53-deficient tumor cells can induce WNT-secreted ligands, increase circulating neutrophils and stimulate TAMs to produce IL-1β, thereby triggering CXCR4 systemic inflammation and leading to breast cancer cell metastasis. In summary, loss of function or mutation of TP53 reprogrammed TAMs to promote tumor immune escape through different regulatory mechanisms (Fig. 3).
Fig. 3.
Schematic diagram of tumor cells lacking WT p53 or expressing Mut p53 against NK cell killing. WT p53 can directly up-regulate the expression levels of ULBP1 and ULBP2, members of the ligand family of NKG2D on the surface of tumor cells and enhance the anti-tumor immune response mediated by NK cells. JQ1, a BET-bromine domain inhibitor of oncogene MYCN, can down-regulate the levels of c-MYC and p53, and then down-regulate the expression of NKG2DL, making tumor cells resistant to NK cell-mediated killing. Dichloroacetate (DCA), a glycolytic inhibitor, can also down-regulate the expression of MICA, MICB and ULBP-1 of NKG2DL family in tumor cells lacking WT p53 or expressing Mut p53, thus mediating immune escape
TP53 regulates the cell proliferation and differentiation of immunosuppressed cells
MDSCs, which are a diverse group of myeloid-like cells in the TME, play a crucial role in anti-tumor activities by suppressing CTL activation [61]. The Fas–FasL cell death pathway was initially identified as a key regulator of CTL activity, and Fas-mediated apoptosis has been found to regulate MDSC homeostasis. In human colon cancer cells, activation of p53 upregulates Fas on the surface of MDSCs, which increases the sensitivity of MDSCs to FasL-induced cell apoptosis, that is, by activating the intrinsic p53–Fas–FasL pathway of MDSCs, promotes cell apoptosis, and reactivates CTL-mediated antitumor immunity [62]. DLEC1, a tumor suppressor linked to p53, interacts with the oncogenic signaling molecule, STAT3, in response to IL-6 stimulation, blocks the JAK2/STAT3 signaling pathway, and inhibits STAT3 phosphorylation, thereby controlling tumor progression [63]. Although Katz et al. [64] found that the GM–CSF/JAK2/STAT3 axis drives the cell proliferation of liver-associated MDSCs, and inhibition of STAT3 activates cell apoptotic signaling pathways in MDSCs, including the upregulation of the pro-apoptotic factor Bax and downregulation of the expression of the anti-apoptotic factor BCL-2. Thus, Mut p53 induces the cell proliferation by promoting STAT3 phosphorylation, leading to tumor immune escape. Ferroptosis is an iron,and ROS-dependent regulation of cell death induced by the degradation of heme oxygenase 1 (Hmox1), a downstream molecule of p53, and the release of free iron to produce ROS in the mitochondrial membrane [65]. Reportedly, ASAH2, a molecule highly expressed on the surface of tumor cells in a mouse colon cancer model, was also upregulated on the surface of MDSCs in TME and affected ferroptosis to promote the survival and accumulation of MDSCs by inhibiting the p53-Hmox1 pathway, consistent with the finding that silencing p53 reduced ROS levels in MDSCs and inhibited ferroptosis in MDSCs [66]. Thus, Mut p53 leads to the accumulation of immunosuppressive cellular MDSCs in the TME, mainly by blocking the apoptotic signaling pathway of MDSCs (Fig. 4).
Fig. 4.
Schematic diagram of Mut p53 reprogramming TAMs. Over-expressed histone deacetylase SIRT1 in tumor cells promotes the expression of CXCL12 on the surface of tumor cells, promotes the high expression of CXCR4 on the surface of macrophages, and promotes the recruitment of M2-like TAMs. Mut p53 tumor cells selectively release exosomes rich in miR-1246. Macrophages ingest these exosomes and trigger miR-1246-dependent self-reprogramming, forming M2-like TAMs
Treg cells, an immunosuppressive subset of CD4 + T cells, primarily ensure immune balance and self-tolerance. They are abundant in the TME and inhibit CTL-driven anti-tumor responses.Currently, there are fewer studies on the mechanisms through which p53 regulates Tregs to influence antitumor immunity; however, in recent years, increasing evidence has suggested that Mut p53 promotes tumor immune escape by affecting Treg cell differentiation. p53 interacts with non-coding RNAs (lncRNAs and miRNAs) to regulate tumor immune escape. LncRNA MEG3 suppresses tumor immune escape by upregulating the expression of miR-149-3p through MDM2-mediated p53 and decreasing the expression of FOXP3, which ultimately reduces the differentiation and maturation of Treg cells [67]. In addition, miR-34, which is positively regulated by p53, can take part in the feedback loop of TGF-β. Elevated TGF-β activity inhibits miRNA-34a expression, leading to enhanced production of chemokine CCL22, which recruits Treg cells to promote immune escape [68]. p53 deficiency in prostate, ovarian, and pancreatic cancers increases the number of Tregs in TME. In conclusion, Mut p53 can promote tumor immune escape by regulating Treg cell differentiation and recruiting Treg cells, and immunotherapy targeting Mut p53 and Treg cells plays an essential role in suppressing tumor immune escape.
In summary, Mut p53 mediates tumor immune escape by regulating immune cells through different mechanisms in different tumor types, providing a basis for the development of different immunotherapeutic strategies. However, Mut p53 ultimately promotes tumor immune escape by regulating the secretion of different cytokines by tumor and immune cells, making it an immunosuppressive signaling molecule. The literature sources of the molecules involved in the above mechanisms are shown in Table 1.
Table 1.
The action mechanism of some molecules described in this paper and its literature sources
| Immune cells regulated by Mut p53 | Mutp53-regulated protein | Regulation by Mut p53 | Fuction | References |
|---|---|---|---|---|
| CTL cell | BCL-2 | Interact with BCL-2 | Pro-cancer: can reduced GzmB-mediated CTL killing | Ben Safta et al. [42] |
| DAB2IP | Down-regulating DAB2IP | Pro-cancer: can promote the activation of NF-κB and inhibited the activation of TNF-α on ASK1/JNK, and finally promoted the survival of tumor cells | Di Minin et al. [44], Sorrentino et al. (2022)[112] | |
| OX40 | Down-regulating OX40 | Pro-cancer:can reduce the number of CTL and Th cells | Buchan et al. [47] | |
| ICOS | Up-regulating ICOS | Pro-cancer: can increase the infiltration of Treg cells | Amatore et al. [48] | |
| NK cell | ULBP1/2 | Down-regulating ULBP1/2 | Pro-cancer:can attenuat NK cell-mediated anti-tumor immune responses | Duan et al. [51] |
| MDSCs cell | STAT3 | Promote STAT3 phosphorylation | Pro-cancer:can induce the cell proliferation of MDSCs | Li et al. [51], Guha et al. [64] |
| ROS | Down-regulating ROS | Inhibit ferroptosis in MDSCs | Chang et al. [65], Zhu et al. [66] | |
| TAMs cell | miR-1246 | Up-regulating miR-1246 | Pro-cancer:can promote polarization of M2 macrophages | Cooks et al. [59] |
| CSF1R | Up-regulating CSF1R | Pro-cancer: can promote the first step of STAT3-mediated tumor cell metastatic cascade epithelial-mesenchymal transformation | Shi et al. [57] | |
| IL-1β | Up-regulating IL-1β | Trigger CXCR4 systemic inflammation and lead to breast can Pro-cancer: can cer cell metastasis | Wellenstein et al. [60] | |
| Treg cell | miR-149-3p | Down-regulating miR-149-3p | Pro-cancer: can promote Treg cell differentiation and maturation | Xu et al. [67] |
| miR-34 | Down-regulating miR-34 | Pro-cancer: can recruit Treg cells to promote immune escape | Yang et al. [68] |
Mut p53 promotes immune escape by regulating non-immune cell components
In the TME, CAFs are the main components of the tumor stroma and exert immunosuppressive effects by secreting growth factors, extracellular matrix proteins and inflammatory ligands, thereby promoting non-restrictive tumor cell growth, angiogenesis and therapeutic resistance [69]. Mut p53 can affect tumor progression by altering the activity and function of CAFs. Mut p53 binds to STAT3 to promote STAT3 phosphorylation and upregulates the expression of α-smooth muscle actin, fibroblast-derived factor 10 and CXCL12 to activate CAFs [70]. CAFs of Mut p53 significantly affect the composition and function of immune cells in the TME by secreting cytokines, including CXCL12, stromal cell-derived factor 1 (SDF-1) and IL-6, mainly manifesting tumor-suppressive effects [71]. In addition, in colorectal cancer, ROS produced by mutant p53 tumor cells facilitate CAF-secreted vascular endothelial growth factor to regulate angiogenesis and ultimately promote tumor growth [72]. Therefore, Mut p53 promotes tumor immune escape by activating CAFs and regulating their secretion of growth factors. In recent years, CAFs have emerged as novel targets for cancer therapy, and mutated p53, a key regulator of CAF activation, has shown potential as a key pathway for novel therapeutic interventions. mesenchymal stem cells (MSCs) are also important cellular components of the TME that contribute to tumor growth and metastasis [73]. Mut p53 promotes tumor growth and proliferation by recruiting MSCs to the TME through a CXCL12-dependent mechanism [74]. Meanwhile, MSCs with Mut p53 are considered the cells of origin of bone tumors, affecting osteogenic differentiation and influencing the properties of osteosarcoma TME components such as inducible nitric oxide synthase, CCL5, IL-6 and TGF-β expression at higher levels, ultimately promoting osteosarcoma development [75].
In summary, Mut p53 induces the production of an immunosuppressive TME by promoting the secretion of various cytokines by non-immune cells in the TME, interacting with tumor cells, and ultimately promoting immune escape.
Strategies for targeting Mut p53
The current mechanism of therapeutic action targeting Mut p53 involves the activation or restoration of WT p53 function in tumor cells. As mentioned above, Mut p53 forms an immunosuppressive microenvironment through different pathways. Therefore, WT p53 reactivation in the TME represents a promising therapeutic strategy to reverse immunosuppression and reshape the immunological landscape to support antitumor immunity.
p53-based vaccines and specific antibodies
P53-derived peptides have been investigated as targets for various immunotherapeutic strategies, including vaccines, bipotent antibodies and TCR-like antibodies. Researchers have tested a modified cowpox virus (MVA) vaccine encoding WT p53 in combination with gemcitabine in patients with platinum-resistant ovarian cancer, and five of eleven subjects were able to induce CD8 + and CD4 + T cell responses [76]. In addition, studies on the p53 MVA vaccine in combination with immune checkpoint inhibitors, including PD-1/PD-L1 and CTLA-4 antibodies, are also underway. Although the vaccine was safe, induced an anti-Mut p53 immune response and achieved disease stabilization in some patients, there were no clinically significant benefits. Bispecific antibodies are innovative cancer immunotherapies with dual specificity for tumor antigens and TCR-CD3 complexes [77]. A Mut p53-based bispecific antibody recognizes neoantigens from the TP53 R175H mutation site and TCR-CD3 complex, overcoming the lack of neoantigen presentation and selectively redirecting T cells to recognize Mut p53 tumor cells [78]. Because of its intracellular localization p53, it is not recognized by classical therapeutic antibodies. TCR-like antibodies mimic the ability of T cells to recognize MHC I-presenting peptides and are specific to pMHCs that present WT and Mut p53 antigens [79]. TCR-like antibodies have shown promising in vitro and in vivo antitumor effects in animal models [80]. In conclusion, most vaccines and antibodies against Mut p53 are currently only validated in animal models but provide avenues for future tumor therapy. Simultaneously, current antibody therapies have overcome the previous limitation of not recognizing intracellular antigens, which provides new evidence for further development of antibodies targeting solid tumors.
Molecular delivery targeting Mut p53
Although p53-based vaccines and specific antibodies have shown antitumor activity, they do not show clinical benefits; therefore, therapeutic strategies for Mut p53 proteins in tumor cells remain limited. Identifying effective and safe therapeutic strategies for Mut p53 is of great significance. Marco et al. [81] found that the delivery of oligonucleotides through modified nanomaterials can overcome the cancer resistance of refractory tumors carrying Mut p53 apoptosis because these nanostructures can inhibit the mTOR signaling pathway and anti-apoptotic protein Bcl-2. Resistance to temozolomide in the treatment of glioblastoma (GBM) is related to the upregulation of O6-methylguanine-DNA methyltransferase (MGMT), whereas the expression of MGMT in tumor cells is negatively regulated by WT p53 [82–84]. Researchers have developed a systemic nanodelivery platform (scL) for tumor-specific targeting that can deliver WT p53 through the blood–brain barrier, effectively target GBM and cancer stem cells (CSC), downregulate MGMT and induce GBM cell apoptosis. Simultaneously, the combination of scL-p53 and TMZ increased the antitumor efficacy of TMZ [85]. Therefore, scLs may provide a novel antitumor therapeutic approach that targets the delivery of molecules. In addition, researchers have developed a redox-responsive nanoparticle (NP) platform that can effectively deliver p53-encoded mRNA to induce cell cycle arrest and apoptosis, significantly delay the proliferation of p53-null hepatocellular carcinoma and non-small cell lung cancer cells, and considerably improve the sensitivity of tumor cells to mTOR inhibitors [86]. The mechanism of NP-mediated p53 gene delivery to decrease tumor progression is to inhibit tumor angiogenesis [87]. KRAS-TP53 co-mutation is closely related to poor prognosis of gastrointestinal tumors. Researchers have proposed a novel double-targeted HA-TPP/A nanocomplex that leads to the ubiquitin-dependent proteasomal degradation of Mut p53 by targeting mitochondrial damage, destroying its GOF activity, increasing the sensitivity of AMG510-induced tumor cell killing, and thus reducing the proliferation and migration of gastrointestinal cancer cells with KRAS-TP53 co-mutation [88]. Therefore, the delivery of molecular-targeted Mutp53 through nanocomposites has become a novel antitumor therapeutic approach, and the study of combined drug transformation based on its different mechanisms will also be focused on further improving the antitumor efficacy.
WT p53 reactivation synergistic immunotherapy
Currently, combination drug therapy is an effective antitumor treatment strategy. In response to the variability in immune cell abundance in different tumor cells, it is crucial to explore immunotherapy for WT p53 reactivation in combination with other effective immunotherapeutic approaches to enhance immune cell recruitment and maintain the normal function of antitumor immune cells. Several p53 activators have been reported, but most studies are in the preclinical stage owing to the lack of specificity. The ubiquitin ligase mouse double minute 2 homolog (MDM2) is a major negative regulator of p53 and closed-loop negative feedback regulation of MDM2-p53 in normal cells maintains a dynamic balance between MDM2 and p53 expression levels [89]. Data from cancer cell lines, a cholera mouse model, and patients with melanoma suggest that MDM2 inhibitor-mediated pharmacological p53 reactivation triggers the ERV-dsRNA-IFN pathway in tumor cells, thereby altering the TME to a therapeutically responsive phenotype and triggering tumor immune surveillance [90]. Immune checkpoint PD-L1 on the surface of tumor cells binds to PD-1 on the surface of CTL to inhibit CTL activation and induce tumor immune escape [91]. The MDM2 inhibitor HDM201 triggered antitumor adaptive immunity in WT p53 tumors alone and further enhanced adaptive immunity by blocking the PD-1/PD-L1 pathway [92]. It suggests that WT p53 is required for the antitumor immune response, which explains the potential mechanism of MDM2 inhibitor combined with immune checkpoint inhibitor therapy. In addition, the team of professors Duda DG and Shi J jointly developed an mRNA tumor-targeting nanoparticle strategy to restore the function of WT p53, which inhibited the proliferation of Mut p53 hepatocellular carcinoma cells while inhibiting tumor growth in combination with anti-PD-1 therapies and significantly enhanced antitumor immunity in hepatocellular carcinoma [93]. This finding reaffirms the importance of WT p53 activators in combination with PD-1/PD-L1 antibodies against tumors. Among the patients with diffuse large B-cell lymphoma receiving CD19 chimeric antigen receptor (CAR) therapy, those with Mut p53 have poorer overall survival rates than those with WT p53; therefore, TP53 is a valuable prognostic biomarker [94]. This suggests that the impact of TP53 abnormalities on CAR-T cell therapy and the need for further research on the related signaling pathways should be considered when assessing the likelihood of success of CAR-T therapies in future and designing the clinical trials for high-risk patients.
In conclusion, Mut p53 can affect the immune ecological site of the TME, WT p53 expression is beneficial for immunotherapy, and the current therapeutic strategy for WT p53 reactivation in synergistic immunotherapy has shown promising antitumor efficacy in animal experiments. Therefore, combining small-molecule drugs that restore WT p53 activity with immunotherapy for tumor patients may improve the success rate of antitumor therapy.
Discussion
At present, the TME, which is composed of the ECM, stromal cells, immune cells and blood vessels, plays a key role in tumor development and chemotherapy resistance [95]. It is usually associated with the evasion of the immune system. The loss of function of WT p53 is key to the formation of an immunosuppressive microenvironment, which not only suppresses the antitumor immune response but is also beneficial to the proliferation of tumor cells [33]. Mut p53 can produce not only a proinflammatory but also an antiinflammatory environment. Mut p53 subtypes can have a profound impact on gene expression patterns, many of which lead to tumor cell proliferation and chemotherapy resistance in various ways [96]. Mut p53 regulates immune escape by increasing the expression of programmed death ligand 1 (PD-L1) in tumor cells [97]. Mut p53 decreases the expression of miR-34/miR-200 during mesenchymal transformation, resulting in an increase in PD-L1 expression in NSCLC cells [98]. Higher PD-L1 levels show significant benefits for PD-1/PD-L1 blocking therapy, highlighting Mut p53 as a potential target for immunotherapy. In cervical cancer, PD-L1 levels can be increased by miR-18a by targeting SOX6 to activate the Wnt/β-catenin pathway and inactivate p53 signaling [99]. Therefore, immune checkpoint inhibitors have potential efficacy in Mut p53 tumors. In addition, inflammation may promote the occurrence and development of cancer and induce the formation of immunosuppressive microenvironment, which is affected by the coordination of several proinflammatory cytokines and carcinogenic pathways, which include NF-kB, MAPK, mTOR and STAT3 signaling pathways. These carcinogenic pathways promote the stability of Mut p53, thus blocking the interaction between the carcinogenic pathways and proinflammatory cytokines to reduce carcinogenic inflammation and Mut p53 expression [100]. Therefore, inhibitors targeting these carcinogenic pathways, such as AG490 and WP1066 [101, 102], mTOR inhibitors [103] targeting the STAT3 pathway, LY3007113 [104] targeting p38MAPK and 17-AAG [105] targeting HSP90, may have promising antitumor efficacy alone or in combination. Many of these inhibitors are already in preclinical or clinical trials and have shown promising results. In addition, patients with Mut p53 appear to be more sensitive to immunosuppressant therapy. Mut p53 can affect immune cell infiltration, cytokine secretion, and inflammatory pathways in the TME and thus significantly affect the antitumor immune response. Mutated p53 cells increase the load of novel antigens and improve the inhibitory response to immune checkpoints. The recovery of WT p53 can induce an antitumor immune response in immune cold tumors, but antitumor immune activation and tumor regression of Mut p53 are heterogeneous in cancer types [106]. At present, other immunotherapies such as adoptive cell therapy, monoclonal antibodies, oncolytic viruses, and immune system regulators have shown efficacy in various types of cancer; however, their efficacy needs to be explored in combination with WT p53 reactivation therapy. In summary, exploring the mechanism through which Mut p53 promotes tumor immune escape is highly significant to further improve the efficacy of antitumor immunity.
Cell-to-cell interactions are crucial for cellular communication; therefore, choosing appropriate tools to evaluate the role of p53 is a prerequisite for exploring the mechanism of tumor immune escape promoted by Mutp53. Bulk RNA-sequencing (RNA-seq), single-cell RNA sequencing (scRNA-seq), and other RNA-seq techniques have emerged to reveal transcriptome heterogeneity. Bulk RNA-sequencing involves transcriptome sequencing of a large number of mixed cells; however, this method could only obtain the average level of gene expression [107]. ScRNA-seq enables the study of gene expression at the single-cell level and has shown advantages in discovering novel cell types and revealing cell heterogeneity; however, single-cell sequencing has lost tissue spatial location information [108]. The latest developments in the spatial transcriptome (ST) can indicate both the level of gene expression and the spatial location of cells [109]. Researchers have tested 16 cell–cell interaction methods by combining scRNA-seq and ST data and have shown that the interactions predicted using different tools are highly random [110]. This tool, named CellTrek, can map single cells to the spatial coordinates of tissue sections according to scRNA-seq and ST data, and study single-cell data with spatial information more flexibly and directly [111].
In summary, there is a very high probability of TP53 mutations in tumors. We described the specific mechanism through which Mut p53 mediates tumor immune escape by forming an immunosuppressive microenvironment, highlighting the key tumor-suppressive role of p53. p53 is considered an important target for antitumor immunotherapy, and various small-molecule drugs have been developed. Targeting Mut p53 therapy in combination with other immunotherapeutic approaches is also being explored. However, many questions remain unanswered, such as the types of TP53 mutations and factors affecting their mutation profiles, the specific mechanisms regulating TP53 mutations, and the mechanisms through which the p53 pathway interacts with other pathways to affect tumor progression. Currently, therapeutic agents targeting Mut p53 are under experimental and clinical investigations. In addition to drug combination therapy, WT p53 drug resistance, complex changes within tumor cells after drug administration to cope with off-target effects and toxicity, and interspecies variability between animals and humans leading to the need for in vitro models to verify drug efficacy are all issues that need attention in future research.
Author contributions
K.Z. and R.W. and M.C. designed the manuscript. K.Z. and F.S. and J.Y. wrote the manuscript. K.Z. and R.X. prepared Figs. 1–4. R.X. and X.L. and T.Z. revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by [National Natural Science Foundation of China] (Grant Number:81802269 and 82260518), [Construction Project of Clinical Medical Research Center from Gansu Provincial Department of Science and Technology] (Grant Number: 21JR7RA390), [Key research and development plan from Gansu Provincial Department of Science and Technology] (Grant Number: 22YF7FA086), [Natural Science Foundation of Gansu Province] (Grant Number: 20JR5RA352 and 22JR5RA918),[Medical Innovation and Development Project of Lanzhou University] (Grant Number: lzuyxcx-2022-183), [Scientific and Technological Development Guiding Plan Project of Lanzhou City] (Grant Number: 2020-ZD-74), [Study on the Optimization, popularization and application of whole process management of lung cancer] (Grant Number:2022LQTD24), Gansu Natural Science Foundation Project (Grant Number:21JR11R076), Lanzhou Chengguan District Scince and Technology Project (Grant Number:2021SHFZ006), Xinrui Tumor Supportive Therapy Research Project (Grant Number: cphcf-2022-013).
Data availability
No Data associated in the manuscript.
Declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kai-li Zhu, Fei Su and Jing-ru Yang have contributed equally to this work.
Contributor Information
Xiao-ling Ling, Email: ldyy_lingxl@lzu.edu.cn.
Tao Zhang, Email: zhangt18@lzu.edu.cn.
References
- 1.O'Donnell JS, Teng MWL, Smyth MJ. Cancer immunoediting and resistance to T cell-based immunotherapy. Nat Rev Clin Oncol. 2019;16(3):151–167. doi: 10.1038/s41571-018-0142-8. [DOI] [PubMed] [Google Scholar]
- 2.Lei X, Lei Y, Li JK, et al. Immune cells within the tumor microenvironment: biological functions and roles in cancer immunotherapy. Cancer Lett. 2020;470:126–133. doi: 10.1016/j.canlet.2019.11.009. [DOI] [PubMed] [Google Scholar]
- 3.Riaz N, Havel JJ, Makarov V, et al. Tumor and microenvironment evolution during Immunotherapy with Nivolumab. Cell. 2017;171(4):934–49.e16. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Q, Wu W, Franca TCC, et al. Immune evasion, a potential mechanism of Trichothecenes: new insights into negative immune regulations. Int J Mol Sci. 2018;19(11):3307. doi: 10.3390/ijms19113307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arneth B. Tumor microenvironment. Medicina. 2019;56(1):15. doi: 10.3390/medicina56010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rui R, Zhou L, He S. Cancer immunotherapies: advances and bottlenecks. Front Immunol. 2023;14:1212476. doi: 10.3389/fimmu.2023.1212476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodis E, Torlai Triglia E, Kwon JYH, et al. Stepwise-edited, human melanoma models reveal mutations' effect on tumor and microenvironment. Science. 2022;376(6592):eabi8175. doi: 10.1126/science.abi8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaur RP, Vasudeva K, Kumar R, et al. Role of p53 gene in breast cancer: focus on mutation spectrum and therapeutic strategies. Curr Pharm Des. 2018;24(30):3566–3575. doi: 10.2174/1381612824666180926095709. [DOI] [PubMed] [Google Scholar]
- 9.Mantovani F, Collavin L, Del Sal G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2019;26(2):199–212. doi: 10.1038/s41418-018-0246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown K, Jenkins LMM, Crooks DR, et al. Targeting mutant p53–R248W reactivates WT p53 function and alters the onco-metabolic profile. Front Oncol. 2022;12:1094210. doi: 10.3389/fonc.2022.1094210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X, Zhang T, Su W, et al. Mutant p53 in cancer: from molecular mechanism to therapeutic modulation. Cell Death Dis. 2022;13(11):974. doi: 10.1038/s41419-022-05408-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou X, Hao Q, Lu H. Mutant p53 in cancer therapy-the barrier or the path. J Mol Cell Biol. 2019;11(4):293–305. doi: 10.1093/jmcb/mjy072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walerych D, Lisek K, Del Sal G. Mutant p53: one, no one, and one hundred thousand. Front Oncol. 2015;5:289. doi: 10.3389/fonc.2015.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang C, Liu J, Xu D, et al. Gain-of-function mutant p53 in cancer progression and therapy. J Mol Cell Biol. 2020;12(9):674–687. doi: 10.1093/jmcb/mjaa040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Agostino S, Sorrentino G, Ingallina E, et al. YAP enhances the pro-proliferative transcriptional activity of mutant p53 proteins. EMBO Rep. 2016;17(2):188–201. doi: 10.15252/embr.201540488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klemke L, Fehlau CF, Winkler N, et al. The gain-of-function p53 R248W mutant promotes migration by STAT3 deregulation in human pancreatic cancer cells. Front Oncol. 2021;11:642603. doi: 10.3389/fonc.2021.642603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kreuzaler P, Panina Y, Segal J, et al. Adapt and conquer: metabolic flexibility in cancer growth, invasion and evasion. Mol Metab. 2020;33:83–101. doi: 10.1016/j.molmet.2019.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lackey DE, Olefsky JM. Regulation of metabolism by the innate immune system. Nat Rev Endocrinol. 2016;12(1):15–28. doi: 10.1038/nrendo.2015.189. [DOI] [PubMed] [Google Scholar]
- 19.Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci. 2016;41(3):211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang C, Liu J, Liang Y, et al. Tumour-associated mutant p53 drives the Warburg effect. Nat Commun. 2013;4:2935. doi: 10.1038/ncomms3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Z, Liu Z, Li M, et al. Increased glycolysis correlates with elevated immune activity in tumor immune microenvironment. EBioMedicine. 2019;42:431–442. doi: 10.1016/j.ebiom.2019.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong X, He X, Wang Y, et al. Warburg effect in colorectal cancer: the emerging roles in tumor microenvironment and therapeutic implications. J Hematol Oncol. 2022;15(1):160. doi: 10.1186/s13045-022-01358-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torrens-Mas M, Cordani M, Mullappilly N, et al. Mutant p53 induces SIRT3/MnSOD axis to moderate ROS production in melanoma cells. Arch Biochem Biophys. 2020;679:108219. doi: 10.1016/j.abb.2019.108219. [DOI] [PubMed] [Google Scholar]
- 24.Wu L, Jin Y, Zhao X, et al. Tumor aerobic glycolysis confers immune evasion through modulating sensitivity to T cell-mediated bystander killing via TNF-α. Cell Metab. 2023;35(9):1580–96.e9. doi: 10.1016/j.cmet.2023.07.001. [DOI] [PubMed] [Google Scholar]
- 25.Juarez D, Fruman DA. Targeting the mevalonate pathway in cancer. Trends Cancer. 2021;7(6):525–540. doi: 10.1016/j.trecan.2020.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parrales A, Thoenen E, Iwakuma T. The interplay between mutant p53 and the mevalonate pathway. Cell Death Differ. 2018;25(3):460–470. doi: 10.1038/s41418-017-0026-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt V, Nagar R, Martinez LA. Control of nucleotide metabolism enables mutant p53's oncogenic gain-of-function activity. Int J Mol Sci. 2017;18(12):2759. doi: 10.3390/ijms18122759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Q, Boire A, Jin X, et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature. 2016;533(7604):493–498. doi: 10.1038/nature18268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gomes AS, Ramos H, Inga A, et al. Structural and drug targeting insights on mutant p53. Cancers. 2021;13(13):3344. doi: 10.3390/cancers13133344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang B, Niu D, Lai L, et al. p53 increases MHC class I expression by upregulating the endoplasmic reticulum aminopeptidase ERAP1. Nat Commun. 2013;4:2359. doi: 10.1038/ncomms3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh M, Saha S, Bettke J, et al. Mutant p53 suppresses innate immune signaling to promote tumorigenesis. Cancer Cell. 2021;39(4):494–508.e5. doi: 10.1016/j.ccell.2021.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asl ER, Rostamzadeh D, Duijf PHG, et al. Mutant P53 in the formation and progression of the tumor microenvironment: friend or foe. Life Sci. 2023;315:121361. doi: 10.1016/j.lfs.2022.121361. [DOI] [PubMed] [Google Scholar]
- 33.Blagih J, Buck MD, Vousden KH. p53, cancer and the immune response. J Cell Sci. 2020 doi: 10.1242/jcs.237453. [DOI] [PubMed] [Google Scholar]
- 34.Agupitan AD, Neeson P, Williams S, et al. P53: a guardian of immunity becomes its saboteur through mutation. Int J Mol Sci. 2020;21(10):3452. doi: 10.3390/ijms21103452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stein Y, Aloni-Grinstein R, Rotter V. Mutant p53-a potential player in shaping the tumor-stroma crosstalk. J Mol Cell Biol. 2019;11(7):600–604. doi: 10.1093/jmcb/mjz071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Addadi Y, Moskovits N, Granot D, et al. p53 status in stromal fibroblasts modulates tumor growth in an SDF1-dependent manner. Cancer Res. 2010;70(23):9650–9658. doi: 10.1158/0008-5472.CAN-10-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cordani M, Pacchiana R, Butera G, et al. Mutant p53 proteins alter cancer cell secretome and tumour microenvironment: involvement in cancer invasion and metastasis. Cancer Lett. 2016;376(2):303–309. doi: 10.1016/j.canlet.2016.03.046. [DOI] [PubMed] [Google Scholar]
- 38.McCubrey JA, Yang LV, Abrams SL, et al. Effects of TP53 mutations and miRs on immune responses in the tumor microenvironment important in Pancreatic cancer progression. Cells. 2022;11(14):2155. doi: 10.3390/cells11142155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farhood B, Najafi M, Mortezaee K. CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: a review. J Cell Physiol. 2019;234(6):8509–8521. doi: 10.1002/jcp.27782. [DOI] [PubMed] [Google Scholar]
- 40.Wang W, Green M, Choi JE, et al. CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569(7755):270–274. doi: 10.1038/s41586-019-1170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Golstein P, Griffiths GM. An early history of T cell-mediated cytotoxicity. Nat Rev Immunol. 2018;18(8):527–535. doi: 10.1038/s41577-018-0009-3. [DOI] [PubMed] [Google Scholar]
- 42.Ben Safta T, Ziani L, Favre L, et al. Granzyme B-activated p53 interacts with Bcl-2 to promote cytotoxic lymphocyte-mediated apoptosis. J Immunol. 2015;194(1):418–428. doi: 10.4049/jimmunol.1401978. [DOI] [PubMed] [Google Scholar]
- 43.Thiery J, Safta TB, Ziani L, et al. Mechanisms of cytotoxic lymphocyte-mediated apoptosis and relationship with the tumor suppressor p53. Crit Rev Immunol. 2015;35(6):433–449. doi: 10.1615/CritRevImmunol.2016015691. [DOI] [PubMed] [Google Scholar]
- 44.Di Minin G, Bellazzo A, Dal Ferro M, et al. Mutant p53 reprograms TNF signaling in cancer cells through interaction with the tumor suppressor DAB2IP. Mol Cell. 2014;56(5):617–629. doi: 10.1016/j.molcel.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 45.Yue X, Wu F, Li Y, et al. Gain of function mutant p53 protein activates AKT through the Rac1 signaling to promote tumorigenesis. Cell Cycle. 2020;19(11):1338–1351. doi: 10.1080/15384101.2020.1749790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sallman DA, McLemore AF, Aldrich AL, et al. TP53 mutations in myelodysplastic syndromes and secondary AML confer an immunosuppressive phenotype. Blood. 2020;136(24):2812–2823. doi: 10.1182/blood.2020006158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buchan SL, Rogel A, Al-Shamkhani A. The immunobiology of CD27 and OX40 and their potential as targets for cancer immunotherapy. Blood. 2018;131(1):39–48. doi: 10.1182/blood-2017-07-741025. [DOI] [PubMed] [Google Scholar]
- 48.Amatore F, Gorvel L, Olive D. Inducible co-stimulator (ICOS) as a potential therapeutic target for anti-cancer therapy. Expert Opin Ther Targets. 2018;22(4):343–351. doi: 10.1080/14728222.2018.1444753. [DOI] [PubMed] [Google Scholar]
- 49.Melaiu O, Lucarini V, Cifaldi L, et al. Influence of the tumor microenvironment on NK cell function in solid tumors. Front Immunol. 2019;10:3038. doi: 10.3389/fimmu.2019.03038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sivori S, Pende D, Quatrini L, et al. NK cells and ILCs in tumor immunotherapy. Mol Aspects Med. 2021;80:100870. doi: 10.1016/j.mam.2020.100870. [DOI] [PubMed] [Google Scholar]
- 51.Duan S, Guo W, Xu Z, et al. Natural killer group 2D receptor and its ligands in cancer immune escape. Mol Cancer. 2019;18(1):29. doi: 10.1186/s12943-019-0956-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Veneziani I, Fruci D, Compagnone M, et al. The BET-bromodomain inhibitor JQ1 renders neuroblastoma cells more resistant to NK cell-mediated recognition and killing by downregulating ligands for NKG2D and DNAM-1 receptors. Oncotarget. 2019;10(22):2151–2160. doi: 10.18632/oncotarget.26736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uddin MB, Roy KR, Hill RA, et al. p53 missense mutant G242A subverts natural killer cells in sheltering mouse breast cancer cells against immune rejection. Exp Cell Res. 2022;417(1):113210. doi: 10.1016/j.yexcr.2022.113210. [DOI] [PubMed] [Google Scholar]
- 54.Pan Y, Yu Y, Wang X, et al. Tumor-associated macrophages in tumor immunity. Front Immunol. 2020;11:583084. doi: 10.3389/fimmu.2020.583084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boutilier AJ, Elsawa SF. Macrophage polarization states in the tumor microenvironment. Int J Mol Sci. 2021;22(13):6995. doi: 10.3390/ijms22136995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu K, Lin K, Li X, et al. Redefining tumor-associated macrophage subpopulations and functions in the tumor microenvironment. Front Immunol. 2020;11:1731. doi: 10.3389/fimmu.2020.01731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi X, Kaller M, Rokavec M, et al. Characterization of a p53/miR-34a/CSF1R/STAT3 feedback loop in colorectal cancer. Cell Mol Gastroenterol Hepatol. 2020;10(2):391–418. doi: 10.1016/j.jcmgh.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fang H, Huang Y, Luo Y, et al. SIRT1 induces the accumulation of TAMs at colorectal cancer tumor sites via the CXCR4/CXCL12 axis. Cell Immunol. 2022;371:104458. doi: 10.1016/j.cellimm.2021.104458. [DOI] [PubMed] [Google Scholar]
- 59.Cooks T, Pateras IS, Jenkins LM, et al. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat Commun. 2018;9(1):771. doi: 10.1038/s41467-018-03224-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wellenstein MD, Coffelt SB, Duits DEM, et al. Loss of p53 triggers WNT-dependent systemic inflammation to drive breast cancer metastasis. Nature. 2019;572(7770):538–542. doi: 10.1038/s41586-019-1450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tian X, Shen H, Li Z, et al. Tumor-derived exosomes, myeloid-derived suppressor cells, and tumor microenvironment. J Hematol Oncol. 2019;12(1):84. doi: 10.1186/s13045-019-0772-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang Y, Zhang M, Zhang Y, et al. 5-Fluorouracil suppresses colon tumor through activating the p53-Fas pathway to sensitize myeloid-derived suppressor cells to FasL(+) cytotoxic T lymphocyte cytotoxicity. Cancers. 2023;15(5):1563. doi: 10.3390/cancers15051563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li L, Xu J, Qiu G, et al. Epigenomic characterization of a p53-regulated 3p22.2 tumor suppressor that inhibits STAT3 phosphorylation via protein docking and is frequently methylated in esophageal and other carcinomas. Theranostics. 2018;8(1):61–77. doi: 10.7150/thno.20893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guha P, Gardell J, Darpolor J, et al. STAT3 inhibition induces Bax-dependent apoptosis in liver tumor myeloid-derived suppressor cells. Oncogene. 2019;38(4):533–548. doi: 10.1038/s41388-018-0449-z. [DOI] [PubMed] [Google Scholar]
- 65.Chang LC, Chiang SK, Chen SE, et al. Heme oxygenase-1 mediates BAY 11–7085 induced ferroptosis. Cancer Lett. 2018;416:124–137. doi: 10.1016/j.canlet.2017.12.025. [DOI] [PubMed] [Google Scholar]
- 66.Zhu H, Klement JD, Lu C, et al. Asah2 represses the p53-Hmox1 axis to protect myeloid-derived suppressor cells from ferroptosis. J Immunol. 2021;206(6):1395–1404. doi: 10.4049/jimmunol.2000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu QR, Tang J, Liao HY, et al. Long non-coding RNA MEG3 mediates the miR-149-3p/FOXP3 axis by reducing p53 ubiquitination to exert a suppressive effect on regulatory T cell differentiation and immune escape in esophageal cancer. J Transl Med. 2021;19(1):264. doi: 10.1186/s12967-021-02907-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang P, Li QJ, Feng Y, et al. TGF-β-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell. 2012;22(3):291–303. doi: 10.1016/j.ccr.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Biffi G, Tuveson DA. Diversity and biology of cancer-associated fibroblasts. Physiol Rev. 2021;101(1):147–176. doi: 10.1152/physrev.00048.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu Q, Yu B, Tian Y, et al. P53 mutant p53(N236S) regulates cancer-associated fibroblasts properties through Stat3 pathway. Onco Targets Ther. 2020;13:1355–1363. doi: 10.2147/OTT.S229065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cui Y, Guo G. Immunomodulatory function of the tumor suppressor p53 in host immune response and the tumor microenvironment. Int J Mol Sci. 2016;17(11):1942. doi: 10.3390/ijms17111942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fang Z, Meng Q, Xu J, et al. Signaling pathways in cancer-associated fibroblasts: recent advances and future perspectives. Cancer Commun. 2023;43(1):3–41. doi: 10.1002/cac2.12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oh IR, Raymundo B, Kim M, et al. Mesenchymal stem cells co-cultured with colorectal cancer cells showed increased invasive and proliferative abilities due to its altered p53/TGF-β1 levels. Biosci Biotechnol Biochem. 2020;84(2):256–267. doi: 10.1080/09168451.2019.1676692. [DOI] [PubMed] [Google Scholar]
- 74.Lin SY, Dolfi SC, Amiri S, et al. P53 regulates the migration of mesenchymal stromal cells in response to the tumor microenvironment through both CXCL12-dependent and -independent mechanisms. Int J Oncol. 2013;43(6):1817–1823. doi: 10.3892/ijo.2013.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Velletri T, Xie N, Wang Y, et al. P53 functional abnormality in mesenchymal stem cells promotes osteosarcoma development. Cell Death Dis. 2016;7(1):e2015. doi: 10.1038/cddis.2015.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hardwick NR, Frankel P, Ruel C, et al. p53-Reactive T cells are associated with clinical benefit in patients with platinum-resistant epithelial ovarian cancer after treatment with a p53 vaccine and gemcitabine chemotherapy. Clin Cancer Res. 2018;24(6):1315–1325. doi: 10.1158/1078-0432.CCR-17-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van de Donk N, Zweegman S. T-cell-engaging bispecific antibodies in cancer. Lancet. 2023;402(10396):142–158. doi: 10.1016/S0140-6736(23)00521-4. [DOI] [PubMed] [Google Scholar]
- 78.Hsiue EH, Wright KM, Douglass J, et al. Targeting a neoantigen derived from a common TP53 mutation. Science. 2021;371(6533):10. doi: 10.1126/science.abc8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shi D, Jiang P. A different facet of p53 function: regulation of immunity and inflammation during tumor development. Front Cell Dev Biol. 2021;9:762651. doi: 10.3389/fcell.2021.762651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chasov V, Zaripov M, Mirgayazova R, et al. Promising new tools for targeting p53 mutant cancers: humoral and cell-based immunotherapies. Front Immunol. 2021;12:707734. doi: 10.3389/fimmu.2021.707734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.García-Garrido E, Cordani M, Somoza Á. Modified gold nanoparticles to overcome the chemoresistance to gemcitabine in mutant p53 cancer cells. Pharmaceutics. 2021;13(12):2067. doi: 10.3390/pharmaceutics13122067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ma J, Murphy M, O'Dwyer PJ, et al. Biochemical changes associated with a multidrug-resistant phenotype of a human glioma cell line with temozolomide-acquired resistance. Biochem Pharmacol. 2002;63(7):1219–1228. doi: 10.1016/S0006-2952(02)00876-6. [DOI] [PubMed] [Google Scholar]
- 83.Fruehauf JP, Brem H, Brem S, et al. In vitro drug response and molecular markers associated with drug resistance in malignant gliomas. Clin Cancer Res. 2006;12(15):4523–4532. doi: 10.1158/1078-0432.CCR-05-1830. [DOI] [PubMed] [Google Scholar]
- 84.Srivenugopal KS, Shou J, Mullapudi SR, et al. Enforced expression of wild-type p53 curtails the transcription of the O(6)-methylguanine-DNA methyltransferase gene in human tumor cells and enhances their sensitivity to alkylating agents. Clin Cancer Res. 2001;7(5):1398–1409. [PubMed] [Google Scholar]
- 85.Kim SS, Rait A, Kim E, et al. A nanoparticle carrying the p53 gene targets tumors including cancer stem cells, sensitizes glioblastoma to chemotherapy and improves survival. ACS Nano. 2014;8(6):5494–5514. doi: 10.1021/nn5014484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kong N, Tao W, Ling X, et al. Synthetic mRNA nanoparticle-mediated restoration of p53 tumor suppressor sensitizes p53-deficient cancers to mTOR inhibition. Sci Transl Med. 2019 doi: 10.1126/scitranslmed.aaw1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Prabha S, Sharma B, Labhasetwar V. Inhibition of tumor angiogenesis and growth by nanoparticle-mediated p53 gene therapy in mice. Cancer Gene Ther. 2012;19(8):530–537. doi: 10.1038/cgt.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mei Y, Qin X, Yang Z, et al. Engineered a dual-targeting HA-TPP/A nanoparticle for combination therapy against KRAS-TP53 co-mutation in gastrointestinal cancers. Bioact Mater. 2024;32:277–291. doi: 10.1016/j.bioactmat.2023.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Koo N, Sharma AK, Narayan S. Therapeutics targeting p53-MDM2 interaction to induce cancer cell death. Int J Mol Sci. 2022;23(9):5005. doi: 10.3390/ijms23095005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou X, Singh M, Sanz Santos G, et al. Pharmacologic activation of p53 triggers viral mimicry response thereby abolishing tumor immune evasion and promoting antitumor immunity. Cancer Discov. 2021;11(12):3090–3105. doi: 10.1158/2159-8290.CD-20-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Han Y, Liu D, Li L. PD-1/PD-L1 pathway: current researches in cancer. Am J Cancer Res. 2020;10(3):727–742. [PMC free article] [PubMed] [Google Scholar]
- 92.Wang HQ, Mulford IJ, Sharp F, et al. Inhibition of MDM2 promotes antitumor responses in p53 wild-type cancer cells through their interaction with the immune and stromal microenvironment. Cancer Res. 2021;81(11):3079–3091. doi: 10.1158/0008-5472.CAN-20-0189. [DOI] [PubMed] [Google Scholar]
- 93.Xiao Y, Chen J, Zhou H, et al. Combining p53 mRNA nanotherapy with immune checkpoint blockade reprograms the immune microenvironment for effective cancer therapy. Nat Commun. 2022;13(1):758. doi: 10.1038/s41467-022-28279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shouval R, Alarcon Tomas A, Fein JA, et al. Impact of TP53 genomic alterations in large B-cell lymphoma treated with CD19-chimeric antigen receptor T-cell therapy. J Clin Oncol. 2022;40(4):369–381. doi: 10.1200/JCO.21.02143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhu G, Pan C, Bei JX, et al. Mutant p53 in cancer progression and targeted therapies. Front Oncol. 2020;10:595187. doi: 10.3389/fonc.2020.595187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fiorini C, Cordani M, Padroni C, et al. Mutant p53 stimulates chemoresistance of pancreatic adenocarcinoma cells to gemcitabine. Biochim Biophys Acta. 2015;1853(1):89–100. doi: 10.1016/j.bbamcr.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 97.Dong ZY, Zhong WZ, Zhang XC, et al. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin Cancer Res. 2017;23(12):3012–3024. doi: 10.1158/1078-0432.CCR-16-2554. [DOI] [PubMed] [Google Scholar]
- 98.Li Y, Zhang MC, Xu XK, et al. Functional diversity of p53 in human and wild animals. Front Endocrinol. 2019;10:152. doi: 10.3389/fendo.2019.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dong P, Xiong Y, Yu J, et al. Control of PD-L1 expression by miR-140/142/340/383 and oncogenic activation of the OCT4-miR-18a pathway in cervical cancer. Oncogene. 2018;37(39):5257–5268. doi: 10.1038/s41388-018-0347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.D'Orazi G, Cordani M, Cirone M. Oncogenic pathways activated by pro-inflammatory cytokines promote mutant p53 stability: clue for novel anticancer therapies. Cell Mol Life Sci. 2021;78(5):1853–1860. doi: 10.1007/s00018-020-03677-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xiu W, Ma J, Lei T, et al. AG490 reverses phenotypic alteration of dendritic cells by bladder cancer cells. Oncol Lett. 2018;16(3):2851–2856. doi: 10.3892/ol.2018.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Groot J, Ott M, Wei J, et al. A first-in-human phase I trial of the oral p-STAT3 inhibitor WP1066 in patients with recurrent malignant glioma. CNS Oncol. 2022;11(2):Cns87. doi: 10.2217/cns-2022-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cai Z, Wang J, Li Y, et al. Overexpressed Cyclin D1 and CDK4 proteins are responsible for the resistance to CDK4/6 inhibitor in breast cancer that can be reversed by PI3K/mTOR inhibitors. Sci China Life Sci. 2023;66(1):94–109. doi: 10.1007/s11427-021-2140-8. [DOI] [PubMed] [Google Scholar]
- 104.Goldman JW, Rosen LS, Tolcher AW, et al. Phase 1 and pharmacokinetic study of LY3007113, a p38 MAPK inhibitor, in patients with advanced cancer. Invest New Drugs. 2018;36(4):629–637. doi: 10.1007/s10637-017-0532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Talaei S, Mellatyar H, Asadi A, et al. Spotlight on 17-AAG as an Hsp90 inhibitor for molecular targeted cancer treatment. Chem Biol Drug Des. 2019;93(5):760–786. doi: 10.1111/cbdd.13486. [DOI] [PubMed] [Google Scholar]
- 106.Carlsen L, Zhang S, Tian X, et al. The role of p53 in anti-tumor immunity and response to immunotherapy. Front Mol Biosci. 2023;10:1148389. doi: 10.3389/fmolb.2023.1148389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Thind AS, Monga I, Thakur PK, et al. Demystifying emerging bulk RNA-Seq applications: the application and utility of bioinformatic methodology. Brief Bioinform. 2021 doi: 10.1093/bib/bbab259. [DOI] [PubMed] [Google Scholar]
- 108.Lu J, Sheng Y, Qian W, et al. scRNA-seq data analysis method to improve analysis performance. IET Nanobiotechnol. 2023;17(3):246–256. doi: 10.1049/nbt2.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Asp M, Bergenstråhle J, Lundeberg J. Spatially resolved transcriptomes-next generation tools for tissue exploration. BioEssays. 2020;42(10):e1900221. doi: 10.1002/bies.201900221. [DOI] [PubMed] [Google Scholar]
- 110.Liu Z, Sun D, Wang C. Evaluation of cell-cell interaction methods by integrating single-cell RNA sequencing data with spatial information. Genome Biol. 2022;23(1):218. doi: 10.1186/s13059-022-02783-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wei R, He S, Bai S, et al. Spatial charting of single-cell transcriptomes in tissues. Nat Biotechnol. 2022;40(8):1190–1199. doi: 10.1038/s41587-022-01233-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sorrentino A, Menevse AN, Michels T, Volpin V, Durst FC, Sax J, Xydia M, Hussein A, Stamova S, Spoerl S, Heuschneider N, Muehlbauer J, Jeltsch KM, Rathinasamy A, Werner-Klein M, Breinig M, Mikietyn D, Kohler C, Poschke I, Purr S, Reidell O, Martins Freire C, Offringa R, Gebhard C, Spang R, Rehli M, Boutros M, Schmidl C, Khandelwal N, Beckhove P. Salt-inducible kinase 3 protects tumor cells from cytotoxic T-cell attack by promoting TNF-induced NF-κB activation. J Immunother Cancer. 2022;10(5):e004258. doi: 10.1136/jitc-2021-004258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No Data associated in the manuscript.