Abstract
At least three Sarcocystis species (S. falcatula, S. halieti and S. wobeseri–like) have been detected infecting raptorial birds. By histopathology and PCR-sequencing of the ITS1 marker, S. halieti was detected in a bearded vulture (Gypaetus barbatus) and a black kite (Milvus migrans) from the Catalonia region in North Spain. The 241 bp-long sequences obtained from the Sarcocystis organisms detected in both raptors showed 97.5–99.6% and 97.9–100% similarity with those of previously identified S. halieti; also, the phylogenetic trees generated placed the identified sequences together with other sequences of S. halieti available in GenBank. In sum, the description of the bearded vulture as a new intermediate host for S. halieti adds new insights on the complex epidemiology of the genus involving avian hosts.
Keywords: Apicomplexa, Sarcocystis, Raptorial birds, ITS1, Phylogeny
Introduction
Sarcocystis (Apicomplexa) genus comprises more than 200 species of cyst-forming obligate intracellular protozoan parasites that require a two-host prey-predator life cycle. Chronic stage as tissue cysts are developed mostly in muscular or nervous tissues of the intermediate hosts (IH); carnivore definitive hosts (DH) pray on IH and sexual phase occurs in their small intestine, giving place to oocysts/sporocysts that constitute the environmentally resistant stages and consequently serve as the source of infection for the IH (Dubey et al. 2016).
Due to the predatory and scavenger role of raptorial birds they greatly contribute to the health of ecosystems (Donázar et al. 2016); raptors are involved in the life cycle of several Sarcocystis species both as DH and IH (Dubey et al. 2016; Máca and González-Solís 2021). Around 30 Sarcocystis spp. use birds as IH, among them, at least three (S. falcatula, S. halieti, S. wobeseri–like) have been recognized infecting raptorial birds (Dubey et al. 2016; Prakas et al. 2021; Shadbolt et al. 2021).
While most of Sarcocystis infections are asymptomatic, a number of lethal cases mostly related to neurological sarcocystosis with the presence of schizonts and merozoites in such lesions have been described in raptors (Dubey et al. 2016; Maier-Sam et al. 2021); nevertheless, the global occurrence and impact of Sarcocystis genus in the health status of birds of prey is unknown.
Until date the only available data on the occurrence of Sarcocystis infections in avian species in Spain was reported by Cardells-Peris et al. (2020) who described Sarcocystis spp. infection by histological examination in 3 song thrushes (Turdus philomelos) without additional characterization in Castellón province (eastern), and by Prakas et al. (2021) who molecularly identified S. halieti infection in a western marsh harrier (Circus aeruginosus) and a black kite (Milvus migrans) in the northern region of Navarre.
In the present study, we identified the bearded vulture (Gypaetus barbatus) as a new intermediate host for S. halieti, a threatened species which population in the Iberian Peninsula is nowadays mainly restricted to the Pyrenees, even if numerous conservation actions have been implemented the last decades (BirdLife International, 2021).
Materials and methods
Following the procedures included under the health surveillance program for protected species in Catalonia (NE Spain), the carcass of a black kite recovered in May 2018, and a bearded vulture found dead in December of the same year were submitted to Vallcalent Rehabilitation Center (Lleida, Catalonia). The black kite corpse (MMi18002) was found in the village of “Les Garrigues” (Lleida), with a good body condition, burns in the plantar surface of both feet, and extensive hemorrhages in the coelomic cavity, consistent with electrocution. The bearded vulture (GB18001), found in the village of “Les” (Lleida), had bilateral cataracts, dehydration, poor body condition, and a proventricular perforation caused by a bone fragment. From both animals, samples of brain, lung, heart, liver, spleen, intestine, kidney and skeletal muscle were fixed in 4% neutral buffered formalin for histopathology.
Histopathology
Fixed tissue samples were routinely processed and 4 µm sections were stained with haematoxylin & eosin (H&E) for microscopic evaluation. In both cases, advanced autolysis harmed the evaluation of tissue sections, especially for the digestive tract, in which assessment was not possible. Nevertheless, no relevant microscopic alterations were described in the rest of the studied organ samples, except for the presence of protozoan-like tissue cysts in the skeletal muscle, which motivated the present study.
Genetic characterization
DNA extraction from paraffin block-embedded tissues
Portions (10 mg in average) of paraffin block where at least 3 sarcocysts had been observed by H&E-staining were excised using sterile blades and placed in empty 1.5 mL vials. Five cycles of incubation of each sample in 1 mL of xylol at room temperature for 5 min, centrifugation at 15871 xg for 5 min, and the removal of the supernatant were conducted to remove the paraffin wax. This was followed by two cycles of washing in isopropanol, followed by centrifugation at 15871 xg for 5 min, and the removal of the supernatant. The sample was then left to dry out overnight at room temperature. DNA was extracted applying a protocol designed for skin, hair, and feathers using a lysis buffer (0.1 M Tris, 0.005 M EDTA, 0.2% SDS, 0.2 M NaCl; pH = 8.5) (Laird et al. 1991). Briefly, each tissue was incubated in 100 µL of lysis buffer with 3 µL proteinase K (Thermo Fisher Scientific, Vilnius, Lithuania) for 3 h at 56°C followed by centrifugation for 10 min at 9391 xg. The supernatant was transferred to a new tube and 96% EtOH was added to it. The tubes were again centrifuged for 10 min at 9391 xg and the 96% EtOH was replaced by 70% EtOH. After 5 min of centrifugation at 9391 xg, supernatant was removed, then the samples were air-dried overnight at room temperature and dissolved in 100 µL 1X TE buffer.
PCR and sequencing
The nested PCR approach with several different primer pairs targeting ITS1 genomic region was used for the identification of Sarcocystis spp. (Table 1). DNA from individual sarcocysts acquired during our previous investigations (Juozaitytė-Ngugu and Prakas 2023) and nuclease-free water were used as positive and negative controls, respectively. For the amplification DreamTaq PCR Master Mix (Thermo Fisher Scientific Baltics, Vilnius, Lithuania) was used according to the manufacturer’s instructions. Cycling conditions were as: initial denaturation step for 5 min at 95°C, 35 cycles of 35 s at 94°C, 45 s of annealing at 51–65°C depending on the primer pair, 60 s at 72°C, and final extension for 5 min at 72°C. PCR products were observed in 1% agarose gel and purified using Exonuclease I and FastAP Thermosensitive Alkaline Phosphatase (Thermo Fisher Scientific Baltics). Amplified products of the second nested PCR step were sequenced directly with the 3500 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) using the same forward and reverse primers as for PCR. The obtained ITS1 sequences were truncated discarding nucleotides at the binding sites of the primers and deposited in GenBank with accession numbers OR059466–OR059469.
Table 1.
The list of primers used for the amplification of ITS1 region of Sarcocystis spp
| Primer name | Sequence | Nested PCR | Annealing temperature (ºC) | Product size (bp) | Target species | |
|---|---|---|---|---|---|---|
| NITSpauk1c | TGTCCGGAATGGGAAGTTTT | I | External pair | 51 | ~ 475–500 | Sarcocystis spp. |
| NITSpauk2c | ACACCATCCDAAATTCTCAG | |||||
| NITSpauk3c | GGAAGGATCATTCACACGTT | Internal pair | 53 | ~ 260–280 | ||
| NITSpauk4c | ATCACTGCAAGTTCCAACCA | |||||
| SU1Fa | GATTGAGTGTTCCGGTGAATTATT | II | External pair | 59 | ~ 1050–1150 | |
| 5.8SR2a | AAGGTGCCATTTGCGTTCAGAA | |||||
| GsShalFb | GATAATTGACTTTACGCGCCATTAC | Internal pair | 65 | 643–647 | S. halieti | |
| GsShalR2b | CCATCCCTTTTTCTAAAGGAGGTC | |||||
| SU1Fa | GATTGAGTGTTCCGGTGAATTATT | III | External pair | 59 | ~ 1050–1150 | |
| 5.8SR2a | AAGGTGCCATTTGCGTTCAGAA | |||||
| ShalBFc | TTTGTGGTTGGAACTTGCAG | Internal pair | 53 | 260–264 | ||
| ShalBRc | GACCTCCCCTCGACGATAAT | |||||
| ShalCFc | AACAACTGAATCCCCCGATA | IV | External pair | 53 | 490–494 | |
| ShalCRc | CCACTGCTAATTTCATCCCTCT | |||||
| ShalBFc | TTTGTGGTTGGAACTTGCAG | Internal pair | 53 | 260–264 | ||
| ShalBRc | GACCTCCCCTCGACGATAAT | |||||
| SahalD1ac | ATGGTGTAACCAGGGGATCA | V | External pair | 53 | 473–474 | |
| SahalD1bc | CAAGACATCCATCGCTGAAA | |||||
| SahalD1cc | GGTCGTTCTCCTCTTTTCAGG | Internal pair | 55 | 282 | ||
| SahalD1dc | TGAACAGCTTCGTTGAGACG | |||||
| SahalD2ac | CGGTGGCATCATCCTTTTT | VI | External pair | 51 | 454–455 | |
| 5.8SR2a | AAGGTGCCATTTGCGTTCAGAA | Sarcocystis spp. | ||||
| SahalD2bc | GAGGGATGAAATTAGCAGTGG | Internal pair | 53 | 283 | S. halieti | |
| SahalD2cc | CAAGACATCCATCGCTGAAA | |||||
Phylogenetic analyses
The resulted sequences of the ITS1 region were compared with those of various Sarcocystis spp. by Nucleotide BLAST analysis (http://blast.ncbi.nlm.nih.gov/, accessed on 20 May 2023). Different haplotypes of ITS1 of previously identified Sarcocystis species were determined using FaBox v. 1.5 (Villesen 2007). Multiple sequence alignments were generated using ClustalW algorithm implemented into MEGA7 (Kumar et al. 2016). Phylogenetic trees were constructed using maximum likelihood (ML) method. The bootstrap test with 1000 replicates was used to test the robustness of the suggested phylogeny.
Results
Histopathological findings
Sarcocystis-like mature tissue cysts were observed in the tissues of both the black kite (3.4 sarcocysts/cm2 of tissue) and the bearded vulture (4.2 sarcocysts/cm2 of tissue). In both specimens, all examined cysts (n = 10) had thin (< 2 µm) wall without visible protrusions by light microscopy, apparent absence of septae and contained numerous bradyzoites (Fig. 1). No myositis nor other histological lesions were observed.
Fig. 1.
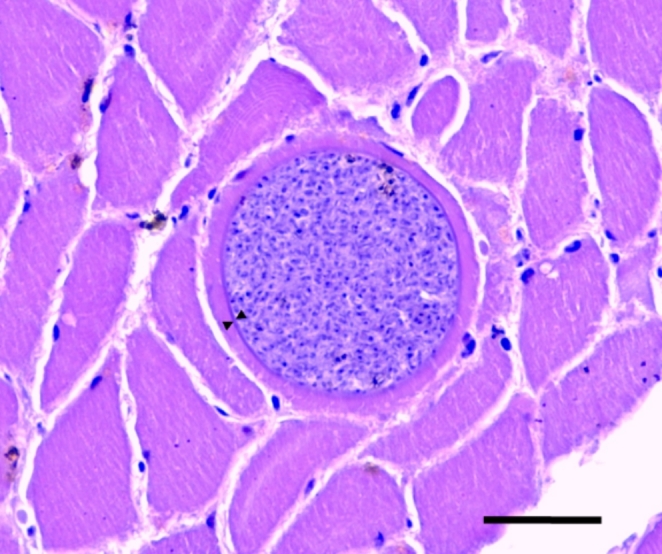
Micrograph of mature Sarcocystis sarcocyst in the breast muscle of a bearded vulture from Spain. Arrowheads point to the thin wall. Bar = 20 μm
Molecular results
Amplification was successful only using primer sets I and V (Table 1). In both cases, using external and internal primers the length of resulted fragments was no longer than 500 bp and 300 bp, respectively. The comparison of 221 bp and 241 bp sequences, obtained using NITSpauk3/NITSpauk4 and SahalD1c/SahalD1d pairs, confirmed the presence of S. halieti in paraffin block-embedded tissues of the bearded vulture and the black kite examined.
The 221 bp sequences obtained from both hosts were 100% identical, while 241 bp sequences differed by one SNP which was caused by A ↔ G transition. In case of 221 bp sequences, the BLAST analysis displayed 96.4–100% similarity with S. halieti, 95.0% similarity with Sarcocystis sp. (MW160469) from cinnamon skua (Stercorarius chilensis), 94.6% similarity with Sarcocystis sp. (KY348755) from Cooper’s hawk (Accipiter cooperii), 94.1% similarity with Sarcocystis sp. (MZ707151) from common raven (Corvus corax), 93.3% similarity with S. corvusi from western jackdaw (Coloeus monedula), and 92.4–92.8% similarity with S. columbae from birds of family Columbidae and Laridae. The comparison of 241 bp sequences from the bearded vulture and the black kite demonstrated 97.5–99.6% and 97.9–100% similarity with those of S. halieti. Higher than 90% similarity was observed when in present study generated 241 bp sequences were compared with Sarcocystis sp. (MZ707151) (96.3–96.7%), with Sarcocystis sp. (MW160469) (95.8–96.2%), with S. columbae (92.1–93.4%), with Sarcocystis sp. (KY348755) (92.6–93.0%) and with S. corvusi (90.2–90.6%).
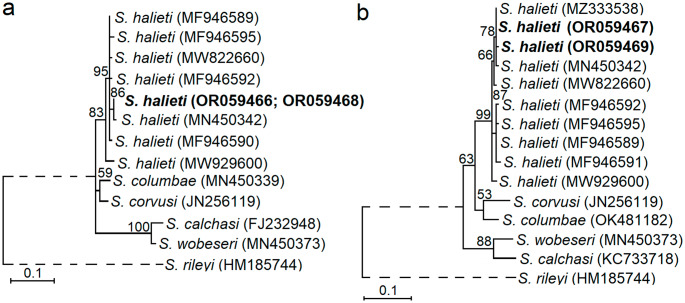
Based on the phylogenetic trees generated using ITS1 sequences and ML method, S. halieti isolates obtained from the bearded vulture and the black kite were placed together with other sequences of S. halieti available in GenBank (Fig. 2). Furthermore, despite the short-length sequences of S. halieti obtained, a high support (83, 99 bootstrap values) was given for the grouping of S. halieti isolates into one cluster.
Fig. 2.
Phylogenetic analysis showing the placement of Sarcocystis halieti isolated from histological samples of birds of prey in Catalonia region of Spain. All possible haplotypes of S. halieti were included in the analysis. Trees were generated using ITS1 fragments amplified with NITSpauk3/NITSpauk4 (a) and SahalD1c/SahalD1d (b) primer pairs. Sequences were obtained from the bearded vulture (OR059466-OR059467) and the black kite (OR059468-OR059469). Based on analysis “Find Best-Fit Substitution Model (ML)” conducted in MEGA7, Tamura 3-parameter + I and Tamura 3-parameter evolutionary models were set for phylogenetic investigation
Discussion
Present paper constitutes the first report of Sarcocystis infection in bearded vulture and add new insights into the complex epidemiology of the genus involving avian hosts. A previous paper had reported the presence of S. halieti in the muscle tissues of black kite in Spain (Prakas et al. 2021). Sarcocystis falcatula and S. calchasi have a recognized unusual wide host rage (Dubey et al. 2016); this might be the case of S. halieti, given the number of recent papers reporting its occurrence in tissues of black headed gull (Larus ridibundus), black kite (Milvus migrans), common gull (Larus canus), common raven (Corvus corax), common starling (Sturnus vulgaris), great cormorant (Phalacrocorax carbo), herring gull (Larus argentatus), hooded crow (Corvus cornix), Kelp gull (Larus dominicanus), little owl (Athene noctua), Manx shearwater (Puffinus puffinus), Neotropic cormorant (Phalacrocorax brasilianus), sharp-shinned hawk (Accipiter striatus), western marsh harrier (Circus aeruginosus), and potentially the Chilean skuas (Stercorarius chilensis), which belong to different avian orders (Accipitriformes, Charadriiformes, Passeriformes, Procellariiformes, Strigiformes and Suliformes) (Prakas et al. 2018; Acosta et al. 2021; Juozaitytė-Ngugu et al. 2022; Llano et al. 2022; Máca and González-Solís 2022; Prakas et al. 2021; Sato et al. 2022; Juozaitytė-Ngugu and Prakas 2023). In addition, recent molecular results suggest the role of predatory birds of the Accipitridae family as DH of S. halieti (Šukytė et al. 2023).
Microscopical examination of the slides revealed the presence of thin-walled sarcocysts (Fig. 1), findings in agreement with the observations by light microscopy carried out by Prakas et al. (2020) in herring gull tissue samples. Thin-walled sarcocysts are formed by several Sarcocystis species characterized by a bird-bird life cycle; therefore, microscopical discrimination between these Sarcocystis spp. is unfeasible (Sneideris et al. 2022). In the present study we did not perform ultrastructural examination given the detailed TEM description provided by Prakas et al. (2018) for S. halieti in great cormorant from Lithuania. Despite the short ITS1 fragments used for molecular analysis, S. halieti was successfully identified in this study (Fig. 2). Notably, S. halieti has high intraspecific variation within ITS1, which might be related with a wide host range of this species (Juozaitytė-Ngugu and Prakas 2023; Šukytė et al. 2023). However, comparing ITS1 sequences of S. halieti with some closely related Sarcocystis spp. relatively low interspecific differences are observed (Juozaitytė-Ngugu and Prakas 2023). Therefore, for the within-species genetic discrimination of S. halieti more variable genetic markers should be identified.
Conclusion
The description of the bearded vulture as a new intermediate host for S. halieti adds new insights on the complex epidemiology of the genus involving avian hosts and warrant the interest of further investigations to unravel the life cycles of Sarcocystis spp. infecting avian hosts and their impact in the health of raptorial bird populations.
Acknowledgements
Authors thank Forestal Catalana, Ministry of Climate Action, Food and Rural Agenda (Government of Catalonia), for funding the rehabilitation and postmortem (gross) examination of the cases included in this study.
Authors’ contributions
Conceptualization: P.P. and R.C.B. Methodology: P.P. Formal analysis: P.P., M.I., D.S. and R.C.B. Investigation: P.P., J.E., R.V., and R.C.B. Resources: P.P., J.E., R.V., and O.N.-F. Data curation: P.P., M.I., and D.S. Writing—original draft preparation: P.P. and R.C.B. Writing—review and editing: P.P., J.E., R.V., O.N.-F., M.I., D.S., I.M. and R.C.B. Supervision: P.P. and R.C.B. Funding acquisition: P.P. and I.M. All authors have read and agreed to the published version of the manuscript.
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature. No specific funding was received for the present study.
Data Availability
DNA sequences of the ITS1 marker have been deposited in GenBank with accession numbers OR059466–OR059469.
Declarations
Ethics approval and consent to participate
No approval of research ethics committees was required for this study because procedures were conducted using samples taken from avian carcasses submitted for post mortem examination at the Vallcalent Rehabilitation Center and Servei d’Ecopatologia de Fauna Salvatge (SEFaS) from the Autonomous University of Barcelona.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Conflict of interest
The authors declare no conflicts of interest.
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Acosta ICL, Gennari SM, Llano HAB, Muñoz-Leal S, Soares RM. Molecular characterization of New Haplotype of Genus Sarcocystis in Seabirds from Magdalena Island, Southern Chile. Anim (Basel) 2021;11(2):245. doi: 10.3390/ani11020245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BirdLife I (2021) Gypaetus barbatus. The IUCN Red List of Threatened Species 2021 eT22695174A154813652. 10.2305/IUCN.UK.2021-3.RLTS.T22695174A154813652.en
- Cardells-Peris J, Gonzálvez M, Ortega-Porcel J, Ruiz de Ybáñez MR, Martínez-Herrero MC, Garijo-Toledo MM. Parasitofauna survey of song thrushes (Turdus philomelos) from the eastern part of Spain. Parasitol Int. 2020;79:102176. doi: 10.1016/j.parint.2020.102176. [DOI] [PubMed] [Google Scholar]
- Donázar JA, Cortés-Avizanda A, Fargallo JA, Margalida A, Moleón M, Morales-Reyes Z, Moreno-Opo R, Pérez-García JM, Sánchez-Zapata JA, Zuberogoitia I, Serrano D. Roles of raptors in a changing world: from flagships to providers of key ecosystem services. Ardeola. 2016;63(1):181–234. doi: 10.13157/arla.63.1.2016.rp8. [DOI] [Google Scholar]
- Dubey JP, Calero-Bernal R, Rosenthal BM, Speer CA, Fayer R. Sarcocystosis of animals and humans. 2. Boca Raton: CRC Press; 2016. [Google Scholar]
- Gjerde B. Molecular characterisation of Sarcocystis rileyi from a common eider (Somateria mollissima) in Norway. Parasitol Res. 2014;113(9):3501–3509. doi: 10.1007/s00436-014-4062-y. [DOI] [PubMed] [Google Scholar]
- Juozaitytė-Ngugu E, Prakas P. The richness of Sarcocystis species in the common gull (Larus canus) and black-headed Gull (Larus ridibundus) from Lithuania. Parasitologia. 2023;3(2):172–180. doi: 10.3390/parasitologia3020018. [DOI] [Google Scholar]
- Juozaitytė-Ngugu E, Švažas S, Šneideris D, Rudaitytė-Lukošienė E, Butkauskas D, Prakas P. The role of birds of the family Corvidae in transmitting Sarcocystis protozoan parasites. Animals. 2021;11(11):3258. doi: 10.3390/ani11113258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juozaitytė-Ngugu E, Butkauskas D, Švažas S, Prakas P. Investigations on Sarcocystis species in the leg muscles of the bird family Corvidae in Lithuania. Parasitol Res. 2022;121(2):703–711. doi: 10.1007/s00436-021-07409-z. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird PW, Zijderveld A, Linders K, Rudnicki MA, Jaenisch R, Berns A. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19(15):4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano HAB, Zavatieri Polato H, Borges Keid L, Ferreira de Souza Oliveira TM, Zwarg T, de Oliveira AS, Sanches TC, Joppert AM, Gondim LFP, Martins Soares R. Molecular screening for Sarcocystidae in muscles of wild birds from Brazil suggests a plethora of intermediate hosts for Sarcocystis falcatula. Int J Parasitol Parasites Wildl. 2022;17:230–238. doi: 10.1016/j.ijppaw.2022.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Máca O, González-Solís D, Aves Musophagidae. Sarcocystis cristata sp. nov. (Apicomplexa, Sarcocystidae) in the imported great blue turaco Corythaeola cristata. Parasit Vectors. 2021;14(1):56. doi: 10.1186/s13071-020-04553-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Máca O, González-Solís D (2022) Role of three bird species in the life cycle of two Sarcocystis spp. (Apicomplexa, Sarcocystidae) in the Czech Republic. Int J Parasitol Parasites Wildl. 17:133–137. doi: 10.1016/j.ijppaw.2022.01.002.Maier-Sam K, Kaiponen T, Schmitz A, Schulze C, Bock S, Hlinak A, Olias P. (2021) Encephalitis Associated with Sarcocystis halieti Infection in a Free-Ranging Little Owl (Athene noctua). J Wildl Dis. 57(3):712–714. doi: 10.7589/JWD-D-20-00184 [DOI] [PubMed]
- Maier-Sam K, Kaiponen T, Schmitz A, Schulze C, Bock S, Hlinak A, Olias P. Encephalitis associated with Sarcocystis halieti infection in a free-ranging little owl (Athene noctua) J Wildl Dis. 2021;57(3):712–714. doi: 10.7589/JWD-D-20-00184. [DOI] [PubMed] [Google Scholar]
- Prakas P, Butkauskas D, Švažas S, Stanevičius V. Morphological and genetic characterisation of Sarcocystis halieti from the great cormorant (Phalacrocorax carbo) Parasitol Res. 2018;117(11):3663–3667. doi: 10.1007/s00436-018-6083-4. [DOI] [PubMed] [Google Scholar]
- Prakas P, Butkauskas D, Juozaitytė-Ngugu E. Molecular identification of four Sarcocystis species in the herring gull, Larus argentatus, from Lithuania. Parasit Vectors. 2020;13(1):2. doi: 10.1186/s13071-019-3869-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakas P, Bea A, Juozaitytė-Ngugu E, Olano I, Villanúa D, Švažas S, Butkauskas D. Molecular identification of Sarcocystis halieti in the muscles of two species of birds of prey from Spain. Parasit Vectors. 2021;14(1):414. doi: 10.1186/s13071-021-04921-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato AP, da Silva TCE, de Pontes TP, Sanches AWD, Prakas P, Locatelli-Dittrich R. Molecular characterization of Sarcocystis spp. in seabirds from southern Brazil. Parasitol Int. 2022;90:102595. doi: 10.1016/j.parint.2022.102595. [DOI] [PubMed] [Google Scholar]
- Shadbolt T, Pocknell A, Sainsbury AW, Egerton-Read S, Blake DP. Molecular identification of Sarcocystis wobeseri-like parasites in a new intermediate host species, the white-tailed sea eagle (Haliaeetus albicilla) Parasitol Res. 2021;120(5):1845–1850. doi: 10.1007/s00436-021-07103-0. [DOI] [PubMed] [Google Scholar]
- Sneideris D, Stalpes M, Butkauskas D, Prakas P. A novel RFLP method for identification of morphologically similar avian sarcocystis species. Parasitol Res. 2022;121(7):2161–2166. doi: 10.1007/s00436-022-07553-0. [DOI] [PubMed] [Google Scholar]
- Šukytė T, Butkauskas D, Juozaitytė-Ngugu E, Švažas S, Prakas P. Molecular confirmation of Accipiter birds of prey as definitive hosts of numerous Sarcocystis species, including Sarcocystis sp., closely related to pathogenic S. calchasi. Pathogens. 2023;12(6):752. doi: 10.3390/pathogens12060752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villesen P. FaBox: an online toolbox for Fasta sequences. Mol Ecol Notes. 2007;7(6):965–968. doi: 10.1111/j.1471-8286.2007.01821.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
DNA sequences of the ITS1 marker have been deposited in GenBank with accession numbers OR059466–OR059469.