Abstract
Understanding plant immune responses is complex because of the high interdependence among biological processes in homeostatic networks. Hence, the integration of environmental cues causes network rewiring that interferes with defense responses. Similarly, plants retain molecular signatures configured under abiotic stress periods to rapidly respond to recurrent stress, and these can alter immunity. Metabolome changes imposed by abiotic stressors are persistent, although their impact on defense remains to be clarified. In this study, we profiled metabolomes of Arabidopsis plants under several abiotic stress treatments applied individually or simultaneously to capture temporal trajectories in metabolite composition during adverse conditions and recovery. Further systemic analysis was performed to address the relevance of metabolome changes and extract central features to be tested in planta. Our results demonstrate irreversibility in major fractions of metabolome changes as a general pattern in response to abiotic stress periods. Functional analysis of metabolomes and co-abundance networks points to convergence in the reconfiguration of organic acid and secondary metabolite metabolism. Arabidopsis mutant lines for components related to these metabolic pathways showed altered defense capacities against different pathogens. Collectively, our data suggest that sustained metabolome changes configured in adverse environments can act as modulators of immune responses and provide evidence for a new layer of regulation in plant defense.
Key words: abiotic stress, Arabidopsis, biotic stress, combinatorial stress, metabolomics, systems biology
Plants exposed to adverse environments configure molecular signatures that modulate immune responses against subsequent pathogen infections. Unlike transcriptomic changes, metabolome reconfigurations after periods of abiotic stress tend to persist, and the central features of this response make important contributions to sustaining plant defense.
Introduction
Plant immunity relies on sophisticated interplays among multiple and diverse features that act coordinately to counteract pathogen progression (Hillmer et al., 2017; Delplace et al., 2022; Ngou et al., 2022; Jia et al., 2023). Defense mechanisms are well characterized in isolation; however, pathogen infections frequently occur in combination with other abiotic stressors, a threat aggravated by climate change, producing responses that differ from those evoked by individual stressors (Atkinson et al., 2013; Prasch and Sonnewald, 2013; Suzuki et al., 2014; Son and Park, 2022). Systemically, components that mediate defense are entangled with the signaling and regulatory networks that operate to balance plant homeostasis and, moreover, occupy central positions as hubs (Mukhtar et al., 2011; Weßling et al., 2014). Because of the high connectivity of immune features, network reconfigurations that activate signaling pathways, fine-tune transcript and protein expression levels, or adjust metabolite abundance to mediate acclimation to environmental perturbations can ultimately interfere with plants’ capacity to cope with pathogen infection (Saijo and Loo, 2020; Zarattini et al., 2021). In the model plant Arabidopsis thaliana (hereafter Arabidopsis), defense responses to Pseudomonas syringae (hereafter Psto) are hindered by elevated temperatures (Wang et al., 2009) but enhanced under water shortage (Gupta et al., 2016). Thus, the outcome of plant–pathogen interactions depends largely on the complex interaction between defense mechanisms and responses to environmental cues.
Combinatorial stress refers to the application of multiple stressors not only simultaneously but also sequentially. For instance, drought episodes, temperature shifts, or pathogen infections occur in transient and uncoordinated waves. In this case, responses to intermittent stressors do not rely on recurrent transitions from basal to acclimation states owing to high energy costs that potentially trade off with plant fitness (Heil, 2002; van Hulten et al., 2006). Instead, plants retain physiological and molecular signatures configured under unfavorable conditions that allow rapid and enhanced responses in the case of subsequent stress events, a process termed “priming” or “stress memory” (Charng et al., 2023). However, the primed state of plants after abiotic stress periods may hamper immune responses, enhancing susceptibility to subsequent pathogen infection or, conversely, boost defense and confer cross-tolerance. As an example, adult Arabidopsis that had recovered from heat shock displayed increased susceptibility to Psto (Janda et al., 2019), whereas cold pre-treatments led to cross-tolerance (Griebel et al., 2022). Therefore, deciphering the molecular mechanisms underlying stress memory is of utmost importance for identifying features that mediate the cross-talk between defense and acclimation to abiotic stressors and understanding new layers of complexity in immune responses.
In general terms, stress memory consists of either the sustained activation of responsive components after stressor removal or the acquired capacity to escalate stress responses in the case of a second event. However, the operating signatures that confer plant tolerance to stress recurrence remain far from completely understood. For priming, the concept of “transcriptional memory” has motivated a large number of studies in the field of plant stress (Hannan Parker et al., 2022; Charng et al., 2023; Wilkinson et al., 2023). Transcriptional memory consists of epigenetic modifications at promoters of stress-related genes in the form of histone modifications and changes in nucleosome occupancy after stress periods that increase chromatin accessibility (Charng et al., 2023). Among other strategies for the persistent activation of stress defense mechanisms, it is well known that, after exposure to elevated temperatures, plants accumulate chaperones that assist in protein refolding to prevent recurrent disruptions in protein homeostasis (Yeh et al., 2012; Fernández-Bautista et al., 2018). Because abiotic stress conditions generally cause cellular damage due to oxidative bursts, plants frequently retain the battery of antioxidant enzymes active upon stress removal (Foyer et al., 2009; Juszczak et al., 2016). Another strategy found in plants that have recovered from conditions that cause cellular disruption, such as cold, drought, or salinity, is the accumulation of osmoprotectant solutes such as mannitol, raffinose, or galactinol (Taji et al., 2002).
The necessity of deciphering mechanisms of stress memory has recently motivated increasing interest in monitoring the recovery dynamics of the physiological and molecular state of plants after adverse conditions. System-wide analyses of de-acclimated plants under moderate stress conditions have consistently shown reversibility of transcriptome changes, whereas metabolomes tend to be steady over time (Kaplan et al., 2004; Zuther et al., 2015; Coolen et al., 2016; Pagter et al., 2017; Vyse et al., 2019; Garcia-Molina et al., 2020). Plant metabolic state can be interpreted as the consequence of the integration of the functionality of all biological processes, and metabolites can therefore act as efficient homeostatic signals. However, the potential role of metabolites in mediating cross-tolerance has mostly been overlooked. To fill this gap, we profiled the dynamics of transcriptome and metabolome changes in Arabidopsis under a wide array of single and combined abiotic stressors: high light intensity, humidity, water availability, and temperature fluctuations. Under our conditions, metabolome changes tended to be irreversible, whereas transcriptional responses returned to the basal state. Further integrative analysis revealed metabolites involved in pathways for carbon metabolism and other pigments as common elements in the sustained response, and pathogen bioassays with mutant lines proved relevant for defense responses in planta. Thus, our data contribute toward generalizing the durability of metabolome reconfigurations during recovery from diverse single and multiple abiotic stress factors and propose central metabolites accumulated during acclimation as modulators of immune responses to a wide variety of pathogens. These observations are fundamental to establishing biotechnological strategies to reinforce plant tolerance under real scenarios.
Results
Arabidopsis immune responses after sequences of abiotic–biotic stress depend on the lag phase between stressors and pathogen
Under sequential stress conditions, physiological and molecular reconfigurations of plants in response to a first stressor can confer increased susceptibility or tolerance in advance of a second stress event. In this regard, we asked how Arabidopsis immune responses were modulated by moderate environmental fluctuations in light, humidity, and temperature. High-light (HL) treatments consisted of a 2.5-fold increase in light intensity relative to standard conditions (300 vs. 120 μmol photons m−2 s−1); high humidity was achieved by watering plants in excess in sealed trays; and drought periods were imposed by withholding water. For heat conditions, the diurnal and nocturnal temperatures were increased by 5°C (27°C day/23°C night), whereas for cold, they were lowered by 10°C (12°C day/8°C night) and the diurnal light intensity was set to 80 μmol photons m−2 s−1. Combinations of heat and drought (heat-drought), heat and HL (heat-HL), heat, drought, and HL (heat-drought-HL), and cold and HL (cold-HL) were also considered in order to evaluate the effects of multiple simultaneous stressors. Pathogen infections were performed using two pathogens with distinct lifestyles: the hemibiotrophic bacterium Psto and the necrotrophic fungus Botrytis cinerea (hereafter Bot).
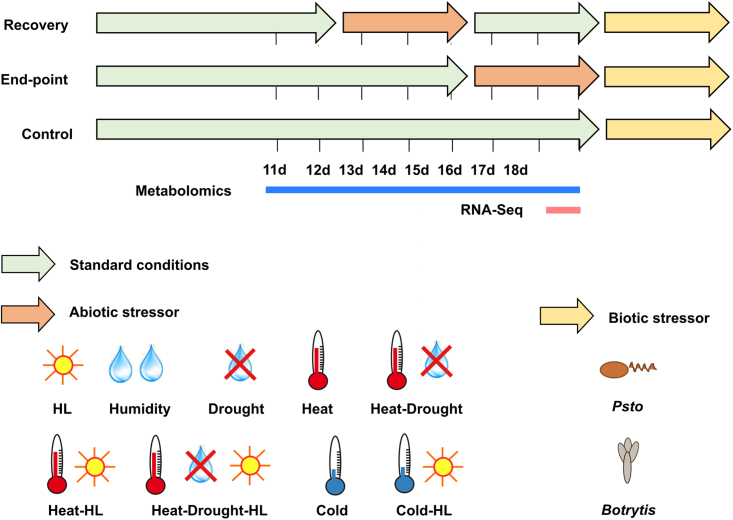
To evaluate Arabidopsis immune responses after abiotic stress periods in a comprehensive manner, two sequential stress setups were designed. First, 12-day-old seedlings cultivated under standard conditions were exposed to abiotic stressors for 3 days and then recovered for an additional 3 days under standard conditions to establish a lag phase before pathogen infection (recovery setup, Figure 1 and Supplemental Figure 1). Second, another batch of plants was treated with abiotic stressors at day 15 and subsequently infected with pathogens (endpoint setup, Figure 1 and Supplemental Figure 1).
Figure 1.
Setup for sequential stress treatments.
Arabidopsis plants were cultivated under 12 h/12 h day–night cycles (22°C at 120 μmol m−2 s−1 and 18°C in the dark) (green arrows) and treated with moderate abiotic stress conditions consisting of high light (HL), humidity, drought, heat, cold, or their combinations (orange arrows) as described in the section “Methods” before Pseudomonas syringae (Psto) or Botrytis cinerea (Bot) challenge (yellow arrows). For the recovery design, 12-day-old Arabidopsis seedlings exposed to abiotic stressors for 3 days were returned to standard conditions for three additional days before pathogen challenge. For the endpoint design, the abiotic stressors were applied to 15-day-old Arabidopsis plantlets for 3 days immediately before pathogen challenge. Plants not exposed to abiotic stressors were used to evaluate the innate immune response (control). Harvest time points for molecular analysis are indicated.
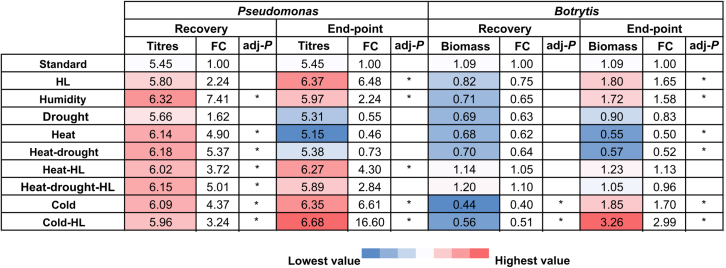
The outcome of the pathogen bioassays was quantified as Psto titer or Bot fungal biomass in leaves at 3 days post infection (dpi) (Figure 2 and Supplemental Figure 2). Plants in the recovery setup showed opposite trends in immune phenotype depending on the pathogen. Plants recovered from abiotic stressors showed an overall enhanced susceptibility to Psto: bacterial titers were approximately 1.6 to 7.5 times higher compared with controls, depending on the treatment. Conversely, a general increase in Bot tolerance was observed: fungal biomass in leaves was approximately 35%–60% lower than under standard conditions (Figure 2 and Supplemental Figure 2). However, the same abiotic stressors applied in the endpoint setup produced similar immune responses to both Psto and Bot infection. HL-, high-humidity-, cold-, and cold-HL-treated plants accumulated approximately 2–16 times higher Psto titers and approximately 60%–300% more Bot fungal biomass compared with those under standard conditions, whereas bacterial titers under drought and heat and fungal biomass under heat and heat-drought were both halved (Figure 2 and Supplemental Figure 2). Collectively, these observations support the existence of molecular footprints after abiotic stressors that affect general features involved in immune defense networks.
Figure 2.
Overview of plant immune phenotypes under sequential abiotic–biotic stress.
Binary map summarizing immune phenotypes of Arabidopsis plantlets upon Psto (Pseudomonas) and Bot (Botrytis) infection in the recovery and endpoint designs in Supplemental Figure 2. Color code depicts the average colony titer (Psto) or fungal biomass (Botrytis). Fold change (FC) was calculated in comparison to standard conditions (standard). Asterisks indicate significant differences relative to control (standard) conditions (adjusted P 0.05, Duncan’s post hoc test on linear mixed models, n = 3 independent biological experiments).
Abiotic stressors produce reversible plant transcriptome responses but persistent metabolome reconfigurations
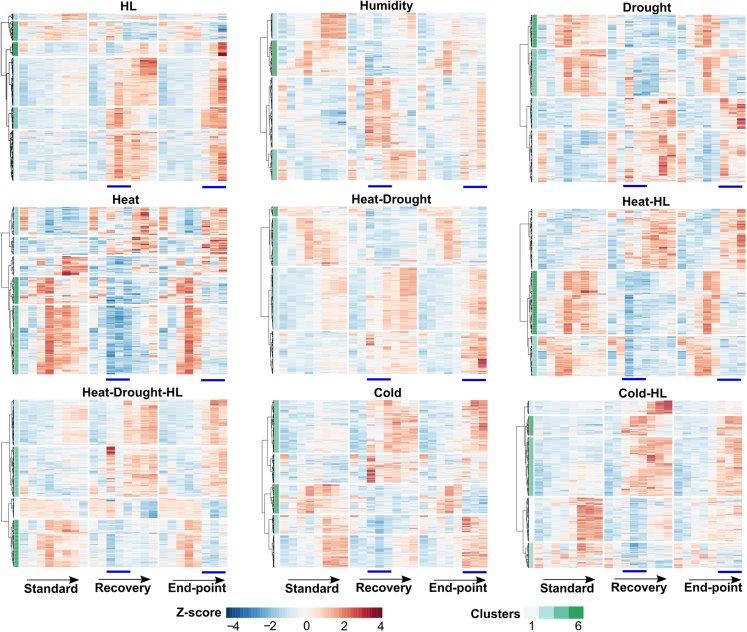
Because our results demonstrated that Arabidopsis immune responses can be modulated by previous exposure to abiotic stressors, we asked which molecular features might serve as signatures for stress memory. Metabolites attracted our interest because previous systemic studies in plants recovered from unfavorable environments revealed the reversibility of transcriptome changes but the durability of metabolome reconfigurations (Kaplan et al., 2004; Zuther et al., 2015; Coolen et al., 2016; Pagter et al., 2017; Vyse et al., 2019; Garcia-Molina et al., 2020). Thus, to assess the dynamics of metabolome responses to abiotic stressors under our conditions, the metabolite composition of Arabidopsis rosettes was monitored daily by untargeted liquid chromatography–tandem mass spectrometry (LC–MS/MS) metabolomics. Stringent deconvolution of chromatograms resulted in a coverage of 10 414 potential metabolites; those that displayed differential accumulation over time in each time course compared with standard conditions were defined as significantly altered metabolites (SAMs) (adjusted P 0.05, Dunnett’s post hoc test on linear mixed models) (Supplemental Table 1). Accordingly, approximately 400 SAMs were identified under heat and heat-HL; approximately 500 under HL, drought, and heat-drought-HL; and approximately 600 under humidity, heat-drought, cold, and cold-HL (Supplemental Figure 3). Z means of SAM abundances detected for each abiotic stressor were used to draw heatmaps with hierarchical clustering to compare the dynamics of stress-responsive metabolites under standard conditions and both recovery and endpoint time courses. Unsupervised k-means heatmap partitioning was used to extract sets of compounds that displayed similar temporal fluctuations (Figure 3). Overall, SAMs either remained steady under standard conditions or changed in abundance toward the end of the time course, likely owing to developmental and/or growth events (Figure 3). By contrast, SAM abundances shifted from the moment of exposure to abiotic stressors in both recovery and endpoint setups (from day 13 or 16, respectively) (Figure 3). Interestingly, the trends in SAM composition changes in response to stress treatments looked identical in both time courses (Figure 3). Next, correlations between SAM abundances in pre-treated (days 11 and 12) and stress-treated plants in the recovery setup (days 13–15) and the corresponding time points in the endpoint setup (days 11 and 12 and 16–18) were computed for each treatment to quantify the similarity of metabolome reconfigurations in both cases. Density plots in Supplemental Figure 4 (orange lines) depicted distributions of Pearson correlation coefficients (PCCs, r) centered on the highest positive values (r = 0.8–1.0) in all treatments, except for heat-drought-HL (r = 0.5–0.8). These data indicated that SAMs qualified as changing compounds strictly because of stress responsiveness and, moreover, demonstrated that our treatments provoked almost identical metabolome reconfigurations in plants during intermediate developmental stages.
Figure 3.
Temporal fluctuations in metabolite abundance under abiotic stress periods.
Untargeted LC–MS/MS was used to profile metabolite abundance in plants exposed to multiple abiotic stress conditions in the recovery and endpoint setups. SAMs detected in each abiotic stress treatment relative to control conditions (adjusted P 0.05, Dunnett’s test on linear mixed models, n = 5 independent biological experiments) were used to draw heatmaps with hierarchical clustering according to the WardD2 method. Metabolite abundances were Z-score normalized to visualize temporal patterns in metabolome changes from days 11–18 in plants cultivated under standard conditions (standard) and in the recovery and endpoint setup designs. Horizontal breaks in the heatmaps represent groups of metabolites that displayed similar patterns according to unsupervised k-means clustering. The directions of the arrows and the blue bars indicate the order of time points (11–18 days) and the duration of abiotic stress treatments, respectively.
Heatmaps also revealed that metabolome changes during stress exposure could either return to the initial state or persist upon stress removal in the recovery setup, the fraction of metabolites that followed each pattern being dependent on the treatment (Figure 3). Thus, to estimate the degree of reversibility of metabolome responses to stress treatments, SAM abundances in pre-treated and recovered plantlets (days 11–12 and 16–18, recovery setup) were compared to those at the corresponding time points under control conditions. In addition, comparisons to the same time points in the endpoint setup were considered in order to evaluate irreversibility, given the conservation of metabolome reconfigurations when stressors were applied in both time courses. PCC distributions for metabolomes of HL, high humidity, heat-drought, and cold-recovered plantlets and those under control conditions, as well as endpoint setups, were centered on the highest positive values (r = 0.8–1.0) with almost the same density, indicating that equal fractions of reversible and irreversible metabolome changes took place under these conditions (Supplemental Figure 4, red vs. blue lines). SAM abundances in heat-, heat-HL-, heat-drought-HL- and cold-HL-recovered plantlets were poorly correlated with those under standard conditions, as PCCs were more broadly distributed. However, PCC distribution was more skewed toward the highest positive values in comparison with endpoint setups (Supplemental Figure 4, red vs. blue lines), meaning that metabolic changes under heat, heat-HL, heat-drought-HL, and cold-HL treatments were predominantly irreversible. Finally, PCC distributions for drought treatments revealed important dissimilarities in reconfiguration of metabolome dynamics in recovery from water shortage compared with both standard conditions and endpoint setups (Supplemental Figure 4, red and blue lines). Consequently, our data demonstrate that important fractions of metabolome changes in response to multiple abiotic stress treatments were irreversible under our conditions.
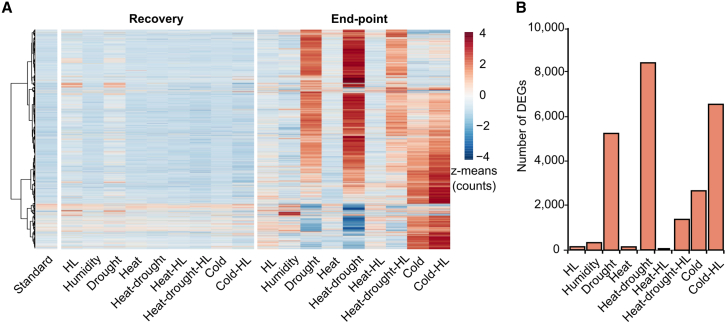
Next, we were interested in determining whether the dynamics of transcriptional responses to abiotic stress followed similar patterns. To that end, normalized counts from RNA sequencing (RNA-seq) of Arabidopsis rosettes at day 18 (Supplemental Table 2) were used to draw a heatmap with hierarchical clustering to visualize the transcriptome under each stressor at the end of the time courses. Transcriptome profiles of plants after the recovery design did not exhibit important divergences compared to standard conditions, whereas the same treatments applied after the endpoint setup revealed important differences, mainly due to increases in transcript levels under drought, heat-drought, heat-drought-HL, cold, and cold-HL conditions (Figure 4A). To quantitatively assess the effects of stressors on transcriptomes, differentially expressed genes (DEGs) were identified based on an absolute fold change (FC) 2 in transcript levels relative to standard conditions (adjusted P 0.05, Wald test). Fewer than 10 DEGs were detected at the end of the recovery phase, except under drought conditions (approximately 500 DEGs) (Supplemental Table 3), and further transcriptome analysis failed to detect significant differences in alternative splicing events relative to standard conditions (see section “methods”). However, the endpoint design led to remarkable changes in transcript abundance, although the number of DEGs depended on the treatment. The lowest DEG fractions were detected after HL, heat, heat-HL (fewer than 60 DEGs) (Figure 4B; Supplemental Table 3). Greater changes took place after high humidity (approximately 250 DEGs), cold, and heat-drought-HL (approximately 1000 DEGs), whereas the highest fractions were found after drought, cold-HL, and heat-drought (approximately 5000, 6600, and 8000 DEGs) (Figure 4B; Supplemental Table 3). Apart from the rapid reversibility of potential early transcriptome events not covered by our strategy, we attributed the differences in DEG fractions among treatments to stress intensity, suggesting that a two-fold increase in light intensity or an overall temperature increase of 5°C, as well as high humidity, were the mildest stressors in our designs. Furthermore, the disparity in the range of alterations in metabolites and transcripts by the end of the endpoint setup (approximately 400–600 SAMs vs. approximately 60–8000 DEGs) indicates additional layers of regulation for metabolic pathways, especially under less adverse conditions.
Figure 4.
Transcriptome changes after abiotic stress periods.
RNA-seq analysis was performed on Arabidopsis plantlets on day 18 of the recovery and endpoint setups.
(A) Identification of patterns in transcriptome changes. Heatmap with hierarchical clustering according to the WardD2 method constructed from z means of normalized counts of samples under standard conditions and abiotic stressors in the recovery and endpoint setups. Normalized counts were filtered (P 0.05, one-way ANOVA) to include genes that changed in at least one treatment.
(B) Fraction of differentially expressed genes (DEGs). DEGs compared with standard conditions (FC 2 and adjusted P 0.05, Wald test, n = 3 independent experiments) were only detected in the endpoint setup. Bar plot depicts the total numbers of DEGs under each stressor.
Collectively, our high-throughput analyses confirm that molecular mechanisms of acclimation to transient exposure to abiotic treatments consist of a reversible reconfiguration of the transcriptome but durable changes in the metabolome after stress removal.
Abiotic stress periods mainly affect carbon metabolism and secondary metabolites independently of late transcriptome responses
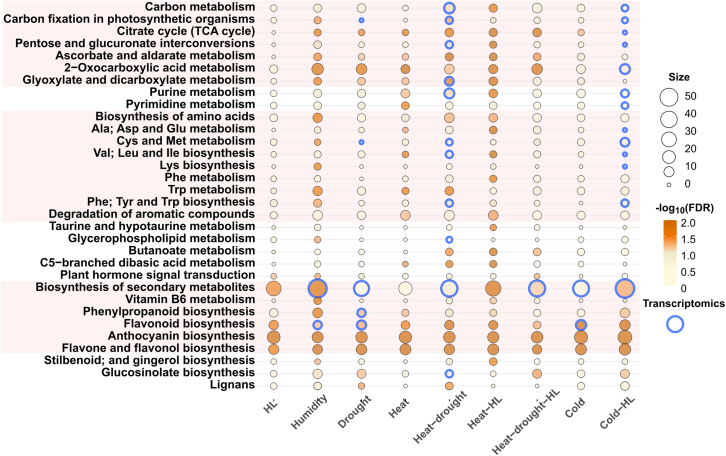
In our attempt to identify metabolites that carry stress memory, we wondered whether acclimation to various abiotic stress treatments could alter common metabolic pathways. Therefore, multiple comparisons among SAMs were performed to investigate commonalities in responses to treatments. As shown in Supplemental Figure 3, approximately 45% of the humidity SAMs were condition specific, whereas the specific percentages of SAMs under HL, drought, heat-drought, heat-drought-HL, cold, and cold-HL were less than 20%, and those under heat and heat-HL were less than 7%. On the other hand, larger numbers of SAMs were found at the intersection between two conditions for cold and cold-HL (186 SAMs), HL and cold-HL (122 SAMs), and heat-drought and heat-drought-HL (72 SAMs). One hundred and forty and 40 SAMs were found at the intersections of three treatments for HL, cold, and cold-HL and HL, heat-drought, and heat-drought-HL, respectively (Supplemental Figure 3). Twenty-four SAMs were common to five treatments (all except heat, drought, heat-drought, heat-HL, heat-drought-HL), 29 SAMs to six (all except HL, humidity, and cold-HL), 44 SAMs to seven (all except HL and cold-HL), 10 SAMs to all except HL, and only five to all treatments (Supplemental Figure 3). Because more than 300 metabolites responded differentially under at least three abiotic stressors compared with standard conditions, commonalities in metabolic responses to abiotic stressors could be expected. Thus, to functionally characterize metabolome changes, SAMs were first annotated based on the Kyoto Encyclopedia of Genes and Genomes (KEGG), BioCyc, and internal libraries (see section “methods”). Despite technical limitations to the proper identification of metabolites in our platform, this approach identified 44.07% of all SAMs as potential markers and annotated 27.16% of them (Supplemental Table 4). Significantly enriched KEGG pathways were identified (adjusted P 0.05, Fisher’s exact test), and the resulting metabolic pathways were grouped into major categories (carbon, nitrogen, nitrogen base, lipid metabolism, etc.) (Figure 5). Biosynthesis of secondary metabolites was the category that included the highest number of tentatively annotated metabolites, and those related to flavonoids and anthocyanins were commonly enriched under all abiotic stress treatments (Figure 5). Abiotic stressors generally altered carbon metabolism, because the tricarboxylic acid (TCA) cycle was enriched under all treatments except HL and cold-HL and the metabolism of ascorbate, 2-oxocarboxylic acids, and glyoxylate was enriched under multiple abiotic stressors (Figure 5). Significant enrichment of pathways involved in the metabolism of several amino acids was found only under humidity, heat, heat-drought, and heat-HL and showed no consistent pattern, suggesting that each stressor provoked a specific reconfiguration of nitrogen metabolism (Figure 5). Other central pathways, such as the metabolism of nitrogen bases, fatty acids, hormones, or glucosinolates, were enriched under a few treatments, and these metabolic perturbations might therefore be condition specific (Figure 5). Together, these results suggest that alterations in secondary metabolites and TCA intermediates are mainly used by plants as a general response to cope with detrimental periods, although specific aspects of metabolome reconfiguration were also found, depending on the nature of the stressor(s).
Figure 5.
Functional relevance of metabolome changes during abiotic stress conditions.
Significant enrichment in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways was determined based on tentative annotations of SAMs under each abiotic stress treatment (adjusted P 0.05, Fisher’s exact test). Circle size is proportional to the number of metabolites included in the pathway, and color code indicates the degree of enrichment significance based on adjusted P as −log10(FDR). KEGG pathways significantly enriched in transcriptome data (adjusted P 0.05, Fisher’s exact test, Supplemental Table 6) for late-changing events are circled in blue. Pink squares highlight groups of pathways involved in the metabolism of carbon, amino acids, and secondary metabolites.
Despite the reversibility of transcriptional responses to abiotic stressors, we questioned whether the alterations observed in metabolic pathways could potentially have been dictated by transcriptomes. Given the high similarity of metabolome reconfigurations in response to abiotic stressors, independent of the developmental stage of plants under our time courses (Figure 3 and Supplemental Figure 4), DEGs detected in each treatment under the endpoint setup were selected as representative for, at least, late transcriptome responsive events. Thus, significantly enriched Gene Ontology (GO) terms and KEGG pathways (adjusted P 0.05, Fisher’s exact test) were identified in the DEGs to assess the impact of abiotic stressors on biological processes and metabolic reactions, respectively, at the transcriptome level (Supplemental Tables 5 and 6). Enriched GO terms supported the activation of general responses to abiotic stressors (light intensity, salt, water deprivation, cold, heat) and only included terms for biotic stressors (fungus, bacteria) under high humidity (Supplemental Table 5). Indeed, steady-state levels of representative abiotic stress markers were consistent with the treatments we applied, whereas selected immune-responsive transcripts did not display remarkable induction (Supplemental Figures 5 and 6). Other significantly enriched GO terms were related to hormones (auxins, cytokinins, jasmonic acid, abscisic acid, gibberellins) and the reconfiguration of central biological processes such as photosynthesis, mitochondrial function, cell division, ubiquitination, or vesicle sorting, although this depended on treatment (Supplemental Table 5). KEGG pathways also reflected changes in fundamental biological processes and, moreover, retrieved metabolic reactions similar to those found for metabolome changes, especially under drought, heat-drought, and cold-HL (Figure 5; Supplemental Table 6). Transcriptional changes in carbon metabolic reactions under heat-drought and in secondary metabolism under high humidity, heat-drought-HL, and cold-HL coincided with SAM enrichments under the same treatments. However, transcriptional changes related to either the metabolism of carbon, amino acids, and nitrogen bases under drought and cold-HL or of amino acids, lipids, and glucosinolates under heat-drought were not observed at the metabolome level (Figure 5). Thus, the overall inconsistency between metabolic reactions enriched in transcriptomes and metabolomes tempted us to speculate that metabolome reconfiguration upon exposure to abiotic stressors may not directly reflect late changes in transcriptional abundance or, alternatively, might be generally mediated by early-responsive transcripts that rapidly recover.
Network analysis of metabolome changes during abiotic stress periods reveals convergent reconfiguration of metabolic pathways mediated by different central components
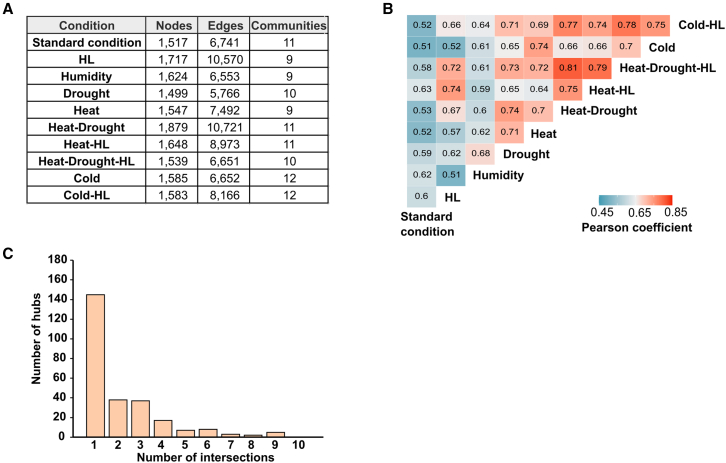
The unique aspects of metabolome rewiring after multiple and moderate abiotic stressors in terms of irreversibility and partial independence from transcriptomes motivated us to gain more insight into systemic responses to treatments. To this end, conditional networks were constructed based on correlations among temporal fluctuations in changing compounds (see section “methods”) (Supplemental Figure 7A). Topologically, metabolome networks were similar in size (approximately 1500–1800 nodes/metabolites) but differed in shape and community composition (5700–10 700 edges and 9–12 modules, depending on the condition) (Figure 6A). Because network topology is determined by the connectivity of metabolites according to co-abundance patterns, we assumed that conserved responses to treatments would lead to sets of metabolites (nodes) with similar degrees (numbers of edges per node) in the networks, whereas functional rewiring would cause changes in the numbers of node connections. Thus, (dis)similarities in metabolome response to abiotic stressors were estimated based on equivalences in node degree distribution among networks. As expected, node connectivity in the standard conditions network differed markedly from that in the abiotic stressor networks (r < 0.63) owing to metabolome reconfigurations during treatments (Figure 6B). Metabolite connectivity in the high-humidity network displayed a low correlation (r < 0.65) with all other treatments except drought (r = 0.68), suggesting a unique metabolome response to elevated humidity in plants (Figure 6B). The network topologies of the remaining treatments presented more similarities (Figure 6B). Interestingly, the topology of the HL network was exclusively comparable to those of other stressors applied simultaneously with HL, i.e., heat-HL, heat-drought-HL, and cold-HL (r = 0.67–0.74), pointing to partial conservation of the metabolic rewiring that occurred under HL in these stressor combinations. Likewise, important similarities (r > 0.65) in topology were found among networks derived from treatments that involved changes in temperature (heat and cold), and the same held true for drought (Figure 6B). Thus, our network analysis suggests that exposure to several abiotic stressors may provoke partially overlapping metabolome reconfigurations under our conditions. Furthermore, changes in metabolic pathways operating in response to single stressors would be expected in combinatorial responses to stressors applied simultaneously.
Figure 6.
Reconfiguration of metabolomes in Arabidopsis plants in different abiotic stress periods.
Compounds that changed in at least one treatment (P 0.05, linear mixed model, n = 5 independent experiments) were used to construct conditional networks (Supplemental Figure 7A).
(A) Network topological metrics in terms of size (number of nodes/metabolites), shape (number of edges), and detected communities (>20 nodes/metabolites) are shown.
(B) Comparison of global metabolome responses among abiotic stress treatments. Pearson correlations between the normalized node degree in each network were calculated to estimate (dis)similarities in the topology of the conditional networks.
(C) Commonalities in the local hub composition of networks. The three highest values for connectivity per community were considered to be local hubs and used for multiple comparisons (Supplemental Figure 7B). Bar plots depict the total number of common local hubs among treatments.
Networks were further investigated for community composition to extract local hubs (i.e., nodes with the highest degree per module). Because of their central position, local hubs can be selected to assess the preservation of the most relevant features among networks. For our purpose, metabolites ranked within the top three degree values were considered to be local hubs (Supplemental Table 7). As a general trend, approximately 55% of all local hubs were specific to each network, approximately 29% were common to two or three networks, and only 7% were shared among half (5) or more of the networks (Figure 6C and Supplemental Figure 7B). In consequence, despite network topologies revealing partial conservation of trends in metabolome changes under a wide range of abiotic stressor periods, differences in local hub composition support the specialization of central features in each response.
Central features involved in metabolome rewiring under abiotic stressors modulate plant responses to pathogen infection
To extract the most relevant components of metabolome responses to abiotic stressors, local hubs were investigated as representatives of the functionality of communities. Among all metabolites that qualified as hubs, our platform provided annotations for 32 of them (Supplemental Table 7). Several anthocyanidins, such as pelargonidin derivatives, coumarin, and cyanidin, appeared as local hubs in the networks for multiple treatments, as did flavonoid derivatives for naringenin and quercetin in specific cases (Supplemental Table 7). Other common hubs at the intersection of multiple networks were fumaric acid, indole-3-acetic acid, 3-butenylglucosinolate, and the nicotinamide derivatives dimethylmaleic acid and NAD+ (Supplemental Table 7). Complementary to this strategy, metabolome datasets were used to construct supervised models to identify orchestrated changes in metabolite composition related to immune phenotypes for Psto and Bot (Figure 2). Decision trees indicated an interplay of sets of unknown compounds with cyanidin derivatives under Psto infection and with pelargonidin and coumaroyl acid under Bot infection to counteract pathogenic infections (Supplemental Figure 8; Supplemental Table 8). Consistent with KEGG pathway enrichment results (Figure 5), our metabolome-wide analysis confirmed the reconfiguration of intermediates in pathways for flavonoid biosynthesis and TCA-organic acids as the most significant changes in response to abiotic stressors.
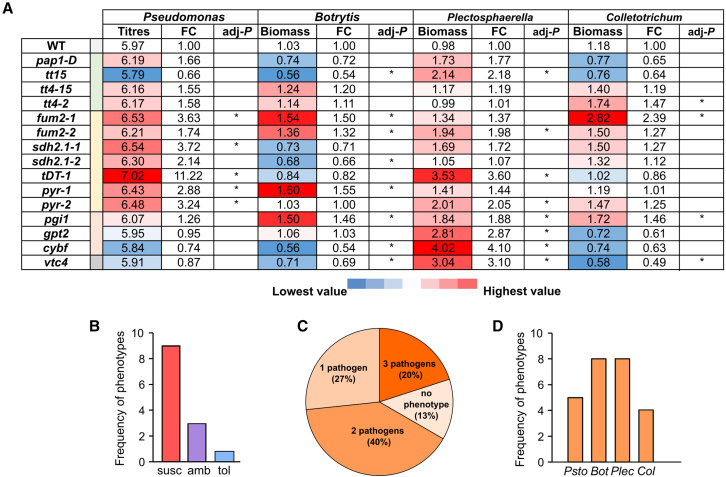
To evaluate whether central features in metabolome responses to abiotic stressors could affect plant defense against pathogens, mutant lines targeting intermediate steps in candidate metabolic pathways were used to test plant–pathogen interactions. For pigment biosynthesis, mutants with increased (pap1-D) or defective (tt4-2 and tt4-15) anthocyanin content (Borevitz et al., 2000; Xu et al., 2020) or reduced flavonoid composition (tt15) were selected. For TCA-organic acids, we used loss-of-function lines for cytosolic fumarase 2 (fum2-1 and fum2-2), succinate dehydrogenase subunit 1 (sdh2.1-1 and sdh2.1-2), and pyruvate kinase (pyr-1 and pyr-2) and the tonoplast dicarboxylate transporter (tDT-1) mutant with altered malate and fumarate accumulation in leaves (Emmerlich et al., 2003). Because KEGG pathway enrichment also highlighted other carbon metabolic pathways as relevant, transfer DNA (T-DNA) lines for phosphoglucose isomerase 1 (pgi1) as the central enzyme in starch production, glucose-6-phosphate/phosphate translocator 2 (gpt2), cytosolic fructose-1,6-bisphosphatase (cybf), and L-galactose-1-phosphate phosphatase (vtc4), which is involved in ascorbate biosynthesis, were also included. Besides Psto and Bot, bioassays were extended to include the necrotrophic pathogen Plectosphaerella cucumerina and the hemibiotrophic pathogen Colletotrichum higginsianum (hereafter Plec and Col) to address the consequences of mutations in response to a broader pathogen repertoire. Despite the different steady-state levels of target genes in mutant lines (Supplemental Figure 9), significant differences from the WT in defense against pathogens (adjusted P 0.05, Duncan’s post hoc test on linear mixed models) were found for all alleles except pap1-D and tt14-15 (Figure 7A and Supplemental Figure 10). Immune phenotypes were largely mutant and pathogen dependent, as Psto and Plec infections tended to show enhanced susceptibility in comparison with the WT, whereas Bot and Col showed heterogeneous patterns (Figure 7A and Supplemental Figure 10). However, the most relevant phenotypes caused by mutations consisted of enhanced susceptibility (nine cases), although ambiguous responses depending on the pathogen were found for three alleles, and only one allele (sdh2.1-2) conferred tolerance (Figure 7B). In terms of convergence, three alleles produced marked phenotypic changes under three pathogens, six under two pathogens, and four under one, causing Bot and Plec challenge in twice as many cases of defense alteration in mutants compared with Psto and Col (Figures 7C and 7D). Remarkably, mutant lines that exhibited an immune phenotype to at least two pathogens showed significant differences from control lines in organic acid composition and secondary metabolites, even for mutants in which steady-state transcript levels of target genes were minimally altered, such as pyr-1 and pig1 (Supplemental Figures 9 and 11). None of these lines displayed noticeable physiological constraints under abiotic stress treatments in our setup (Supplemental Figures 12 and 13). Per group, tt15 was the pigment-production mutant that induced the most significant immune phenotypes, showing an approximately two-fold loss or increase in fungal biomass compared with the WT upon Bot and Plec infection, respectively (adjusted P 0.05, Duncan’s post hoc test on linear mixed models) (Figure 7A). The mutations selected for TCA-organic acids significantly enhanced susceptibility to at least two pathogens, and the defense response of fum2 mutants was significantly impaired under all four pathogens for at least one allele (Figure 7A). Interestingly, pgi1, gpt2, and cybf alleles significantly enhanced Plec susceptibility phenotypes, revealing that balanced carbon metabolism is a particular requisite for counteracting Plec infections (Figure 7A). Finally, the ascorbate deficiency of vtc4 produced different phenotypes depending on the pathogen: enhanced susceptibility to Plec (three-fold increase in fungal biomass vs. WT) but enhanced tolerance to Bot and Col (30%–50% less fungal biomass vs. WT) (Figure 7A). In summary, our results demonstrate that perturbations in central metabolic pathways to cope with multiple abiotic stressors significantly alter the plant’s capacity to defend against pathogens with different lifestyles, revealing them as crucial features that modulate Arabidopsis immunity during sequential stresses.
Figure 7.
Overview of immune phenotypes of selected candidates to pathogen infection.
(A) Binary map summarizing the immune phenotypes of the indicated Arabidopsis mutant lines upon challenge with Psto (Pseudomonas), Bot (Botrytis), Plectosphaerella cucumerina (Plectosphaerella), and Colletotrichum higginsianum (Colletotrichum) in Supplemental Figure 10. Color code depicts average colony titer (Psto) or fungal biomass (Botrytis, Plectosphaerella, and Colletotrichum). FC was calculated in comparison to wild-type (WT) plantlets, and asterisks indicate significant differences from the WT (adjusted P 0.05, Duncan’s post hoc test on linear mixed models, n = 4 independent biological experiments).
(B) Frequency of immune phenotypes for selected mutants: enhanced susceptibility (susc), enhanced tolerance (tol), or ambiguous (amb).
(C) Pie chart depicting the fraction of mutants that displayed significant immune phenotypes to one or several pathogens.
(D) The number of mutant alleles that produced immune phenotypes to Pseudomonas (Psto), Botrytis (Bot), Plaectosphaerella (Plec), and Colletotrichum (Col).
Discussion
The extreme complexity of plant immunity networks hampers the isolation of new components with pivotal roles in modulating defense responses. In this work, we addressed the configuration of stress memory states after abiotic stressors to identify new modulators of plant defense. Our pathogen bioassays demonstrated that plants that had recovered from single and combinatorial fluctuations in light intensity, humidity, water availability, and temperature showed overall greater susceptibility to Psto but cross-tolerance to Bot (Figure 2). Such an antagonistic pattern can be attributed to differences in pathogen lifestyles, because reinforcement of certain aspects of plant homeostasis in terms of primary metabolism or antioxidant barriers after the primary stressor would be beneficial for colonization by the hemibiotrophic Psto but would constrain progression of the necrotrophic Bot.
Despite most attention to priming and stress memory being paid to epigenetic mechanisms (Oberkofler et al., 2021; Hannan Parker et al., 2022; Charng et al., 2023), we wanted to explore the potential contribution of metabolites in detail. Accumulation of osmoprotectants and antioxidants such as raffinose, galactinol, myo-inositol, or ascorbic acid is a well-known strategy for extending responses to counteract recurrent stress (Taji et al., 2002; Foyer et al., 2009; Juszczak et al., 2016). However, our understanding of the impact of abiotic stress on plant metabolomes is limited. Nevertheless, previous studies on plants during recovery from heat, cold, or HL reported sustained changes in primary metabolites (Kaplan et al., 2004; Caldana et al., 2011; Pagter et al., 2017; Garcia-Molina et al., 2020), which could be envisaged as a signature to maintain active stress responses. Here, non-targeted metabolomic profiling of plants under nine abiotic stress factors applied individually or simultaneously was used to obtain a broader coverage of metabolome composition in response to a wide range of stressors. The temporal trajectories of metabolite abundance in our recovery setup confirmed the irreversibility of major metabolite fractions after exposure to stressors (Figure 3). In addition, metabolome changes showed similar patterns regardless of the timing of treatment application, whereas transcriptome responses evoked by abiotic stressor periods were completely absent in recovered plants (Figures 3 and 4). Consequently, our work provides important evidence for the irreversibility of metabolome changes as a general strategy in plant responses to abiotic stress, at least in the short term.
The Arabidopsis metabolome displayed remarkable plasticity in response to environmental cues, even for moderate treatments that did not markedly disturb late transcriptome changes under our experimental conditions (e.g., HL, humidity, heat, and heat-HL; Figures 3 and 4 and Supplemental Figure 4). Systemic comparisons between networks of co-abundant metabolites under each treatment revealed similarities in topology but differences in local hub composition (Figure 6). Thus, abiotic stressors are expected to affect different metabolites with a high hierarchy, and these perturbations might cause subsequent reconfigurations that ultimately converge on similar metabolic pathways. Indeed, despite the residual overlap in SAMs among treatments, functional enrichments of annotated metabolites were consistent with intermediates in the TCA cycle, metabolism of 2-oxocarboxylic acids and ascorbic acid, and secondary metabolites such as flavonoids and anthocyanins (Figure 5). Limitations to the bona fide identification of metabolites detected by untargeted LC–MS/MS metabolomics precluded a more in-depth interpretation of changes in metabolite composition. However, the function of local hubs can be extended to other co-abundant metabolites according to the “guilt-by-association” principle, as network communities were configured based on interconnectivity among metabolites with identical behavior. Interestingly, fumaric acid and flavonoid and anthocyanin derivatives such as quercetins, pelargonidin, or cyanidin were identified as central metabolites in the network communities of several treatments (Supplemental Table 7). Therefore, our results are consistent with the eventual effect of abiotic stress conditions on TCA cycle–derived organic acids and pigment composition in Arabidopsis.
The TCA cycle operates as a versatile metabolic crossroads that integrates energy production and cellular respiration with amino acid metabolism, nitrogen assimilation, and biosynthesis of secondary metabolites, among other processes (Sweetlove et al., 2010). In Brassicaceae, fumaric acid constitutes a major fraction of photosynthate and therefore plays a pivotal role in the partitioning and distribution of carbon skeletons (Chia et al., 2000; Dyson et al., 2016). As stress responses are accompanied by important energy demands, plant acclimation to HL and cold conditions depends largely on a balanced fumaric acid content (Dyson et al., 2016), although we did not observe such dependency under moderate abiotic stress treatments (Supplemental Figures 12 and 13). Furthermore, fumaric acid, as well as other TCA cycle–derived organic acids, has been credited with signaling capacity to induce defense responses against Psto, although at high physiological levels (Less et al., 2011; Finkemeier et al., 2013; Balmer et al., 2018). Similarly, naringenin and other anthocyanins accumulate in adult leaves of Arabidopsis plants under several stressors to promote tolerance as antioxidant barriers and can also trigger defense-related genes (An et al., 2021; Kaur et al., 2023). However, the contribution of such metabolites to defense against multiple pathogens has not been explored in detail.
Here, immune responses were investigated in plants at intermediate developmental stages to avoid advanced differentiation between sink and source tissues and interference caused by metabolites that accumulate to high levels as plants grow or signal for the flowering transition. We observed immune susceptibility for fum2 mutant lines upon Psto, Bot, Plec, or Col challenge, as well as for other mutant lines with abrogated expression of enzymes catalyzing the production of succinate and pyruvate and the import of malate and fumarate into vacuoles, depending on the pathogen (Figure 7A). In line with the apparent dependence on carbon skeletons to ensure plant defense, potential constraints to accumulating normal levels of starch in the pgi1 mutant would also cause alterations in fumaric acid composition and, ultimately, susceptibility to the three fungi, although the steady-state transcript levels of the target gene remained unaltered (Figure 7A, Supplemental Figures 9 and 11). On the other hand, tt4-15 and tt4-2 lines with abrogated anthocyanin content tended to display susceptibility to the four pathogens, whereas tt15 and the over-accumulator pap1-D showed tolerance to Bot and Col (Figure 7A). It is notable that mutant lines that displayed the broadest immune phenotypes exhibited altered patterns for organic acids and secondary metabolite concentrations (Supplemental Figure 11). Thus, manipulation of central metabolites whose accumulation is naturally disrupted in response to multiple abiotic stressors complicates plant defense responses against several pathogens with different lifestyles.
Organization of plant immunity networks obeys evolutionary forces that have fixed defense strategies, as well as regulatory mechanisms, to ensure optimal and efficient transitions from basal to defensive states based on frequently encountered pathogens (Ngou et al., 2022). Metabolites as modulators of plant immunity represent a novel, biologically relevant mechanism that has not been addressed in depth. The resolution of large-scale plant–pathogen protein interaction maps has revealed evolutionarily conserved strategies of pathogen infection, which involve targeting proteins centrally positioned in the network (Mukhtar et al., 2011; Weßling et al., 2014). However, none of these protein hubs are strictly related to metabolic pathways, meaning that pathogens do not prioritize the direct dismantling of metabolomes to colonize hosts. Moreover, metabolic reactions that produce compounds capable of modulating immune responses were not regulated at the transcriptome level after multiple abiotic stress factors. Therefore, we propose that the metabolome is the result of the interplay between multiple biological processes and thus reflects the final homeostatic state of the plant. As a result, the abundance of central metabolites could serve as a systemic signal for Arabidopsis to modulate immunity and adapt defense to the overall state of the plant.
Methods
Plant cultivation
Arabidopsis thaliana ecotype Col-0 was used as the WT control. Mutant lines for PRODUCTION OF ANTHOCYANIN PIGMENT 1 (pap1-D), TRANSPARENT TESTA 14/CHALCONE SYNTHASE (tt14-15), and TONOPLAST DICARBOXYLATE TRANSPORTER (tDT-1) were described previously (Borevitz et al., 2000; Emmerlich et al., 2003; Xu et al., 2020). The lines tt4-2 (CS2111524), TRANSPARENT TESTA 15/UDP-GLUCOSE STEROL GLUCOSYLTRANSFERASE 15 (tt15; SALK_103581), FUMARASE 2 (fum2-1, SALK_025631; fum2-2, GABI_107E05), SUCCINATE DEHYDROGENASE 2.1 (sdh2.1-1, SALK_031100C; sdh2.1-2, SALK_127672C),PYRUVATE KINASE FAMILY PROTEIN (pyr-1, SALK_049481C; pyr-2, SALK_014468C), PHOSPHOGLUCOSE ISOMERASE 1 (pgi1, SALK_107903C), GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2 (gpt2, GK-454H06-018837), CYTOSOLIC FRUCTOSE-1,6-BISPHOSPHATASE (cybf, SALK_064456C), and INOSITOL MONOPHOSPHATASE FAMILY PROTEIN (vtc4, SALK_077222C) were requested from the Nottingham Arabidopsis Stock Centre (NASC, https://arabidopsis.info/). Seeds were stratified at 4°C for 2 days and sown on Jiffy-7 substrate (Jiffy Group International, Zwijndrecht, the Netherlands) under a 12-h light (22°C and 120 μmol m−2 s−1)/12-h dark (18°C) cycle with a relative humidity of 65% in FitoClima 1200 PLH LED cabinets (Aralab, Rio de Mouro, Portugal). Abiotic stress treatments consisted of raising diurnal light to 350 μmol m−2 s−1 (HL), sealing trays with excess water to preserve 100% of soil moisture and high humidity (humidity), water shortage until approximately 30% of soil moisture (drought), increasing temperature cycles to 27°C/23°C (heat), lowering temperature cycles to 12°C/8°C and adjusting light intensity to 80 μmol m−2 s−1 (cold) or 200 μmol m−2 s−1 (cold-HL), and combinations of these conditions. Rosette leaves were harvested 6 h after the onset of light and flash frozen in liquid nitrogen.
Pathogen infection assays
Psto pv. tomato DC3000 colonies cultivated at 28°C for 2 days on King’s medium (Condalab, Madrid, Spain) plates supplemented with antibiotics were resuspended at an optical density 600 (OD600) of 0.05 in 10 mM MgCl2 and Silwet L77 0.02% (v/v) and used to spray Arabidopsis rosettes. Bacterial growth curves at 0 and 3 dpi were quantified on the basis of colony forming units (CFU) from serial dilutions of leaf disk extracts on selection plates.
Fungal material was grown at 25°C in darkness for 15 days. Bot and Plec were cultivated on full or ½ potato dextrose agar medium, respectively, and fungal spores were prepared on ¼ potato dextrose broth at 5 ×·105 spores ml−1. Col (O’Connell et al., 2012) was cultivated on oatmeal plates, and spores were resuspended in water at 5 ×·105 spores ml−1. Fungal biomass was quantified from entire rosette leaves at 3 dpi (Psto, Bot) or 7 dpi (Plec, Col) by real-time qPCR. Amplification reactions were performed with approximately 10 ng of gDNA with SYBR Green I dye using specific fungus primers and the Arabidopsis ACTIN2 (ACT2) gene as a reference (Supplemental Table 9) in a Roche Light Cycler 480 instrument (Roche, Basel, Switzerland). The two-step protocol consisted of an initial cycle at 95°C for 10 min and 45 cycles of 95°C for 10 s and 60°C for 20 s. Fungal biomass was determined according to the 2−Δ(Ct fungus gene – Ct ACT2) method and normalized to the average value for controls in each biological replicate. Relative fungal biomass was determined after outlier imputation, and average values were used to calculate FC relative to control conditions.
Metabolome preparation and profiling
Metabolites were extracted from 1–3 mg of lyophilized plant material in 1 ml of 30% methanol (v/v) supplemented with 0.01% formic acid (v/v). Samples were homogenized with glass beads in a mixer mill for 3 min at 30 Hz and centrifuged at 14 000 rpm for 20 min. Supernatants were filtered through 0.22-μm regenerated cellulose (Phenomenex, Torrance, USA) and frozen at −80°C. Five microliters of metabolite extracts were injected and separated in a reverse Kinetex C18 analytical column (2.6 μm particle size, 50 mm × 2.1 m, Phenomenex) using a gradient of methanol and H2O supplemented with 0.01% formic acid. The flow rate was set to 0.3 ml/min. Samples were ionized in positive and negative ion modes for electrospray ionization (ESI) in a 40–1100 m/z range using an Acquity UPLC I-Class System interfaced to a hybrid quadrupole time-of-flight mass spectrometer, SYNAPT G2-S high-definition tandem mass spectrometry (MS/MS) detector (Waters, Milford, USA).
Raw data were obtained using MassLynx 4.2 software (Waters) and converted to CDF format with the MassLynx DataBridge tool. Peak intensities of compounds were retrieved using the xcms package integrated into RStudio, referred to dry weight, and quantile normalized. Linear mixed models were fitted to temporal fluctuations in metabolite abundances, and significant differences relative to controls were computed using a post hoc Dunnett’s test (adjusted P 0.05). Filtered metabolites were loaded into MarVis-Suite 2.0 software (Kaever et al., 2015) to obtain isotope corrections and identifications using the MarVis pathway interface with a 0.01 tolerance for m/z correction. Metabolite annotations and functional enrichment of metabolic pathways (adjusted P 0.05, Fisher’s exact test) were performed on combined lists of compounds identified in ESI+ and ESI− modes using KEGG (https://www.genome.jp/kegg/), BioCyc (https://biocyc.org/), and internal libraries described in Gamir et al. (2014). Metabolome datasets were deposited at MetaboLights (https://www.ebi.ac.uk/metabolights/) under accession number MTBLS7018.
Transcriptome profiling and analysis
Total RNA was extracted from 18-day-old rosettes with the Maxwell RSC Plant RNA kit (Promega, Wisconsin, USA) according to the manufacturer’s instructions. RNA-seq libraries were prepared and paired-end sequenced (2 × 150 bp) at Sistemas Genómicos (València, Spain) with standard Illumina protocols. RNA-seq datasets were analyzed on the Galaxy platform (Afgan et al., 2022). In brief, adaptors were removed from reads using Trimmomatic (Bolger et al., 2014), and clean reads were mapped to the Arabidopsis reference genome (TAIR10) with STAR (Dobin et al., 2013). Read counts were computed with featureCounts (Liao et al., 2014), and DEGs were identified for pair-wise comparisons between standard conditions and each treatment under the recovery and endpoint setup (absolute log2 FC 1, adjusted P 0.05, Wald test) using DESeq2 (Love et al., 2014). Differences in alternative splicing were evaluated with ASpli (Mancini et al., 2021). GO term and KEGG pathway enrichment (adjusted P 0.05, Fisher’s exact test) were performed in Panther (http://www.pantherdb.org/) and ShinyGO v.0.77 (http://bioinformatics.sdstate.edu/go/). Transcriptome datasets have been deposited at the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE226105.
Construction of co-abundance networks
Co-abundance networks were constructed for metabolites that exhibited significant differences in temporal trends of compound abundance under at least one condition (P 0.05, linear mixed model). Random matrix theory on pair-wise Pearson correlation matrices was used to determine a cutoff that avoided network saturation with a resolution of 0.01 using the RMThreshold package in RStudio. Node connectivity and community composition according to the FastGreedy algorithm were computed using the Igraph package in RStudio. Networks were exported to Cytoscape version 3.9.1 (Shannon et al., 2003) for visualization and manipulation.
Miscellaneous statistical and bioinformatic analyses
Statistical tests and plotting were performed with the basic interface of RStudio or implemented packages. Outliers and missing values in infection assays and metabolome datasets were detected and imputed by the k-nearest neighbors method (k = 5) using the VIM package. Multiple comparisons of linear mixed models were computed according to Duncan’s multiple range or Dunnett’s post hoc tests (adjusted P 0.05). Heatmaps with hierarchical clustering according to the Ward D2 method were drawn using z-means of normalized data with the pheatmap package. UpSet plots were constructed with the UpSetR package. Supervised decision trees were modeled with the rpart package using log10-transformed metabolite abundances of plants during the 3 days of stressor treatment and tags based on cutoffs of two-fold pathogen infection for Psto and 30% for Bot relative tostandard conditions.
Analysis of mutant lines
For gene expression analysis, total RNA from 18-day-old Col-0 and mutant plants grown under standard conditions was prepared as indicated above. RNA was retrotranscribed to cDNA using the NZY First-Strand cDNA Synthesis kit (NZYTech, Lisboa, Portugal). qPCR was performed using specific primers and normalized with ACT2 as the reference gene (Supplemental Table 9) using the protocol described above for fungal biomass.
Organic acid profiling was performed by targeted LC–MS/MS metabolomics as described in Balmer et al. (2018) with modifications. In brief, metabolites were extracted from 1–2 mg of dry tissue with 0.1% formic acid (v/v). Five microliters of the extracts were injected into a Xevo TQS instrument (Waters Micromass, Manchester, UK) equipped with a T-wave device and coupled to a triple quadrupole through an Acquity UPLC HSS T3 2.1 column (1.8 μm, 100 mm) (Waters). The ESI was set to negative for all compounds. All parameters for chromatography and MS/MS analysis were set as described in Balmer et al. (2018). Secondary compounds were prepared and analyzed as described above for untargeted metabolomics. The ChromaLynx function in MassLynx 4.2 software (Waters) was used to identify secondary metabolites against internal libraries of compounds based on retention times and fragments of selected standards.
Photosynthetic measurements were performed on dark-adapted plants after abiotic stress treatments with an IMAGING-PAM M-Series instrument (Walz, Effeltrich, Germany), and the maximum quantum yield of photosystem II (PSII) (Fv/Fm) was calculated as (Fm − F0)/Fm).
Funding
This project received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement no. 101028809 (AG-M), from project RTI2018-094350-B-C33 funded by the Spanish Ministry for Science, Innovation and Universities (VP), from grants SEV-2015-0533 and CEX2019-000902-S funded by MCIN/AEI/10.13039/501100011033, and from the CERCA Programme/Generalitat de Catalunya.
Author contributions
Conceptualization, A.G.-M.; methodology, A.G.-M. and V.P.; formal analysis, A.G.-M. and V.P.; investigation, A.G.-M. and V.P.; resources, A.G.-M. and V.P.; data curation, A.G.-M.; writing – original draft, A.G.-M. and V.P.; writing – review & editing, A.G.-M. and V.P.; project administration, A.G.-M.; funding acquisition, A.G.-M. and V.P.
Acknowledgments
We acknowledge the Greenhouse Facility and Stress Program at CRAG for technical assistance and for providing biological material, and Dr. Cristian Vicent (Servei Central d'Instrumentació Científica at UJI) for technical assistance in metabolomics. Dr. Núria Sánchez-Coll (CRAG) is acknowledged for critically reading our manuscript. We thank Dr. Víctor Manuel González (CRAG) and Dr. Lisa van den Broeck (North Carolina State University, USA) for assistance in data analysis. The mutant lines tDT-1, pap1-D, and tt4-15 were a kind gift from Dr. Ekkehard Neuhaus (Technical University Kaiserslautern, Germany) and Dr. Andreas Richter (University of Rostock, Germany). Helena Kruyer is acknowledged for English editing and proofreading of the manuscript. This work is dedicated to the memory of Dr. María Coca. No conflict of interest is declared.
Published: July 4, 2023
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Supplemental information
Arabidopsis plants were exposed to either standard conditions (control, cont), HL, humidity, drought, heat, cold, or combinations of treatments as described in Figure 1. Representative plants at day 18 in the recovery (A) and endpoint (B) setups are shown. Bar, 1 cm.
Arabidopsis plantlets after recovery and endpoint setups were inoculated with Psto at OD600 0.05 or Bot (Botrytis) at 105 spores/ml. Psto titers were quantified as CFU at 0 dpi (A) and 3 dpi (B). Botrytis relative fungal biomass (C) was determined at 3 dpi by qPCR using specific oligonucleotides. Boxplots summarize the results from three independent experiments, and asterisks indicate significant differences (adjusted P 0.05, Duncan’s post hoc test on linear mixed models, n = 3 independent experiments).
Significantly changed metabolites were filtered to identify SAMs under each abiotic stressor in comparison with standard conditions (adjusted P 0.05, Dunnett’s test on linear mixed models, n = 5 independent experiments). An UpSet plot summarizes the number of condition-specific and non-condition-specific SAMs (left), as well as a fraction of commonly identified SAMs among treatments (right). For simplicity, intersections below seven conditions with fewer than five metabolites are not shown.
Density plots summarize the distribution of PCCs for comparisons of temporal fluctuations in significantly altered metabolite abundance under abiotic stress treatments to assess (dis)similarities in metabolome responses independently of the day of stressor application (changes during days 11–15 in the recovery setup vs. 11, 12, and 16–18 days in the endpoint setup, orange), the reversibility of metabolome changes (changes during time points 11, 12, and 16–18 days in the recovery setup vs. standard conditions, red), or irreversibility (changes during 11, 12, and 16–18 days in the recovery vs. endpoint setup, blue).
Representative marker genes for HL (A), humidity (B), drought (C), heat (D), and cold (E) conditions were selected: EARLY LIGHT-INDUCIBLE PROTEIN 1/2 (ELIP1/2), PYRABACTIN RESISTANCE 1-LIKE 5 (PYL5), PYRABACTIN RESISTANCE 1 (PYR1), DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN 2A (DREB2A), RD29B, HEAT STRESS TRANSCRIPTION FACTORS (HSFA2/1A), INDUCER OF CBF EXPRESSION (ICE1), and COLD REGULATED 15B (COR15B). Regularized log transcript counts from RNA-seq datasets were used to compute log2(FC) relative to controls (standard conditions) under each abiotic stress treatment at day 18. Bars represent mean ± SD of n = 3 independent experiments.
Representative marker genes responsive to pathogen infection were selected: PATHOGENESIS RELATED-GENES 1/2/3 (PR1/2/3), PLANT DEFENSIN 1.1/2.1/2.2 (PDF1.1/2.1/2.2), ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1), and PHYTOALEXIN DEFICIENT 4 (PAD4). Regularized log transcript counts from RNA-seq datasets were used to compute log2(FC) relative to controls (standard conditions) under each abiotic stress treatment at day 18. Bars represent mean ± SD of n = 3 independent experiments.
(A) Conditional networks for abiotic stress treatments. Nodes represent metabolite and edges relevant co-abundances. Nodes are colored based on the community to which they belong. (B) UpSet plot for multiple comparisons between local hubs of each conditional network. The total number of local hubs per condition (left) and a fraction of common hubs among networks (right) are depicted.
Metabolome datasets were filtered for compounds that changed in at least one treatment relative to controls (P 0.05, linear mixed model, n = 5 independent experiments) and log10 transformed. Compound abundance during the period of stress exposure (days 13–15 under standard conditions and recovery setup or 16–18 under endpoint setup) was tagged with the pathogen infection assay outcome as tolerant, unaltered (o), or susceptible according to cutoffs of two-fold for Psto(A) and 30% for Bot(B). The corresponding decision trees are shown with rules for known and unknown compounds (Supplemental Table 8) with a log10 scale.
Total RNA was prepared from 18-day-old WT and mutant lines grown under standard conditions. mRNA levels of indicated genes relative to ACTIN2 were determined by qPCR. Bars represent mean ± SD of n = 3 independent experiments. Statistical differences relative to the WT were assessed using a two-tailed t test (∗P 0.05, ∗∗P 0.01, ∗∗∗P 0.001).
Box plots show colony titers for Psto (A) or fungal biomass for Bot (B), Plect (C), and Col (D) in WT Arabidopsis and the indicated mutant lines. Red dashed lines indicate the average value for the WT. Statistical differences relative to the WT are indicated by letters (adjusted P 0.05, Duncan’s post hoc test on linear mixed models, n = 5 independent experiments).
Metabolites extracted from 18-day-old WT and selected mutant lines cultivated under standard conditions were profiled by LC–MS/MS. Boxplots summarize the concentrations of representative organic acids (A) and coumarins (coumarin), flavonoids (kaempferol and apigenin derivatives), and phenylpropanoids (sinapic acid) (B). Statistical differences relative to the WT are indicated (∗adjusted P 0.05, ∗∗P 0.01, ∗∗∗adjusted P 0.001, one-way ANOVA and Dunnet’s post hoc test, n = 3 independent experiments).
Mutant lines that exhibited immune phenotypes to at least two selected pathogens. Fifteen-day-old plants grown under standard conditions (control) were exposed to HL, humidity, drought, heat, cold, or combinations of treatments for 3 days. Representative pictures of plants at day 18 are shown. Bars, 1 cm.
The maximum quantum yield of photosystem II (PSII) (Fv/Fm) was determined for mutant lines grown as in Supplemental Figure 12. Boxplots summarize Fv/Fm measurements of plants from n = 3 independent experiments.
Data availability
Transcriptome and metabolome data are available at GEO and MetaboLights with accession numbers GSE226105 and MTBLS7018, respectively.
References
- Afgan E., Nekrutenko A., Grüning B.A., Blankenberg D., Goecks J., Schatz M.C., Ostrovsky A.E., Mahmoud A., Lonie A.J., Syme A., et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 2022;50:W345–W351. doi: 10.1093/nar/gkac247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J., Kim S.H., Bahk S., Vuong U.T., Nguyen N.T., Do H.L., Kim S.H., Chung W.S. Naringenin induces pathogen resistance against Pseudomonas syringae through the activation of NPR1 in Arabidopsis. Front. Plant Sci. 2021;12 doi: 10.3389/fpls.2021.672552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson N.J., Lilley C.J., Urwin P.E. Identification of genes involved in the response of Arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiol. 2013;162:2028–2041. doi: 10.1104/pp.113.222372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer A., Pastor V., Glauser G., Mauch-Mani B. Tricarboxylates induce defense priming against bacteria in Arabidopsis thaliana. Front. Plant Sci. 2018;9:1221. doi: 10.3389/fpls.2018.01221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1104/pp.113.222372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz J.O., Xia Y., Blount J., Dixon R.A., Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12:2383–2394. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldana C., Degenkolbe T., Cuadros-Inostroza A., Klie S., Sulpice R., Leisse A., Steinhauser D., Fernie A.R., Willmitzer L., Hannah M.A. High-density kinetic analysis of the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions. Plant J. 2011;67:869–884. doi: 10.1111/j.1365-313X.2011.04640.x. [DOI] [PubMed] [Google Scholar]
- Charng Y.Y., Mitra S., Yu S.-J. Maintenance of abiotic stress memory in plants: Lessons learned from heat acclimation. Plant Cell. 2023;35:187–200. doi: 10.1093/plcell/koac313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia D.W., Yoder T.J., Reiter W.-D., Gibson S.I. Fumaric acid: an overlooked form of fixed carbon in Arabidopsis and other plant species. Planta. 2000;211:743–751. doi: 10.1007/s004250000345. [DOI] [PubMed] [Google Scholar]
- Coolen S., Proietti S., Hickman R., Davila Olivas N.H., Huang P.-P., van Verk M.C., van Pelt J.A., Wittenberg A.H.J., de Vos M., Prins M., et al. Transcriptome dynamics of Arabidopsis during sequential biotic and abiotic stresses. Plant J. 2016;86:249–267. doi: 10.1111/tpj.13167. [DOI] [PubMed] [Google Scholar]
- Delplace F., Huard-Chauveau C., Berthomé R., Roby D. Network organization of the plant immune system: from pathogen perception to robust defense induction. Plant J. 2022;109:447–470. doi: 10.1111/tpj.15462. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson B.C., Miller M.A.E., Feil R., Rattray N., Bowsher C.G., Goodacre R., Lunn J.E., Johnson G.N. FUM2, a cytosolic fumarase, is essential for acclimation to low temperature in Arabidopsis thaliana. Plant Physiol. 2016;172:118–127. doi: 10.1104/pp.16.00852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerlich V., Linka N., Reinhold T., Hurth M.A., Traub M., Martinoia E., Neuhaus H.E. The plant homolog to the human sodium/dicarboxylic cotransporter is the vacuolar malate carrier. Proc. Natl. Acad. Sci. USA. 2003;100:11122–11126. doi: 10.1073/pnas.1832002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Bautista N., Fernández-Calvino L., Muñoz A., Toribio R., Mock H.P., Castellano M.M. HOP family plays a major role in long-term acquired thermotolerance in Arabidopsis. Plant Cell Environ. 2018;41:1852–1869. doi: 10.1111/pce.13326. [DOI] [PubMed] [Google Scholar]
- Finkemeier I., König A.C., Heard W., Nunes-Nesi A., Pham P.A., Leister D., Fernie A.R., Sweetlove L.J. Transcriptomic analysis of the role of carboxylic acids in metabolite signaling in Arabidopsis leaves. Plant Physiol. 2013;162:239–253. doi: 10.1104/pp.113.214114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C.H., Bloom A.J., Queval G., Noctor G. Photorespiratory metabolism: Genes, mutants, energetics, and redox signaling. Annu. Rev. Plant Biol. 2009;60:455–484. doi: 10.1146/annurev.arplant.043008.091948. [DOI] [PubMed] [Google Scholar]
- Gamir J., Pastor V., Kaever A., Cerezo M., Flors V. Targeting novel chemical and constitutive primed metabolites against Plectosphaerella cucumerina. Plant J. 2014;78:227–240. doi: 10.1111/tpj.12465. [DOI] [PubMed] [Google Scholar]
- Garcia-Molina A., Kleine T., Schneider K., Mühlhaus T., Lehmann M., Leister D. Translational components contribute to acclimation responses to high light, heat, and cold in Arabidopsis. iScience. 2020;23:101331. doi: 10.1016/j.isci.2020.101331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griebel T., Schütte D., Ebert A., Nguyen H.H., Baier M. Cold exposure memory reduces pathogen susceptibility in Arabidopsis based on a functional plastid peroxidase system. Mol. Plant Microbe Interact. 2022;35:627–637. doi: 10.1094/MPMI-11-21-0283-FI. [DOI] [PubMed] [Google Scholar]
- Gupta A., Dixit S.K., Senthil-Kumar M. Drought stress predominantly endures Arabidopsis thaliana to Pseudomonas syringae infection. Front. Plant Sci. 2016;7:808. doi: 10.3389/fpls.2016.00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan Parker A., Wilkinson S.W., Ton J. Epigenetics: a catalyst of plant immunity against pathogens. New Phytol. 2022;233:66–83. doi: 10.1111/nph.17699. [DOI] [PubMed] [Google Scholar]
- Heil M. Ecological costs of induced resistance. Curr. Opin. Plant Biol. 2002;5:345–350. doi: 10.1016/S1369-5266(02)00267-4. [DOI] [PubMed] [Google Scholar]
- Hillmer R.A., Tsuda K., Rallapalli G., Asai S., Truman W., Papke M.D., Sakakibara H., Jones J.D.G., Myers C.L., Katagiri F. The highly buffered Arabidopsis immune signaling network conceals the functions of its components. PLoS Genet. 2017;13:e1006639. doi: 10.1371/journal.pgen.1006639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hulten M., Pelser M., van Loon L.C., Pieterse C.M.J., Ton J. Costs and benefits of priming for defense in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2006;103:5602–5607. doi: 10.1073/pnas.0510213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda M., Lamparová L., Zubíková A., Burketová L., Martinec J., Krčková Z. Temporary heat stress suppresses PAMP-triggered immunity and resistance to bacteria in Arabidopsis thaliana. Mol. Plant Pathol. 2019;20:1005–1012. doi: 10.1111/mpp.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia A., Huang S., Ma S., Chang X., Han Z., Chai J. TIR-catalyzed nucleotide signaling molecules in plant defense. Curr. Opin. Plant Biol. 2023;73:102334. doi: 10.1016/j.pbi.2022.102334. [DOI] [PubMed] [Google Scholar]
- Juszczak I., Cvetkovic J., Zuther E., Hincha D.K., Baier M. Natural variation of cold deacclimation correlates with variation of cold-acclimation of the plastid antioxidant system in Arabidopsis thaliana accessions. Front. Plant Sci. 2016;7:305. doi: 10.3389/fpls.2016.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaever A., Landesfeind M., Feussner K., Mosblech A., Heilmann I., Morgenstern B., Feussner I., Meinicke P. MarVis-Pathway: integrative and exploratory pathway analysis of non-targeted metabolomics data. Metabolomics. 2015;11:764–777. doi: 10.1007/s11306-014-0734-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F., Kopka J., Haskell D.W., Zhao W., Schiller K.C., Gatzke N., Sung D.Y., Guy C.L. Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol. 2004;136:4159–4168. doi: 10.1104/pp.104.052142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S., Tiwari V., Kumari A., Chaudhary E., Sharma A., Ali U., Garg M. Protective and defensive role of anthocyanins under plant abiotic and biotic stresses: An emerging application in sustainable agriculture. J. Biotechnol. 2023;361:12–29. doi: 10.1016/j.jbiotec.2022.11.009. [DOI] [PubMed] [Google Scholar]
- Less H., Angelovici R., Tzin V., Galili G. Coordinated gene networks regulating Arabidopsis plant metabolism in response to various stresses and nutritional cues. Plant Cell. 2011;23:1264–1271. doi: 10.1105/tpc.110.082867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini E., Rabinovich A., Iserte J., Yanovsky M., Chernomoretz A. ASpli: integrative analysis of splicing landscapes through RNA-Seq assays. Bioinformatics. 2021;37:2609–2616. doi: 10.1093/bioinformatics/btab141. [DOI] [PubMed] [Google Scholar]
- Mukhtar M.S., Carvunis A.-R., Dreze M., Epple P., Steinbrenner J., Moore J., Tasan M., Galli M., Hao T., Nishimura M.T., et al. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science. 2011;333:596–601. doi: 10.1093/bioinformatics/btab141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngou B.P.M., Ding P., Jones J.D.G. Thirty years of resistance: Zig-zag through the plant immune system. Plant Cell. 2022;34:1447–1478. doi: 10.1093/bioinformatics/btab141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberkofler V., Pratx L., Bäurle I. Epigenetic regulation of abiotic stress memory: maintaining the good things while they last. Curr. Opin. Plant Biol. 2021;61:102007. doi: 10.1016/j.pbi.2021.102007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell R.J., Thon M.R., Hacquard S., Amyotte S.G., Kleemann J., Torres M.F., Damm U., Buiate E.A., Epstein L., Alkan N., et al. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat. Genet. 2012;44:1060–1065. doi: 10.1038/ng.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagter M., Alpers J., Erban A., Kopka J., Zuther E., Hincha D.K. Rapid transcriptional and metabolic regulation of the deacclimation process in cold acclimated Arabidopsis thaliana. BMC Genom. 2017;18:731. doi: 10.1186/s12864-017-4126-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasch C.M., Sonnewald U. Simultaneous application of heat, drought, and virus to Arabidopsis plants reveals significant shifts in signaling networks. Plant Physiol. 2013;162:1849–1866. doi: 10.1016/j.pbi.2021.102007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y., Loo E.P.I. Plant immunity in signal integration between biotic and abiotic stress responses. New Phytol. 2020;225:87–104. doi: 10.1111/nph.15989. [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son S., Park S.R. Climate change impedes plant immunity mechanisms. Front. Plant Sci. 2022;13:1032820. doi: 10.3389/fpls.2022.1032820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Rivero R.M., Shulaev V., Blumwald E., Mittler R. Abiotic and biotic stress combinations. New Phytol. 2014;203:32–43. doi: 10.1111/nph.12797. [DOI] [PubMed] [Google Scholar]
- Sweetlove L.J., Beard K.F.M., Nunes-Nesi A., Fernie A.R., Ratcliffe R.G. Not just a circle: flux modes in the plant TCA cycle. Trends Plant Sci. 2010;15:462–470. doi: 10.1016/j.tplants.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Taji T., Ohsumi C., Iuchi S., Seki M., Kasuga M., Kobayashi M., Yamaguchi-Shinozaki K., Shinozaki K. Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J. 2002;29:417–426. doi: 10.1046/j.0960-7412.2001.01227.x. [DOI] [PubMed] [Google Scholar]
- Vyse K., Pagter M., Zuther E., Hincha D.K. Deacclimation after cold acclimation—a crucial, but widely neglected part of plant winter survival. J. Exp. Bot. 2019;70:4595–4604. doi: 10.1093/jxb/erz229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Bao Z., Zhu Y., Hua J. Analysis of temperature modulation of plant defense against biotrophic microbes. Mol. Plant Microbe Interact. 2009;22:498–506. doi: 10.1094/MPMI-22-5-0498. [DOI] [PubMed] [Google Scholar]
- Weßling R., Epple P., Altmann S., He Y., Yang L., Henz S., McDonald N., Wiley K., Bader K., Gläßer C., et al. Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe. 2014;16:364–375. doi: 10.1094/MPMI-22-5-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson S.W., Hannan Parker A., Muench A., Wilson R.S., Hooshmand K., Henderson M.A., Moffat E.K., Rocha P.S.C.F., Hipperson H., Stassen J.H.M., et al. Long-lasting memory of jasmonic acid-dependent immunity requires DNA demethylation and ARGONAUTE1. Nat. Plants. 2023;9:81–95. doi: 10.1038/s41477-022-01313-9. [DOI] [PubMed] [Google Scholar]
- Xu D., Dhiman R., Garibay A., Mock H.P., Leister D., Kleine T. Cellulose defects in the Arabidopsis secondary cell wall promote early chloroplast development. Plant J. 2020;101:156–170. doi: 10.1111/tpj.14527. [DOI] [PubMed] [Google Scholar]
- Yeh C.-H., Kaplinsky N.J., Hu C., Charng Y.Y. Some like it hot, some like it warm: Phenotyping to explore thermotolerance diversity. Plant Sci. 2012;195:10–23. doi: 10.1016/j.plantsci.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarattini M., Farjad M., Launay A., Cannella D., Soulié M.C., Bernacchia G., Fagard M. Every cloud has a silver lining: how abiotic stresses affect gene expression in plant-pathogen interactions. J. Exp. Bot. 2021;72:1020–1033. doi: 10.1093/jxb/eraa531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuther E., Juszczak I., Lee Y.P., Baier M., Hincha D.K. Time-dependent deacclimation after cold acclimation in Arabidopsis thaliana accessions. Sci. Rep. 2015;5:12199. doi: 10.1038/srep12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Arabidopsis plants were exposed to either standard conditions (control, cont), HL, humidity, drought, heat, cold, or combinations of treatments as described in Figure 1. Representative plants at day 18 in the recovery (A) and endpoint (B) setups are shown. Bar, 1 cm.
Arabidopsis plantlets after recovery and endpoint setups were inoculated with Psto at OD600 0.05 or Bot (Botrytis) at 105 spores/ml. Psto titers were quantified as CFU at 0 dpi (A) and 3 dpi (B). Botrytis relative fungal biomass (C) was determined at 3 dpi by qPCR using specific oligonucleotides. Boxplots summarize the results from three independent experiments, and asterisks indicate significant differences (adjusted P 0.05, Duncan’s post hoc test on linear mixed models, n = 3 independent experiments).
Significantly changed metabolites were filtered to identify SAMs under each abiotic stressor in comparison with standard conditions (adjusted P 0.05, Dunnett’s test on linear mixed models, n = 5 independent experiments). An UpSet plot summarizes the number of condition-specific and non-condition-specific SAMs (left), as well as a fraction of commonly identified SAMs among treatments (right). For simplicity, intersections below seven conditions with fewer than five metabolites are not shown.
Density plots summarize the distribution of PCCs for comparisons of temporal fluctuations in significantly altered metabolite abundance under abiotic stress treatments to assess (dis)similarities in metabolome responses independently of the day of stressor application (changes during days 11–15 in the recovery setup vs. 11, 12, and 16–18 days in the endpoint setup, orange), the reversibility of metabolome changes (changes during time points 11, 12, and 16–18 days in the recovery setup vs. standard conditions, red), or irreversibility (changes during 11, 12, and 16–18 days in the recovery vs. endpoint setup, blue).
Representative marker genes for HL (A), humidity (B), drought (C), heat (D), and cold (E) conditions were selected: EARLY LIGHT-INDUCIBLE PROTEIN 1/2 (ELIP1/2), PYRABACTIN RESISTANCE 1-LIKE 5 (PYL5), PYRABACTIN RESISTANCE 1 (PYR1), DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN 2A (DREB2A), RD29B, HEAT STRESS TRANSCRIPTION FACTORS (HSFA2/1A), INDUCER OF CBF EXPRESSION (ICE1), and COLD REGULATED 15B (COR15B). Regularized log transcript counts from RNA-seq datasets were used to compute log2(FC) relative to controls (standard conditions) under each abiotic stress treatment at day 18. Bars represent mean ± SD of n = 3 independent experiments.
Representative marker genes responsive to pathogen infection were selected: PATHOGENESIS RELATED-GENES 1/2/3 (PR1/2/3), PLANT DEFENSIN 1.1/2.1/2.2 (PDF1.1/2.1/2.2), ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1), and PHYTOALEXIN DEFICIENT 4 (PAD4). Regularized log transcript counts from RNA-seq datasets were used to compute log2(FC) relative to controls (standard conditions) under each abiotic stress treatment at day 18. Bars represent mean ± SD of n = 3 independent experiments.
(A) Conditional networks for abiotic stress treatments. Nodes represent metabolite and edges relevant co-abundances. Nodes are colored based on the community to which they belong. (B) UpSet plot for multiple comparisons between local hubs of each conditional network. The total number of local hubs per condition (left) and a fraction of common hubs among networks (right) are depicted.
Metabolome datasets were filtered for compounds that changed in at least one treatment relative to controls (P 0.05, linear mixed model, n = 5 independent experiments) and log10 transformed. Compound abundance during the period of stress exposure (days 13–15 under standard conditions and recovery setup or 16–18 under endpoint setup) was tagged with the pathogen infection assay outcome as tolerant, unaltered (o), or susceptible according to cutoffs of two-fold for Psto(A) and 30% for Bot(B). The corresponding decision trees are shown with rules for known and unknown compounds (Supplemental Table 8) with a log10 scale.
Total RNA was prepared from 18-day-old WT and mutant lines grown under standard conditions. mRNA levels of indicated genes relative to ACTIN2 were determined by qPCR. Bars represent mean ± SD of n = 3 independent experiments. Statistical differences relative to the WT were assessed using a two-tailed t test (∗P 0.05, ∗∗P 0.01, ∗∗∗P 0.001).
Box plots show colony titers for Psto (A) or fungal biomass for Bot (B), Plect (C), and Col (D) in WT Arabidopsis and the indicated mutant lines. Red dashed lines indicate the average value for the WT. Statistical differences relative to the WT are indicated by letters (adjusted P 0.05, Duncan’s post hoc test on linear mixed models, n = 5 independent experiments).
Metabolites extracted from 18-day-old WT and selected mutant lines cultivated under standard conditions were profiled by LC–MS/MS. Boxplots summarize the concentrations of representative organic acids (A) and coumarins (coumarin), flavonoids (kaempferol and apigenin derivatives), and phenylpropanoids (sinapic acid) (B). Statistical differences relative to the WT are indicated (∗adjusted P 0.05, ∗∗P 0.01, ∗∗∗adjusted P 0.001, one-way ANOVA and Dunnet’s post hoc test, n = 3 independent experiments).
Mutant lines that exhibited immune phenotypes to at least two selected pathogens. Fifteen-day-old plants grown under standard conditions (control) were exposed to HL, humidity, drought, heat, cold, or combinations of treatments for 3 days. Representative pictures of plants at day 18 are shown. Bars, 1 cm.
The maximum quantum yield of photosystem II (PSII) (Fv/Fm) was determined for mutant lines grown as in Supplemental Figure 12. Boxplots summarize Fv/Fm measurements of plants from n = 3 independent experiments.
Data Availability Statement
Transcriptome and metabolome data are available at GEO and MetaboLights with accession numbers GSE226105 and MTBLS7018, respectively.