Abstract
Immunohistochemistry (IHC) is a diagnostic technique used throughout pathology. A machine learning algorithm that could predict individual cell immunophenotype based on hematoxylin and eosin (H&E) staining would save money, time, and reduce tissue consumed. Prior approaches have lacked the spatial accuracy needed for cell-specific analytical tasks. Here IHC performed on destained H&E slides is used to create a neural network that is potentially capable of predicting individual cell immunophenotype. Twelve slides were stained with H&E and scanned to create digital whole slide images. The H&E slides were then destained, and stained with SOX10 IHC. The SOX10 IHC slides were scanned, and corresponding H&E and IHC digital images were registered. Color-thresholding and machine learning techniques were applied to the registered H&E and IHC images to segment 3,396,668 SOX10-negative cells and 306,166 SOX10-positive cells. The resulting segmentation was used to annotate the original H&E images, and a convolutional neural network was trained to predict SOX10 nuclear staining. Sixteen thousand three hundred and nine image patches were used to train the virtual IHC (vIHC) neural network, and 1,813 image patches were used to quantitatively evaluate it. The resulting vIHC neural network achieved an area under the curve of 0.9422 in a receiver operator characteristics analysis when sorting individual nuclei. The vIHC network was applied to additional images from clinical practice, and was evaluated qualitatively by a board-certified dermatopathologist. Further work is needed to make the process more efficient and accurate for clinical use. This proof-of-concept demonstrates the feasibility of creating neural network-driven vIHC assays.
Introduction
Since the 1980s [1], immunohistochemistry (IHC) has been an integral part of anatomic pathology, aiding in the diagnosis of both benign and malignant lesions. Even though there is near ubiquitous use of IHC in modern anatomic pathology labs, it has several flaws. The technique increases turnaround time, consumes tissue, and has a nontrivial cost. In cases where large panels of IHC are required, as well as in small biopsies where tissue is limited, these limitations can be significant. In addition, the financial burden of IHC may be prohibitive in resource-limited regions around the world. While IHC can make important contributions to rendering a diagnosis, a more rapid and more economical alternative would be welcome.
In pathology, machine learning has been gaining prominence. A particular form of machine learning, the convolutional neural network (CNN), has proven to be particularly well-suited for use with whole slide images (WSI) [2]. There are several types of CNNs, and they are typically named after the task which they perform. Categorical CNNs, for example, classify images into predefined categories. Segmentation CNNs are similar, but categorize sub-regions within an image into different predefined categories.
CNNs are created in two steps. The first step is the assembly of annotated (labeled) images, and the second step is inputting these images into the CNN and training it to predict the image labels (Fig. 1a). Once this is complete, the CNN can be empirically evaluated on new images (Fig. 1b).
Fig. 1. Convolutional neural network (CNN) overview.
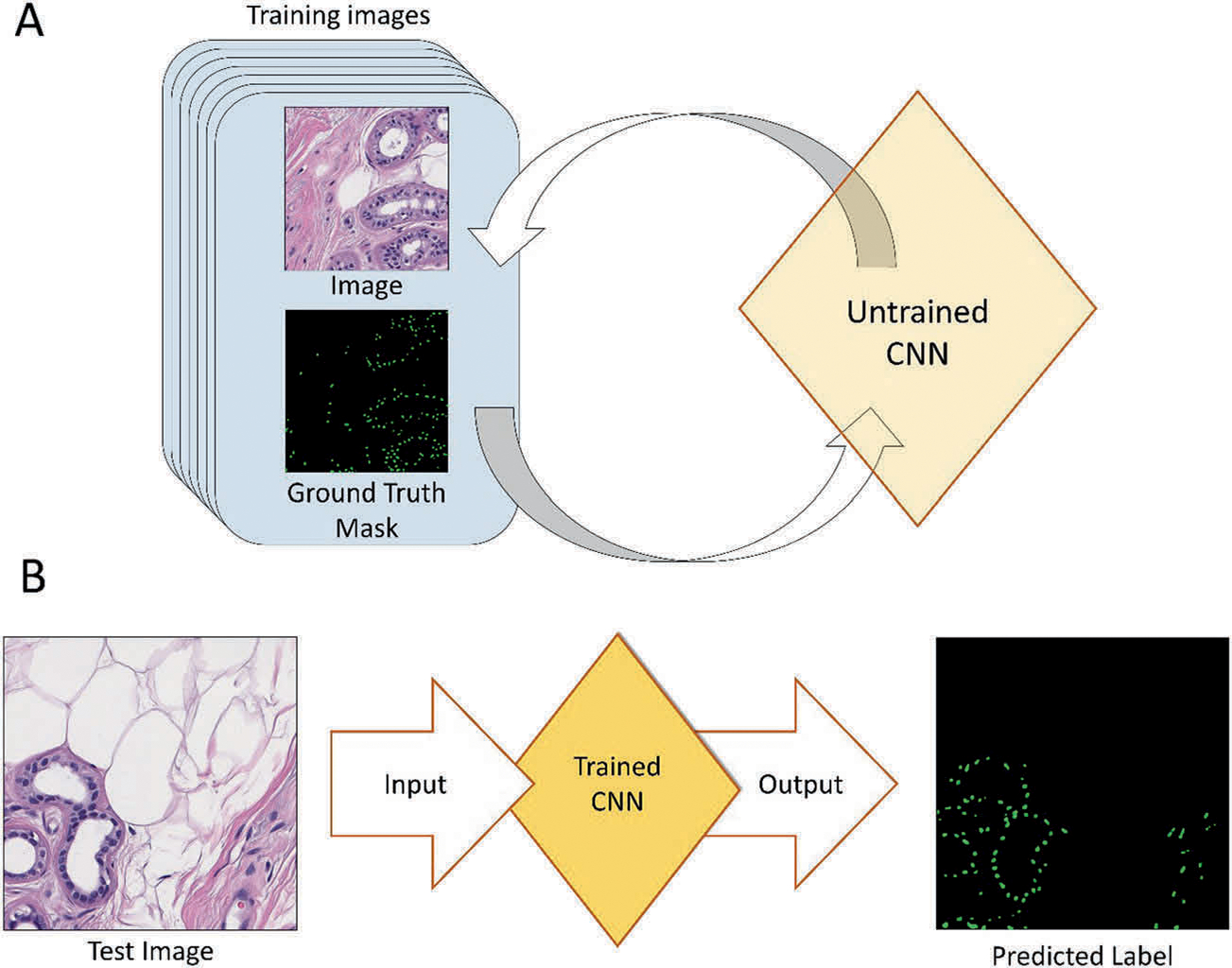
a CNNs are first trained on a set of annotated images. This iterative process is continued until the CNN can accurately predict the image labels. b Once the CNN is trained, it can then be fed new images and output predicted labels.
For segmentation CNNs, the correct image annotation is referred to as the ground-truth mask. A number of methods can be used to generate the ground-truth mask. In digital pathology, these are often created by a pathologist that manually annotates the different regions of an image. This approach has already been used in cervical cancer [3], skin lesions [4, 5], breast cancer [6–8], kidneys [9], lungs [10], and colon cancers [11]. Despite its popularity, this approach is time-intensive and susceptible to human bias.
Significant efforts have focused on creating machine learning algorithms capable of rendering a diagnosis without the aid of a human being [12], but there are barriers to clinical implementation. Substantial regulatory and safety concerns must be addressed when automating a diagnosis without human supervision [13]. In addition, some believe inertia within the field of medicine has already stalled early efforts at using machine learning in clinical practice [14]. But algorithms designed to aide pathologists and clinicians in an ancillary manner may be useful in the near future. Such approaches have been used in the research setting for non-small cell lung cancer prognostication [15], predicting recurrence in early stage colon cancer [16], and predicting molecular aberrations [10, 17].
Unsupervised CNNs have also made inroads into antibody-based ancillary testing. In such approaches, ground-truth masks are generated from antibody-bound slides. These are used with their corresponding H&E images to train a CNN. Recent examples of these used IHC performed on tissue slightly deeper in the tissue block [18, 19]. This method has the advantage of using data, which is already available from clinical practice, and does not require pathologists to laboriously annotate images. In addition, the ground truth is unbiased, and the process can easily be automated. Using the subsequent tissue layer, however, does not yield precise cell-specific classification. Such precision may be vital in cases of microinvasion and micrometastasis, lesions with mixed cell populations, and lesions where individual cell immunophenotype is vital to the diagnosis. Moreover, as this technique cannot classify individual cells, it cannot accurately quantify lesional cells or perform cell-specific analytical tasks that may one day be useful in cancer staging and prognostication [20–23].
An alternative approach was recently developed where antibodies on the same tissue layer as the H&E WSI were used to generate the ground-truth mask. In one approach, immunofluorescence was used instead of IHC [24, 25]. This work achieved an accuracy of 94.5% when using cytokeratin and smooth muscle actin markers in pancreatic tissue. The disadvantages of using immunofluorescence are (I) that it is not commonly used to classify immunophenotype in the clinical setting, (II) it undergoes a loss of signal over time when exposed to light [26], (III) requires specialized equipment to perform, and (IV) is subject to considerable variation between different runs and different laboratories [27]. In an alternative approach, a recent study used phosphohistone-H3 IHC, performed on the same tissue layer as H&E to count mitoses within a cohort of breast cancer cases [28]. This algorithm appeared to have yielded adequate resolution, but has limited benefit since IHC is not required to count mitoses.
We hypothesize that a CNN could provide information that is currently only obtainable from IHC, in a manner that is rapid and inexpensive. Specifically, a CNN could potentially predict cell-specific immunophenotype based on H&E characteristics, and graphically represent this prediction as a WSI. We term the technique virtual IHC (vIHC). Herein, we document a pilot study using the SOX10 nuclear stain. By selecting regions of interest (ROI) in skin and lymph node biopsies, we created a neural network specifically designed to identify cells of melanocytic lineage. We report its successes, limitations, and outline directions for future works.
Material and methods
WSI preparation
A database search at our institution was carried out for recent cases that had been stained with SOX10 IHC as part of their routine workup. Cases that had abundant tissue and represented a range of diagnoses were favored. The set of 12 slides (NN-master set), from both skin (nine cases) and lymph nodes (three cases), consisted of eight melanomas, two in situ melanomas, one neuroma, and one pigmented basal cell carcinoma.
The images in the NN-master set were processed and used to train the vIHC CNN. A diagram depicting the full process is shown in Fig. 2. New tissue sections were first cut from the tissue blocks at 5 μm and stained with H&E. The slides were scanned at 400× (Fig. 2a) using the Leica Aperio AT2 scanner (Buffalo Grove, IL, USA). A washout process was applied to the slides to remove the H&E stain (Fig. 2b). The slides were then stained with SOX10 IHC (Leica, PA0813, pre-diluted) using automated techniques (Leica Bond; Leica Microsystems, Bannockburn, IL, USA) with appropriate controls (Fig. 2c). The resulting SOX10 IHC slides were scanned at 400× (Fig. 2d).
Fig. 2. Diagram summarizing the methodology used to create the virtual immunohistochemistry (vIHC) neural network training set.
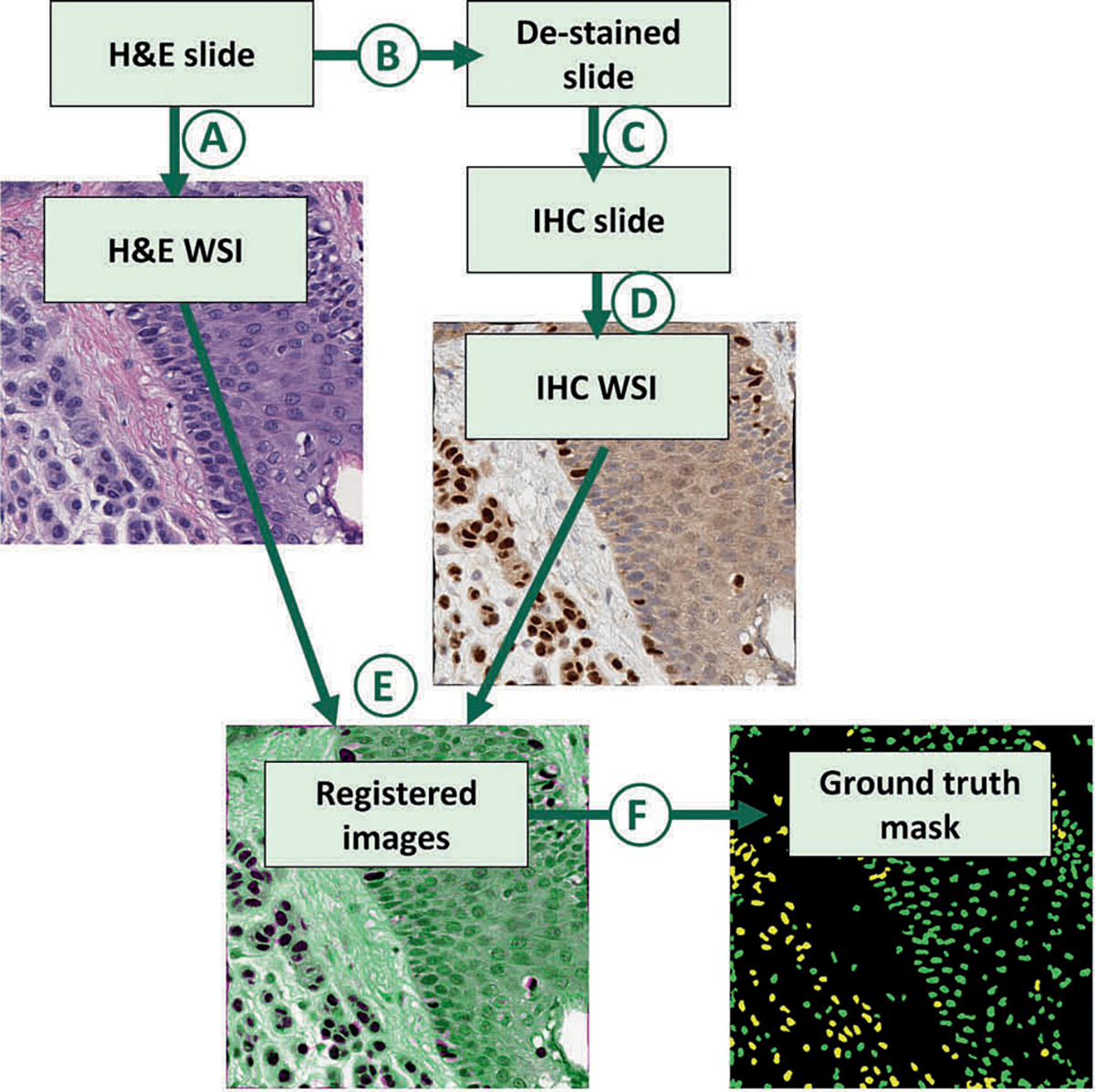
Hematoxylin and eosin (H&E) slides were first prepared from formalin-fixed paraffin-embedded tissue. The H&E slides were first scanned (a) to create digital H&E whole slide images (WSI). The H&E slides were then (b) destained and (c) stained with SOX10 immunohistochemistry (IHC). The resulting IHC slides were scanned (d) to create a digital IHC WSI. The H&E and corresponding IHC WSIs were registered (e), and processed (f) to create a ground-truth mask. In the ground-truth masks, SOX10-positive nuclei are colored yellow and SOX10-negative nuclei are colored green.
Registration
Registration is the process of aligning two images. In our study, the H&E and IHC WSIs were too large to be loaded into RAM at the same time, so they were registered in two steps similar to previously published work [29]. In the first step, the entire images were approximately registered. Then, ROI (see below) were more precisely registered so that corresponding nuclei in both images were superimposed onto one another (Fig. 2d). Specifically, the first step was a multimodal affine registration that allowed for rotation, translation, changes in scale, and shearing. In the second step, the images were divided into 1,000 × 1,000-pixel sub-image, some of which overlapped with selected ROI. The multimodal affine registration was repeated at high magnification for the sub-images that overlapped the ROI. This was followed by a nonrigid transformation function native to MATLAB [30, 31].
It was noted that the above described method resulted in poor nuclear overlap in 34.5% of sub-images. It was assumed that poorly registered images could result in mislabeled nuclei and would be deleterious to training the neural network. To ensure high-quality annotation, a categorical CNN was trained to distinguish well-registered sub-images from poorly registered sub-images. This was used to remove poorly registered sub-images from the NN-master set.
Regions of interest (ROI)
In the present iteration, only ROI were fully registered. A pathologist manually circled the ROI using the paintbrush tool in MS Paint (Microsoft, Redmond, WA, USA) in copies of the IHC WSI. A MATLAB code was created to read the annotated IHC images and extract the annotated regions, which were then used in the registration process. All regions that contained SOX10-positive melanocytes were annotated as ROI, while all regions without SOX10 positivity and regions with nonmelanocytic SOX10-positive cells were purposefully excluded.
Seeding
Seeding techniques were developed to accurately count and localize cells, without painstakingly annotating cellular contours [32]. In these techniques, a segmentation CNN is trained to recognize the center of the nucleus, which is annotated with a single dot [32–35]. Here, 500 H&E images were generated from clinical practice, each having a resolution of 500 × 500 pixels. The center of each nucleus was manually annotated by a pathologist using MS Paint, and was performed by adding a green dot to the center of each nucleus. A MATLAB code was generated to read the dotted images, and convert these into a ground-truth mask. These were used to train a CNN capable of segmenting individual cells, which was used as part of the annotation process described below.
Annotation
Annotation is the process of labeling regions within an image to create a ground-truth mask. Typically, different colors are used to annotate different regions. Here, the training images were annotated using automated techniques. The nuclei from the H&E images were annotated by the seeding CNN described above. In each ROI, a color-thresholding technique was applied to the corresponding registered IHC image to allow for positive staining to automatically be distinguished from negative staining. The individual nuclei were classified as positive or negative, and labeled accordingly. The result was a final ground-truth mask, where SOX10-positive nuclei were labeled as yellow, and SOX10-negative nuclei were labeled as green (Fig. 2e). All nuclei that were outside the ROI were automatically categorized as negative.
In addition, all sub-images with <20 nuclei were discarded. The resulting master neural network set (NN-master) consisted of 18,122 images. Ninety percent of these images were randomly assigned to train the SOX10-vIHC neural network (NN-train set; 16,309 images), and the remaining 10% were used to test it (NN-test set; 1,813 images). All image sets are described in Table 1. The total number of sub-images, the number of well-registered sub-images with more than 20 nuclei, and the number of SOX10-positive and SOX10-negative cells for each WSI in the NN-master, NN-train, and NN-test sets are shown in Table 2. The annotation process was entirely automated using a MATLAB code.
Table 1.
Description of image sets.
| Alias | Lesion types | Number of WSIs | Same-layer IHC | Description |
|---|---|---|---|---|
|
| ||||
| NN-master set | MIS, malignant melanoma, MM, BCC, neuroma | 12 | Yes | All H&E images with corresponding digital IHC masks; see Table 2. The NN-master set was randomly divided into the NN-train set (90%), which was used to train the neural network, and the NN-test set (10%), which was used to test the neural network. |
| IHC-test set | One case of inflamed melanoma | 1 | Yes | Image used for graphical evaluation of vIHC with direct comparison to IHC. |
| Subjective-test set | Invasive melanoma, MM, BCC | 6 | No | Graphical evaluation of vIHC using H&E only with inferred correct IHC-staining pattern. |
BCC basal cell carcinoma, H&E hematoxylin and eosin, IHC immunohistochemistry, MIS melanoma in situ, MM metastatic melanoma, NN neural network, WSI whole slide image.
Table 2.
Histopathologic characteristics of the NN-master set.
| Case | Location Lesion type | Sub-images | Number of cells |
Cell-fraction of data set [%] | |||
|---|---|---|---|---|---|---|---|
| SOX10-positive | SOX10-negative | Percent SOX10-positive [%] | |||||
|
| |||||||
| 1 | Skin | MIS | 1,987 | 654 | 317,261 | 0.2 | 8.4 |
| 2 | Skin | MIS | 1,290 | 594 | 187,875 | 0.3 | 5.0 |
| 3 | Skin | Melanoma | 2,168 | 19,142 | 336,058 | 5.4 | 9.4 |
| 4 | Skin | Melanoma | 2,004 | 19,551 | 264,744 | 6.9 | 7.5 |
| 5 | Skin | BCC | 260 | 1,051 | 58,222 | 1.8 | 1.6 |
| 6 | Skin | Melanoma | 975 | 25,493 | 167,175 | 13.2 | 5.1 |
| 7 | Skin | Neuroma | 361 | 556 | 66,314 | 0.8 | 1.8 |
| 8 | Skin | Melanoma | 488 | 31,210 | 99,420 | 23.9 | 3.4 |
| 9 | LN | MM | 2,796 | 18,402 | 661,343 | 2.7 | 17.9 |
| 10 | LN | MM | 2,249 | 61,901 | 623,796 | 9.0 | 18.1 |
| 11 | Skin | Melanoma | 768 | 10,964 | 91,754 | 10.7 | 2.7 |
| 12 | LN | MM | 2,776 | 116,648 | 522,706 | 18.2 | 16.9 |
| Total | 18,122 | 306,166 | 3,396,668 | 8.3 | 100 | ||
BCC basal cell carcinoma, LN lymph node, MIS melanoma in situ, MM metastatic melanoma.
Once the SOX10-vIHC network was trained, it could be run on new images to generate positive-nucleus scores, negative-nucleus scores, and background score. These scores corresponded to the probability that a given pixel corresponded to a SOX10-positive nucleus, a SOX10-negative nucleus, or a nonnuclear region, respectively.
CNN specifications
All CNNs were created using either a pretrained VGG19 network [36] or InceptionV3 network [37] using MATLAB R2018b (MathWorks, version 9.5.0.944444, Natick, MA, USA). The networks were trained using a Titan Xp (NVi-dia, Santa Clara, CA, USA) graphics processing unit and a Ryzen Threadripper 1950×16-Core CPU (Advanced Micro Devices [AMD], Santa Clara, CA, USA). Each network was trained between 30 and 300 epochs.
Evaluation
The SOX10-vIHC network was first evaluated by processing the NN-test set images. A MATLAB program was created to compare the vIHC output predictions to their corresponding ground-truth masks. This was performed on a per-cell basis by setting all areas with combined SOX10 positivity and negativity scores <0.95 to 0. A connected-components analysis was then performed to delineate each nucleus. The mean SOX10-positivity score and SOX10-negativity score for each segmented nucleus was computed, and the category with the largest score was recorded and compared with the ground truth category. The results for 20,000 SOX10-positive and 20,000 SOX10-negative cells were randomly selected using the randperm function native to MATLAB. These were aggregated and used to calculate the true positive (TP), true negative (TN), false positive, and false negative values (Fig. 3). A cumulative density plot and a receiver operator characteristics (ROC) curve was produced using these scores (Figs. 4, 5).
Fig. 3. Example image from NN-test set.
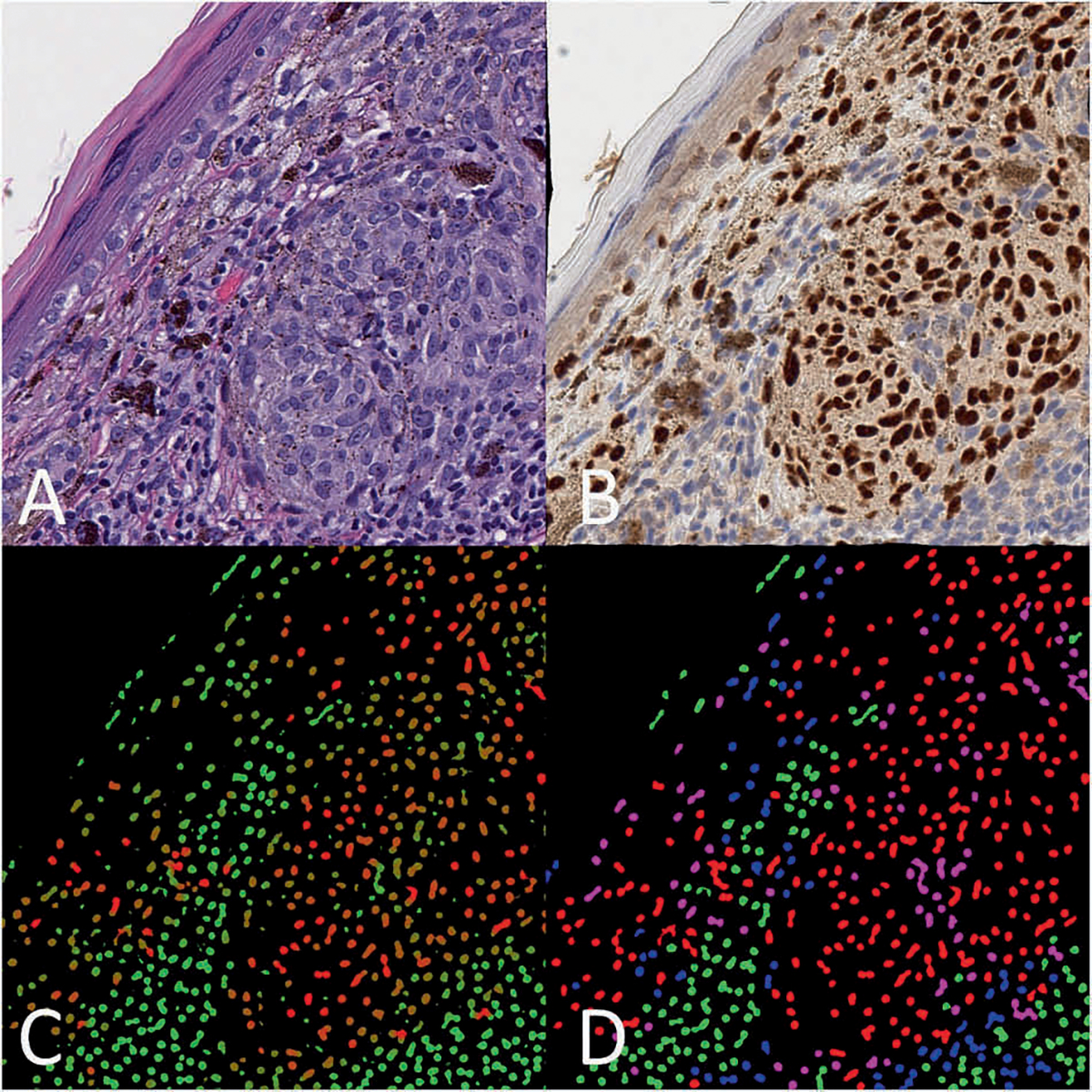
a H&E image of melanoma in a tissue section of skin (H&E, 400×); b corresponding IHC (SOX10, 400×); c raw vIHC output, where the green color channel is scaled to the SOX10-negativity score, and the red color channel is scaled to the SOX10-positivity score. A strongly red nucleus is predicted to be SOX10 positive, while a strongly green nucleus is predicted to be SOX10 negative. d Corresponding color map where nuclei are colored depending on their true SOX10 IHC and predicted vIHC positivity. True positives are colored red, true negatives are colored green, false positives are colored pink, and false negatives are colored blue.
Fig. 4. Receiver operator curve characteristics (ROC) curve for the SOX10 vIHC.
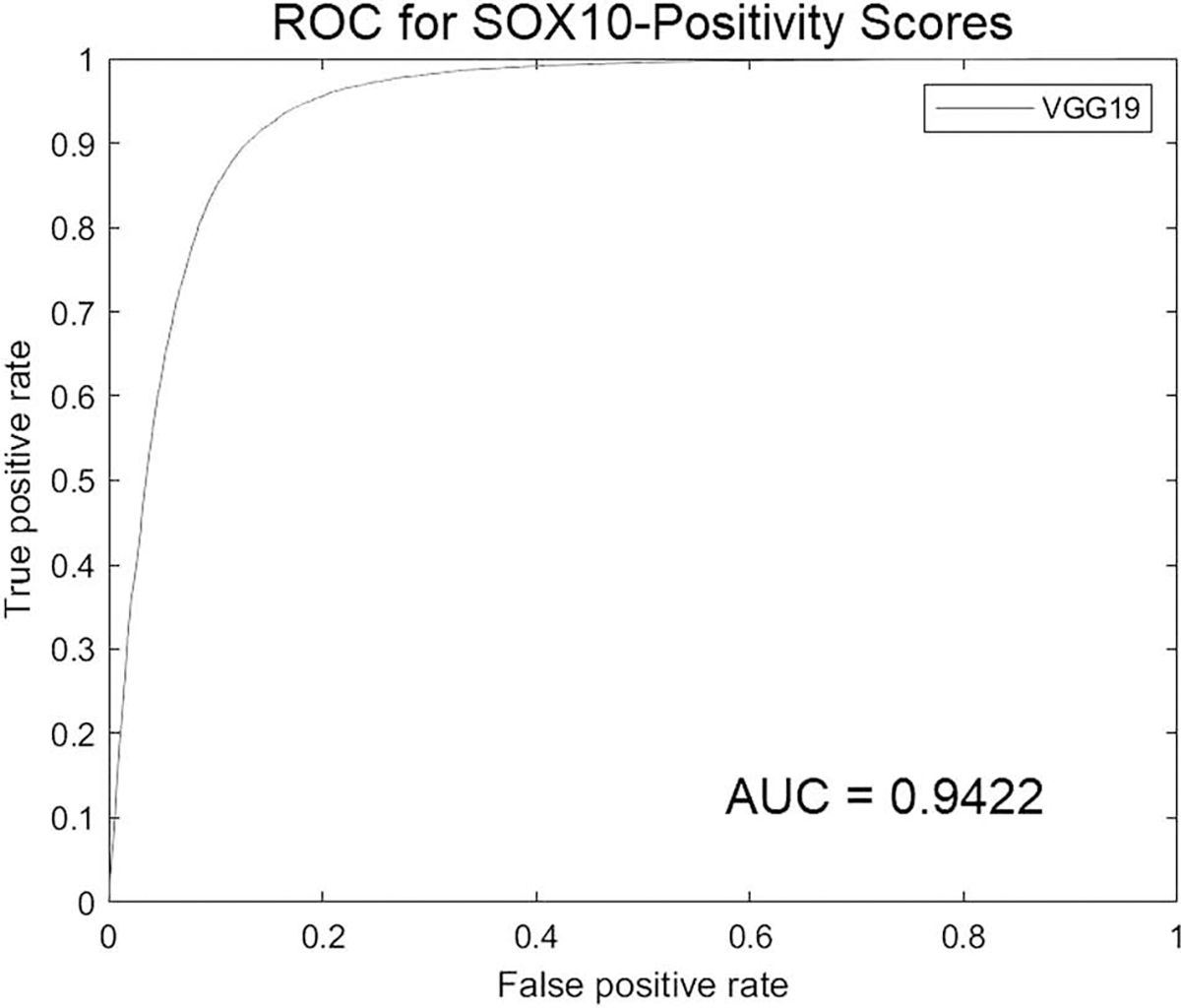
AUC area under the curve.
Fig. 5. Cumulative distribution plot of the SOX10-positivity scores.
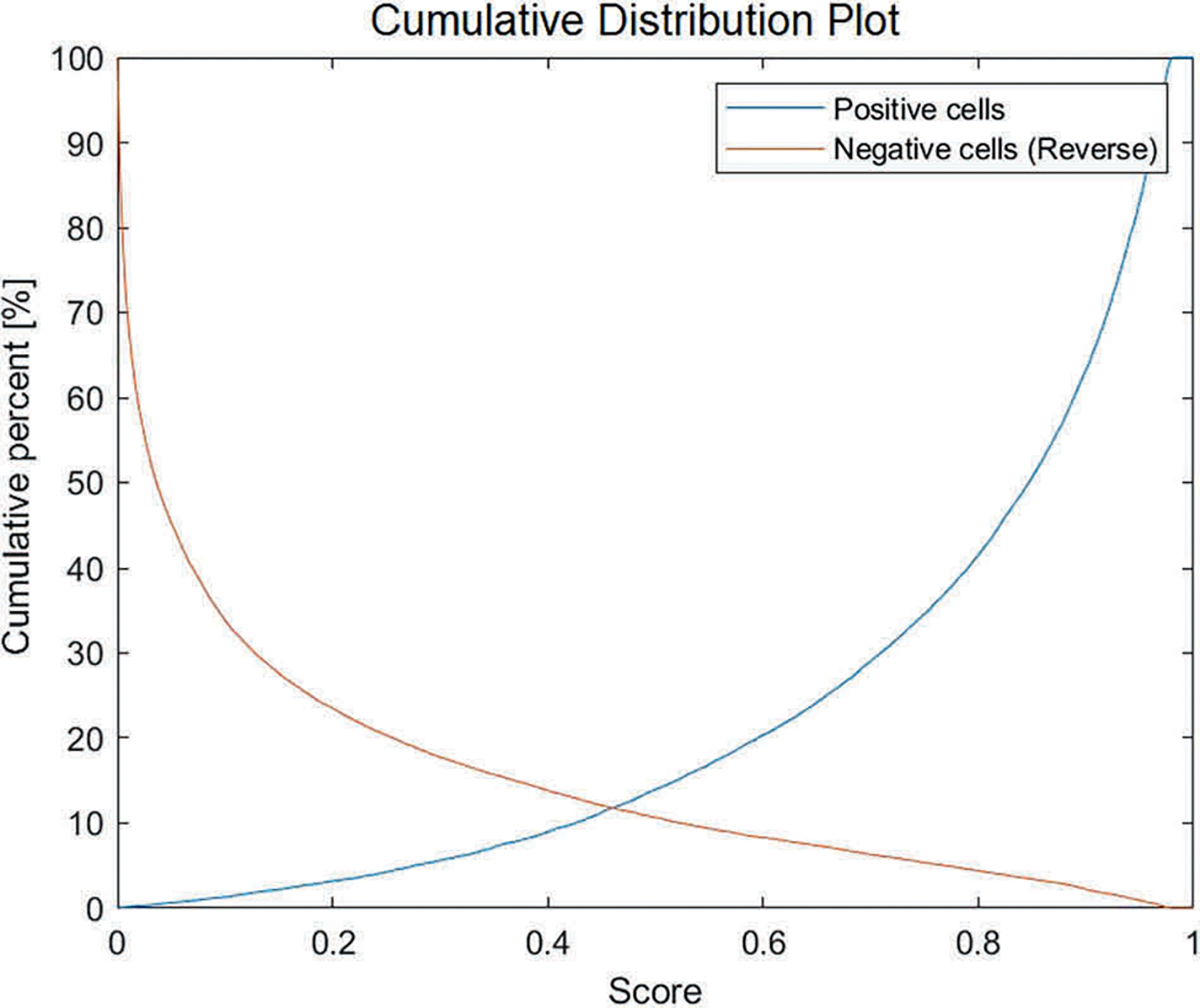
The average SOX10-positivity score is calculated for each cell nuclei within the NN-test set. Blue line: SOX10-positive cells; Orange line: Reverse cumulative distribution plot of SOX10-negative cells.
To comprehensively analyze the vIHC and IHC, the two were directly compared qualitatively by an experienced board-certified dermatopathologist. This allowed for characterization of the cell populations that were overcalled and undercalled as SOX10-positive. An additional set of images was used for this purpose (IHC-test set). Four H&E sub-images from a single case of inflamed melanoma were processed by the CNN. This case was chosen as it was not used in the training set. The four sub-images in the IHC-test set contained areas of normal skin, an area of dysplastic junctional melanocytes, and frankly invasive melanoma interfacing with lymphocytic inflammation. In a manner similar to that described above, the H&E slide were scanned at 400× resolution. The slide was then destained, and stained with SOX10 IHC before it was scanned at 400×. vIHC was applied to the H&E WSI, and the output was converted to a color map that was overlaid on top of the H&E image. In the color map, nuclei predicted to be SOX10-negative were colored blue, while nuclei predicted to be SOX10-postive were colored red. This was directly compared with the SOX10 IHC. A final set of images (subjective-test set) was also graphically evaluated and consisted of a lymph node containing metastatic melanoma, four cases of primary melanoma, and a case of basal cell carcinoma. The appropriate staining pattern in the subjective-training set was inferred by an experienced board-certified dermatopathologist based on either the diagnosis, or SOX10 IHC performed on an adjacent tissue layer.
Results
IHC ground truth results
SOX10 IHC highlighted the nuclei of melanocytes, as well as Schwann cells, and the myoepithelial cells of eccrine glands. One case with a neuroma expressed SOX10. For the purposes of training the vIHC, only melanocytes were included in the ROI.
WSI results
The average vIHC SOX10-positivity and SOX10-negativity scores were calculated for every nucleus in the NN-test set. The TP and TN values were calculated by comparing the ground truth category of each nucleus to the scores computed by the vIHC network (Fig. 3). An ROC curve was created (Fig. 4), and the area under the curve (AUC) was calculated as 0.9422. The resulting sensitivity and specificity were 91.62% and 85.66%, respectively at the optimal point on the ROC curve which was a score of 0.3868. The resulting cumulative distribution plot is shown in Fig. 5.
vIHC graphical evaluation results
The four sub-images from the IHC-test set were evaluated (Fig. 6). SOX10 IHC highlighted several cell populations in the IHC-test set images. Benign melanocytes, dysplastic melanocytes, and malignant melanocytes within the melanoma were all highlighted.
Fig. 6. Melanoma with adjacent lymphocytic inflammation from the IHC-test set.
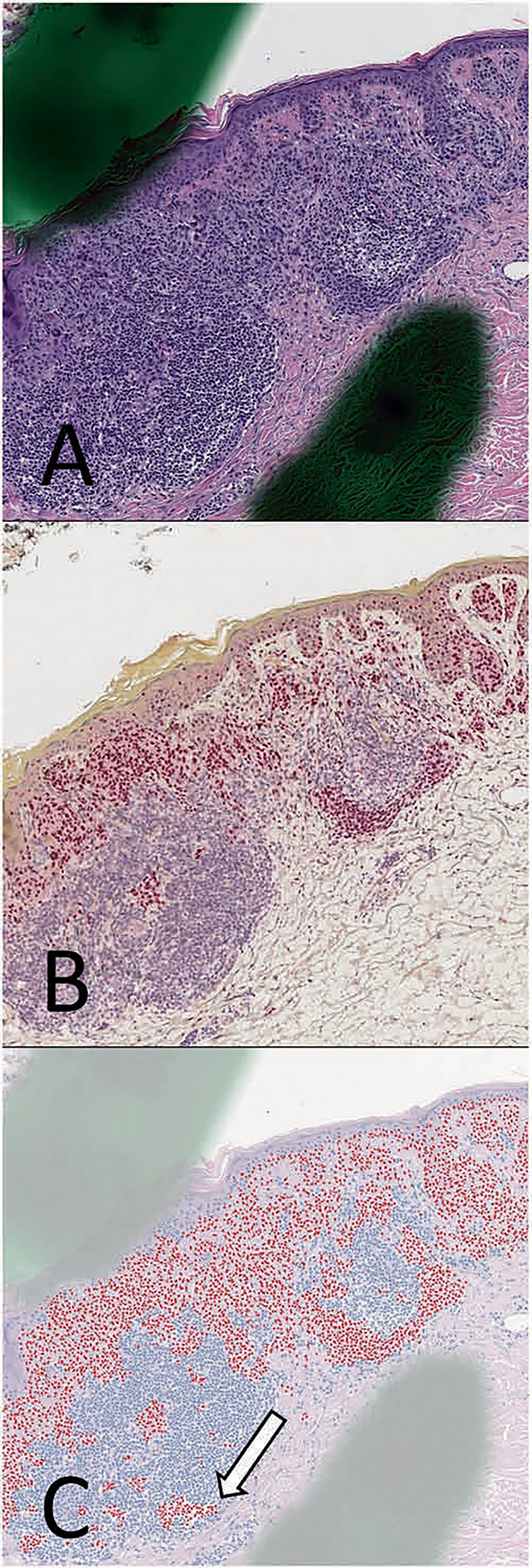
a Hematoxylin and eosin (H&E) stains, along with corresponding b SOX10 immunohistochemistry (IHC) performed on the same tissue layer. c SOX10 virtual immunohistochemistry (vIHC) was performed using the H&E image as an input. Lymphocytes that were erroneously labeled as SOX10-positive by the vIHC are shown (white arrows). Notably, artifacts that will be found in slides used for clinical care, such as dotting marks, do not appear to have a significant adverse effect on the neural network in this case.
The resulting vIHC was compared with the SOX10 IHC stain performed on the same cell layer (Fig. 7). vIHC subjectively highlighted most of the malignant melanoma cells, and appropriately did not highlight most of the inflammatory infiltrates interfacing with the lesion. Nests of dysplastic melanocytes were also appropriately highlighted. The SOX10 vIHC was comparable to the IHC in areas of malignant melanoma, nested melanocytes, and in normal skin, as assessed by an experienced board-certified dermatopathologist. The vIHC was nevertheless imperfect, and highlighted several foci of lymphocytes as well as occasional keratinocytes (Fig. 7c). Rarely, melanoma cells were improperly classified as nonmelanocytic.
Fig. 7. Region of inflammation abutting invasive melanoma from the IHC-test set.
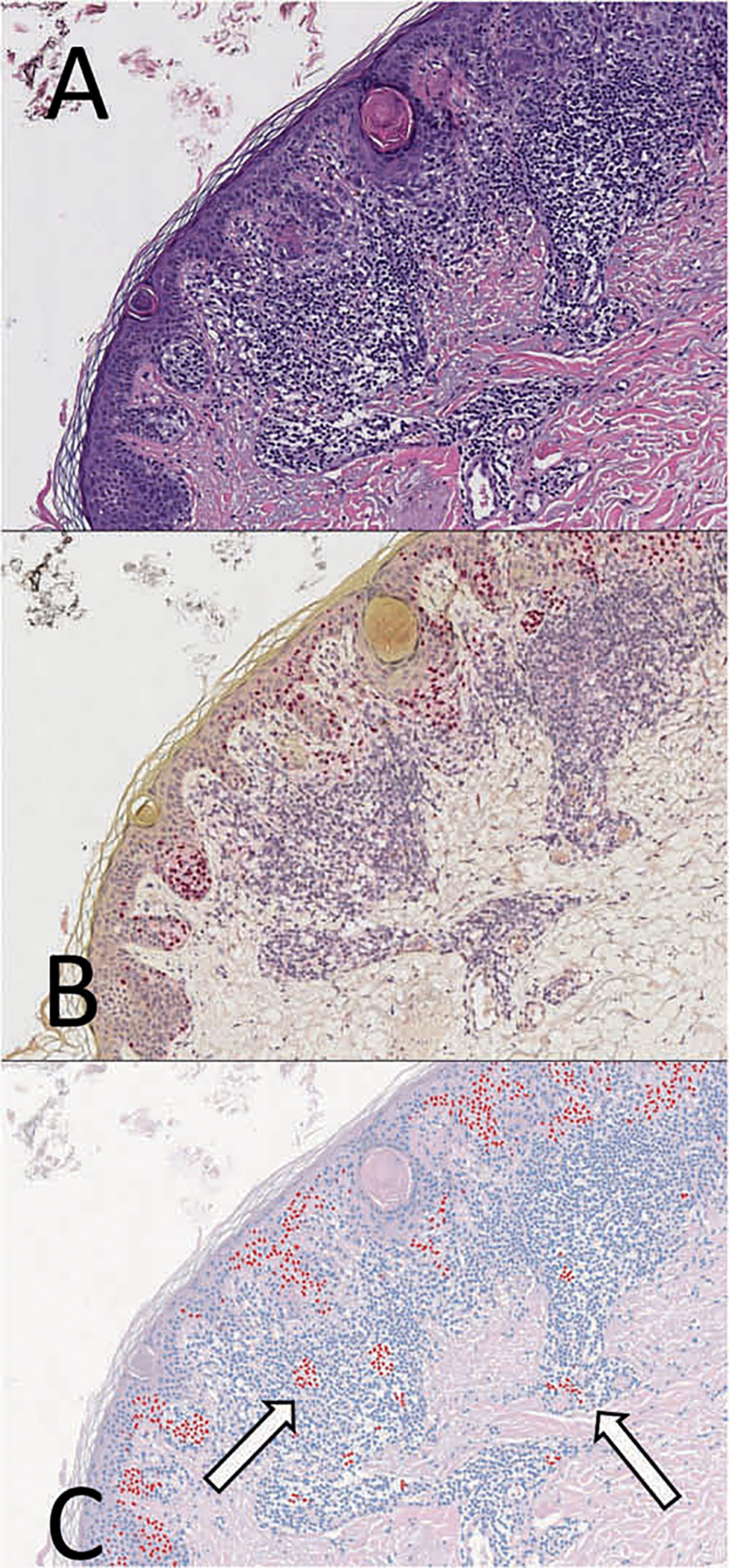
a Hematoxylin and eosin (H&E) stains, along with corresponding b SOX10 immunohistochemistry (IHC) performed on the same tissue layer. c SOX10 virtual immunohistochemistry (vIHC) was performed using the H&E image as an input. Lymphocytes that were erroneously labeled as SOX10-positive by the vIHC are shown (white arrows).
vIHC was also performed on the images in the subjective-test set (Fig. 8). For these images, the appropriate IHC-staining pattern was inferred either based on SOX10 IHC performed on an adjacent tissue layer or based on the pathologic diagnosis. These images included primary melanomas, a metastatic melanoma, and basal cell carcinoma. Most metastatic melanoma cells in the lymph node were appropriately classified as melanocytic (Fig. 8b). Many regions within primary melanomas were appropriately highlighted (Fig. 8d), though not uniformly so. The basal cell carcinoma appropriately had negative SOX10-vIHC expression throughout most of the lesion. Adjacent to the lesion, however, clusters of lymphocytes were inappropriately labeled as SOX10-posivite (Fig. 8f, white arrows). A Melan-A stain was used to verify these results.
Fig. 8. Subjective-test set examples.
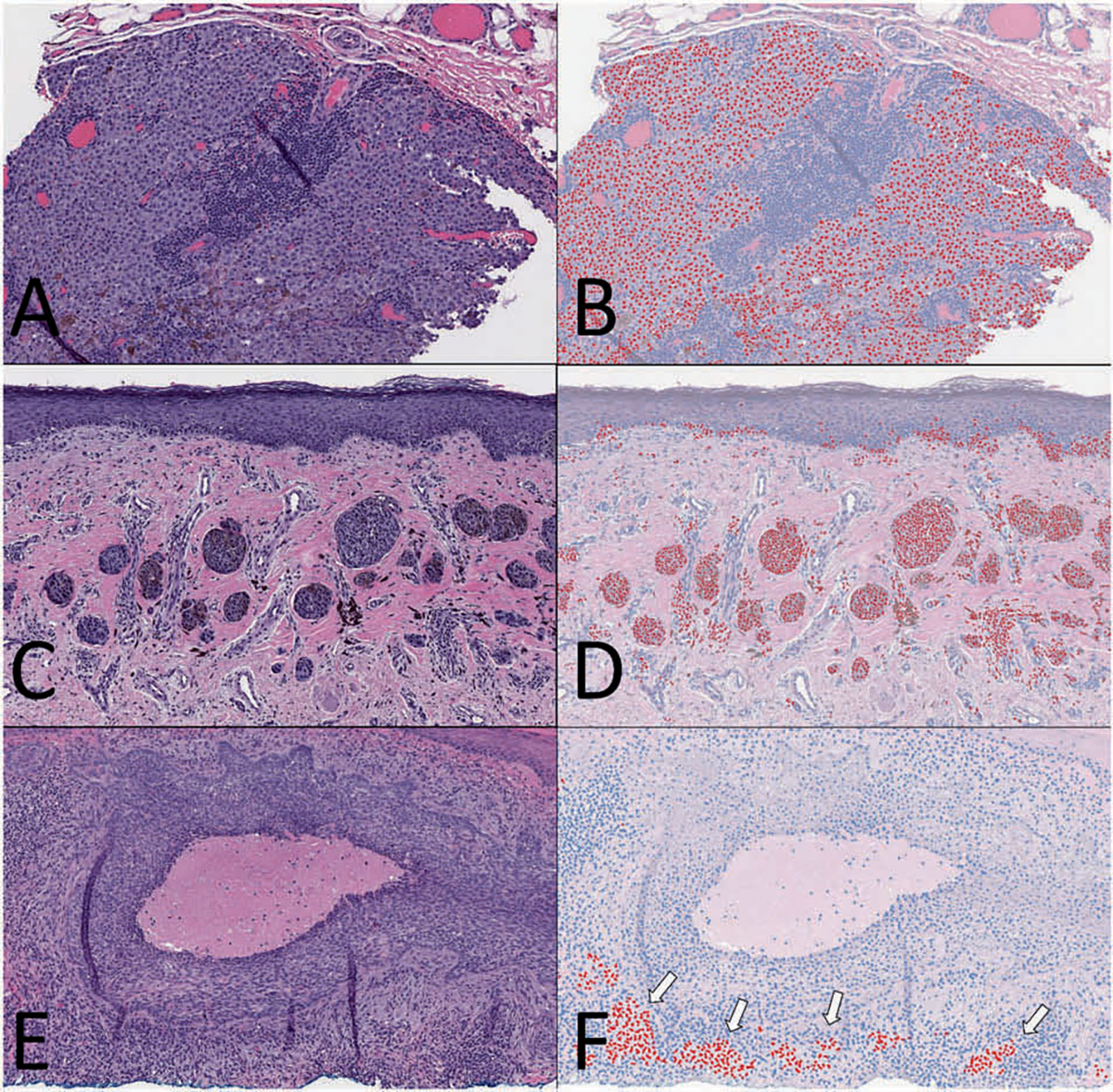
Representative images from three different cases in the subjective-test set comparing H&E (a, c, e) to SOX10 virtual immunohistochemistry (vIHC) (b, d, f). (a, b) A case of melanoma metastasis within a lymph node that was appropriately highlighted as SOX10-positive. (c, d) A case of melanoma with features of superficial spreading and nevoid types. Melanocytic nests were highlighted by the SOX10 vIHC. (e, f) A case of basal cell carcinoma that was shown to be void of melanocytes using a Melan-A stain. vIHC erroneously highlighted a population of lymphocytes near the bottom of the image (white arrows).
Discussion
IHC is highly useful in surgical pathology, but problems remain. It consumes tissue, increases turnaround time, and adds to healthcare cost. Accurate vIHC may improve upon these shortcomings. vIHC could also be used in novel ways, such as counting individual cells and performing cellular morphometrics. A neural network with single-cell resolution capable of providing data only obtainable from IHC has not been made before. We document the development and performance of the first such virtual assays.
In developing the CNN, the nuclei in the H&E and corresponding IHC images required a high degree of overlap for individual cells to be labeled appropriately. Achieving this degree of overlap was difficult because the size of the WSI could exceed 40,000 × 40,000 pixels. For reference, the width of a nucleus was ~28 pixels in diameter. Even if only 50% overlap was required, such large images would require a low tolerance for error, ~0.035% (28 pixels × 50% ÷ 40,000 pixels). This margin for error is significantly smaller than that required in other manipulations of high-resolution images. Next-layer WSI registration, for example, needs only to have segments of tissue overlap and not the nuclei themselves. Likewise, MRI and CT scan image sizes are significantly smaller than those of WSIs, and the required relative registration accuracy is much lower [38]. There are likely several approaches to overcome this problem. One approach would be to manually register the images, but this was beyond the technical resources provided for our study. Moreover, as the goal of the study was to establish a proof-of-concept for a high-throughput method, a method that could be automated was employed. The performance of the categorical CNN used to remove poorly registered images from the data set is an inherent source error for the downstream vIHC network, but allows for a highly automated, reproducible workflow. Future efforts can be directed toward achieving a more efficient and robust registration process.
We assessed the SOX10 vIHC both quantitatively and qualitatively. The quantitative assessment included an ROC analysis (Fig. 3) using conventional IHC WSIs as the gold standard. The computed probability of nuclear SOX10 expression from the neural network was treated as the analyte. An optimal cutoff value of 0.3868 was determined. The AUC was 0.9422 and the sensitivity and specificity at this cutoff were 91.62% and 85.66%, respectively.
The F1 score is a measure of a test’s accuracy, and is defined as:
Although not directly comparable due to the composition of our respective test sets, the F1 score (0.890) was much higher than that of a similar prior approach that used phosphohistone-H3 to detect mitotic figures (0.668) [28]. The improvement may be attributable to a rigorous automated-registration protocol that removed poorly registered images from the data set. The sensitivity and specificity from our study were comparable to a similar work that used immunofluorescence to investigate cytokeratin expression in pancreatic carcinoma [25].
It is worth noting some of the differences between our work and prior studies that used immunofluorescence to annotate H&E images. The first difference is related to biology; melanocyte morphology is notoriously varied [39–41] when compared with the other tumors examined in the small number of related prior studies. In our study, the NN-master set consisted of multiple lesion and tissue types in order to better simulate clinical conditions where the diagnosis is often not known prior to IHC. In contrast, previous work focused on a single cancer type from a single tissue source. It is possible that lesion and tissue diversity in the present study may have improved generalizability, but perhaps at the cost of accuracy. Future work could assess whether using a single lesion type and tissue type improves performance, and quantify how much generalizability, if any, is lost as a result.
A qualitative evaluation was also performed because, unlike other forms of testing, IHC requires correlation with morphology and tissue architecture. Particularly in melanocytic neoplasms, a small number of cells that are inappropriately classified can drastically change the diagnosis if they are in a critical location. For such reasons, all the images were reviewed by a board-certified dermatopathologist (Figs. 6, 7). The majority of the resulting images were not considered absolutely equivalent to IHC. Many, if used blindly and without regard to morphology, could result in a misdiagnosis. Much of the error resulted from the algorithm mislabeling of keratinocytes. Because pagetoid spread can be an important diagnostic feature in the distinction of nevus from melanoma, this type of mislabeling had a significant negative impact on the qualitative assessment. As vIHC outputs images, the errors are generally transparent; trends in mislabeling can be identified. Future work could augment the training set to systematically address these errors.
Our study had multiple limitations worth examining. One of these is imperfect registration which could have led to erroneous nuclear categorization in the NN-master set. Tissue warping during the wash out and IHC-staining steps added to the challenge of image registration. Future studies could design and assess protocols that minimize the degree of warping during wash out. The effect of using different digital scanners, along with different tissue processing and staining protocols, was not examined in this study. Follow-up studies could be performed with slides from different labs, and stained and scanned under different protocols with the goal of documenting the effects that these will have on model performance.
An important potential pitfall of vIHC is that it may not perform in the same manner as the conventional IHC marker on which it is was based, especially in lesions and tissue sites of original not included in the original training set. It is likely that each vIHC model needs to be empirically tested on a multitude of different lesions from different sites of origin, similar to a de novo IHC marker. The lesions included in the test set should include all lesions under diagnostic consideration, and the respective sensitivities and specificities need to be established for each lesion type. Fortunately, once the model is created, there is almost no additional cost to performing such studies on a large scale.
Notably, our approach relied on a pathologist to highlight ROI. By limiting fine registration to the ROI, the time required to register the training set was reduced from 5.5 days to 1.5 days. Although ROI were used, it may be desirable to register the entire WSI in the training set for some applications.
As a pilot study, this work offers numerous directions for useful future research. First, the algorithm’s performance with regard to mislabeling of keratinocytes and lymphocytes can be improved by including larger numbers of cases in the training set. Second, other IHC targets may be developed. Third, vIHC may augment pathology research efforts.
Opportunities abound for future studies in vIHC beyond SOX10. It may prove feasible to design algorithms for other common nuclear and cytoplasmic markers, including those that are lineage-specific. PAX8, PAX5, CD3, CDX2, LCA, TTF1, cytokeratins, CK5/6, OCT3/4, WT1, synaptophysin, desmin, CK7, and CK20, for example, are commonly used in the workup of a number of lesions [42]. vIHC could potentially expedite the workup of these lesions. Ki67 would be valuable as it could be used to automatically calculate the Ki67 index which has diagnostic or prognostic value in several lesions [20–22]. vIHC offers a potential avenue to standardize stain interpretation.
vIHC presents opportunities for basic science research in at least two domains: (I) automated cell quantitation and (II) the study of cellular morphometrics. Automated cell quantitation can be done by selecting a ROI, applying the vIHC, and thereby obtaining a count of the cells that express the marker. The study of cellular morphometrics could be aided by vIHC as this allows for easy isolation of cells based on their immunophenotype. Studies that examine properties of specific cell types could be facilitated. For example, research that examines the nuclear-to-cytoplasm ratio of lesional cells could be automated while nonlesional cells could easily be excluded from analysis.
In conclusion, conventional IHC is expensive, labor-intensive, time-consuming, and can waste precious tissue in small biopsy samples. Nevertheless, the diagnostic information provided by IHC can be extremely useful, and in some cases is indispensable [20, 43, 44]. A rapid and inexpensive method to accurately obtain the same information could have numerous benefits for research and clinical care in both resource-heavy and resource-limited settings. Our work is a proof-of-concept study. It demonstrates that immunohistochemical data with cell-specific resolution can be obtained using artificial intelligence. The advantages in terms of time, labor, and cost are clear. These initial results indicate that accurate vIHC is feasible. Future work can examine methods to optimize image registration and improve accuracy. With such improvements, it is possible that an inexpensive, rapid, accurate tool, capable of yielding indispensable diagnostic information, could become widely used in the diagnosis and treatment of cancer patients.
Supplementary Material
Acknowledgements
This work was supported in part by a grant from the DHMC Norris Cotton Cancer Center, Cancer Fellows Program, the GPU Research Grant, Nvidia (Santa Clara, California), and the Dartmouth Hitchcock Medical Center Department of Pathology and Laboratory Sciences.
Footnotes
Conflict of interest Authors CRJ and LJV have a provisional patent on the approach taken in this manuscript: provisional App. (Ref. 076/0054R): system and method for providing a rapid virtual diagnostic companion for use in diagnosis of cancer and related conditions using immunohistochemistry based upon a neural network.
Supplementary information The online version of this article (https://doi.org/10.1038/s41379-020-0526-z) contains supplementary material, which is available to authorized users.
References
- 1.De Matos LL, Trufelli DC, De Matos MG, da Silva Pinhal MA. Immunohistochemistry as an important tool in biomarkers detection and clinical practice. Biomark Insights. 2010;5:S2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alom MZ, Aspiras T, Taha TM, Asari VK, Bowen T, Billiter D, et al. Advanced deep convolutional neural network approaches for digital pathology image analysis: a comprehensive evaluation with different use cases. Ithica, New York: Cornell University; 2019. http://arxiv.org/abs/1904.09075. [Google Scholar]
- 3.Song Y, Zhang L, Chen S, Ni D, Li B, Zhou Y, et al. A deep learning based framework for accurate segmentation of cervical cytoplasm and nuclei. In: Conference Proceedings of the IEEE Engineering in Medicine and Biology Society. Chicago, IL: Institute of Electrical and Electronics Engineers; 2014;2014:2903–6. [DOI] [PubMed] [Google Scholar]
- 4.Olsen TG, Jackson BH, Feeser TA, Kent MN, Moad JC, Krishnamurthy S, et al. Diagnostic performance of deep learning algorithms applied to three common diagnoses in dermatopathology. J Pathol Inform. 2018;9:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cruz-Roa AA, Ovalle JEA, Madabhushi A, Osorio FAG. A deep learning architecture for image representation, visual interpretability and automated basal-cell carcinoma cancer detection. Adv Inf Syst Eng. 2013. 10.1007/978-3-642-40763-5_50. [DOI] [PubMed] [Google Scholar]
- 6.Albarqouni S, Baur C, Achilles F, Belagiannis V, Demirci S, Navab N. AggNet: deep learning from crowds for mitosis detection in breast cancer histology images. IEEE Trans Med Imaging. 2016;35:1313–21. [DOI] [PubMed] [Google Scholar]
- 7.Cruz-Roa A, Basavanhally A, González F, Gilmore H, Feldman M, Ganesan S, et al. Automatic detection of invasive ductal carcinoma in whole slide images with convolutional neural networks, In: Proceedings of the Medical Imaging 2014: Digital Pathology. San Diego, CA: SPIE - International Society for Optics and Photonics; 2014. 10.1117/12.2043872. [DOI] [Google Scholar]
- 8.Araújo T, Aresta G, Castro E, Rouco J, Aguiar P, Eloy C, et al. Classification of breast cancer histology images using convolutional neural networks. PloS One. 2017;12. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5453426/. [DOI] [PMC free article] [PubMed]
- 9.Marsh JN, Matlock MK, Kudose S, Liu T-C, Stappenbeck TS, Gaut JP, et al. Deep learning global glomerulosclerosis in transplant kidney frozen sections, IEEE Trans Med Imaging. 2018;37. 10.1109/tmi.2018.2851150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, et al. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24:1559–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sirinukunwattana K, Ahmed Raza SE, Tsang Y-W, Snead DRJ, Cree IA, Rajpoot NM. Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images, IEEE Trans Med Imaging. 2016;35. 10.1109/tmi.2016.2525803. [DOI] [PubMed] [Google Scholar]
- 12.Chen H, Qi X, Yu L, Dou Q, Qin J, Heng PA. DCAN: deep contour-aware networks for object instance segmentation from histology images. Med Image Anal. 2017;36:135–46. [DOI] [PubMed] [Google Scholar]
- 13.Challen R, Denny J, Pitt M, Gompels L, Edwards T, Tsaneva-Atanasova K. Artificial intelligence, bias and clinical safety. BMJ Qual Saf. 2019;28:231–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Longoni C, Bonezzi A, Morewedge CK. Resistance to medical artificial intelligence. J Consum Res. 2019;46:629–50. [Google Scholar]
- 15.Hosny A, Parmar C, Coroller TP, Grossmann P, Zeleznik R, Kumar A, et al. Deep learning for lung cancer prognostication: a retrospective multi-cohort radiomics study. PLoS Med. 2018;15:e1002711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yue X, Dimitriou N, Arandjelovic O. Colorectal cancer outcome prediction from H&E whole slide images using machine learning and automatically inferred phenotype profiles. Ithica, New York: Cornell University; 2019. http://arxiv.org/abs/1902.03582. [Google Scholar]
- 17.Rawat RR, Ruderman D, Agus DB, Macklin P. Abstract 540: deep learning to determine breast cancer estrogen receptor status from nuclear morphometric features in H&E images, Bioinform Syst Biol. 2017. 10.1158/1538-7445.am2017-540. [DOI] [Google Scholar]
- 18.Sharma H, Zerbe N, Klempert I, Hellwich O, Hufnagl P. Deep convolutional neural networks for automatic classification of gastric carcinoma using whole slide images in digital histopathology. Comput Med Imaging Graph. 2017;61:2–13. [DOI] [PubMed] [Google Scholar]
- 19.Xu J, Luo X, Wang G, Gilmore H, Madabhushi A. A deep convolutional neural network for segmenting and classifying epithelial and stromal regions in histopathological images. Neurocomputing. 2016;191:214–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amin MB. AJCC cancer staging system. 8th ed. Chicago, IL: American Joint Committee on Cancer; 2017. [Google Scholar]
- 21.Soliman NA, Yussif SM. Ki-67 as a prognostic marker according to breast cancer molecular subtype. Cancer Biol Med. 2016;13: 496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rudolph P, Lappe T, Hero B, Berthold F, Parwaresch R, Harms D, et al. Prognostic significance of the proliferative activity in neuroblastoma. Am J Pathol. 1997;150:133. [PMC free article] [PubMed] [Google Scholar]
- 23.Udall M, Rizzo M, Kenny J, Doherty J, Dahm S, Robbins P, et al. PD-L1 diagnostic tests: a systematic literature review of scoring algorithms and test-validation metrics. Diagn Pathol. 2018;13:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang YH, Burlingame EA, Gray JW, Margolin AA. SHIFT: speedy histopathological-to-immunofluorescent translation of whole slide images using conditional generative adversarial networks. In: Proceedings of the Med Imaging 2018: Digital Pathology. Houston, TX: SPIE - International Society for Optics and Photonics; 2018. 10.1117/12.2293249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang YH, Thibault G, Madin O, Azimi V, Meyers C, Johnson B, et al. Deep learning based nucleus classification in pancreas histological images. In: Proceedings of the 39th Annual International Conference of the IEEE Engineering in Medicine and Biological Society. Seogwipo, South Korea: Institute of Electrical and Electronics Engineers; 2017. 10.1109/embc.2017.8036914. [DOI] [PubMed] [Google Scholar]
- 26.Odell ID, Cook D. Immunofluorescence techniques. J Invest Dermatol. 2013;133:e4. [DOI] [PubMed] [Google Scholar]
- 27.Kivity S, Gilburd B, Agmon-Levin N, Carrasco MG, Tzafrir Y, Sofer Y, et al. A novel automated indirect immunofluorescence autoantibody evaluation. Clin Rheumatol. 2012;31:503–9. [DOI] [PubMed] [Google Scholar]
- 28.Tellez D, Balkenhol M, Otte-Holler I, van de Loo R, Vogels R, Bult P, et al. Whole-slide mitosis detection in H&E breast histology using PHH3 as a reference to train distilled stain-invariant convolutional networks, IEEE Trans Med Imaging. 2018;37. 10.1109/tmi.2018.2820199. [DOI] [PubMed] [Google Scholar]
- 29.Lotz J, Olesch J, Muller B, Polzin T, Galuschka P, Lotz JM, et al. Patch-based nonlinear image registration for gigapixel whole slide images, IEEE Trans Med Imaging. 2016;63. 10.1109/tbme.2015.2503122. [DOI] [PubMed] [Google Scholar]
- 30.Vercauteren T, Pennec X, Perchant A, Ayache N. Diffeomorphic demons: efficient non-parametric image registration. Neuroimage. 2009;45:S61–72. [DOI] [PubMed] [Google Scholar]
- 31.Thirion JP. Image matching as a diffusion process: an analogy with Maxwell’s demons. Med Image Anal. 1998;2:243–60. [DOI] [PubMed] [Google Scholar]
- 32.Malon CD, Cosatto E. Classification of mitotic figures with convolutional neural networks and seeded blob features. J Pathol Inform. 2013;4. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3709419/. [DOI] [PMC free article] [PubMed]
- 33.Xu H, Lu C, Berendt R, Jha N, Mandal M. Automatic nuclear segmentation using multiscale radial line scanning with dynamic programming, IEEE Trans Biomed Eng. 2017;64. 10.1109/tbme.2017.2649485. [DOI] [PubMed] [Google Scholar]
- 34.Rouhi R, Jafari M, Kasaei S, Keshavarzian P. Benign and malignant breast tumors classification based on region growing and CNN segmentation. Expert Syst Appl. 2015;42:990–1002. [Google Scholar]
- 35.Xie Y, Xing F, Kong X, Su H, Yang L. Beyond classification: structured regression for robust cell detection using convolutional neural network, lecture notes in Computer Science, Cham, Switzerland: Springer; 2015. 10.1007/978-3-319-24574-4_43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. Ithica, New York: Cornell University; 2014. 10.1109/cvpr.2016.308. [DOI] [Google Scholar]
- 37.Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the Inception architecture for computer vision, In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Las Vegas, NV: Institute of Electrical and Electronics Engineers; 2016. 10.1109/cvpr.2016.308. [DOI] [Google Scholar]
- 38.Dean CJ, Sykes JR, Cooper RA, Hatfield P, Carey B, Swift S, et al. An evaluation of four CT–MRI co-registration techniques for radiotherapy treatment planning of prone rectal cancer patients. Br J Radio. 2012;85:61–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reed RJ. The histological variance of malignant melanoma: the interrelationship of histological subtype, neoplastic progression, and biological behaviour. Pathology. 1985;17:301–12. [DOI] [PubMed] [Google Scholar]
- 40.Pulitzer DR, Martin PC, Cohen AP, Reed RJ. Histologic classification of the combined nevus. Analysis of the variable expression of melanocytic nevi. Am J Surg Pathol. 1991;15:1111–22. [DOI] [PubMed] [Google Scholar]
- 41.Kapila K, Kharbanda K, Verma K. Cytomorphology of metastatic melanoma—use of S-100 protein in the diagnosis of amelanotic melanoma. Cytopathology. 1991;2:229–37. [DOI] [PubMed] [Google Scholar]
- 42.Rajeev LK, Asati V, Lokesh KN, Rudresh AH, Babu S, Jacob LA, et al. Cancer of unknown primary: opportunities and challenges. Indian J Med Paediatr Oncol. 2018;39:219. [Google Scholar]
- 43.Duraiyan J, Govindarajan R, Kaliyappan K, Palanisamy M. Applications of immunohistochemistry. J Pharm Bioallied Sci. 2012;4(Suppl2):S307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olawaiye AB, Mutch DG. Lymphnode staging update in the American Joint Committee on Cancer 8th edition cancer staging manual. Gynecol Oncol. 2018;150:7–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


