Abstract
Significant amounts of organic carbon in marine sediments are degraded, coupled with sulfate reduction. However, the actual carbon and energy sources used in situ have not been assigned to each group of diverse sulfate-reducing microorganisms (SRM) owing to the microbial and environmental complexity in sediments. Here, we probed microbial activity in temperate and permanently cold marine sediments by using potential SRM substrates, organic fermentation products at very low concentrations (15–30 μM), with RNA-based stable isotope probing. Unexpectedly, SRM were involved only to a minor degree in organic fermentation product mineralization, whereas metal-reducing microbes were dominant. Contrastingly, distinct SRM strongly assimilated 13C-DIC (dissolved inorganic carbon) with H2 as the electron donor. Our study suggests that canonical SRM prefer autotrophic lifestyle, with hydrogen as the electron donor, while metal-reducing microorganisms are involved in heterotrophic organic matter turnover, and thus regulate carbon fluxes in an unexpected way in marine sediments.
Keywords: sulfate-reducing microorganisms, carbon utilization, fermentation products, RNA-SIP
Introduction
Marine sediments are the largest organic matter sink on Earth [1]. Mineralization of buried organic matter is driven by the anaerobic microbial food chain, a network of fermenting and anaerobically respiring microorganisms perched below the seafloor [2, 3], which orchestrates the fate of organic compounds as well as the biogeochemical cycling of elements such as carbon, sulfur, nitrogen, iron and manganese [4, 5].
In the anoxic, sulfate-laden layers below the surface of the sediment, i.e. the sulfate reduction zone (SRZ), sulfate-reducing microorganisms (SRM) are one of the most important players that mediate a large fraction of organic matter degradation [6, 7]. Accordingly, SRM are genetically equipped to utilize divergent organic compounds, such as short-chain fatty acids, alcohols, carbohydrates, organohalogens, and aromatics [2, 6, 8]. Among those compounds, organic fermentation products are believed to be the most crucial substrates, the degradation of which is coupled to sulfate reduction as the terminal electron-accepting process in sediments [6, 9]. Fermentation products originating from organic matter degradation such as acetate, lactate, propionate, butyrate, and ethanol are typically present in micromolar concentrations [9-14]. However, organic fermentation products are not under thermodynamic control for potential degraders such as SRM and metal-reducing microorganisms [14-16], and thus it is not clear which microbes are active for low concentration of fermentation products. It is assumed that SRM is one of the most important microbial groups responsible for the degradation of organic fermentation products. For example, multiple SRM, such as representatives of the Desulfobacteraceae, Desulfocapsaceae, Desulfosarcinaceae, and Desulfovibrionaceae, can degrade short-chain fatty acids and alcohols [17-20]. Iron-reducing microorganisms Desulfuromondales also can use these organic substrates [21]. Sulfate reduction and iron reduction can co-occur in the SRZ of marine sediments [9, 22-25]; however, how fermentation products can be degraded by these microorganisms is not well studied in situ.
SRM have been enriched [15, 26-31] and isolated in pure culture [17, 18] from many different sediments, but using concentrations of fermentation products in the mM range, which is much higher than typically encountered in situ [9-14]. Apparently, enrichments and pure cultures amended with a high level of fermentation products challenge whether these microorganisms are relevant in situ. Detecting the activity modes of SRM in sediments rather than in enrichments or cultures is key to understand their physiological features in the environment as well as their role in elemental cycling. To date, however, much of the work on active SRM has focused on enrichments with known limitations regarding its environmental relevance. Only few studies attempting to capture in situ conditions suggest that the well-known SRM of the family Desulfobacteraceae can use both, acetate or H2 [32-34]. On the other hand, significant discrepancies between sulfate reduction rate and acetate oxidation rate have been reported. Such imbalanced ratios of sulfate reduction and acetate oxidation rates between 1:4 and 20:1 [16, 35-37] suggest that either SRM might use other fermentation products, or other microorganisms present in sediments participate in fermentation product degradation [16, 18, 21, 25, 32, 38-40]; however, the active microbes are not well linked to specific processes when rates are measured.
Given the apparent knowledge gap in understanding the role of SRM in fermentation product degradation in anoxic marine sediments, the following questions require investigation: (i) which fermentation products are utilized by SRM in the SRZ of marine sediment and (ii) what is the impact on biogeochemical cycling therein. We hypothesize that SRM and other microorganisms occupy different but complementary niches for fermentation product utilization. Thus, in the SRZ, the oxidation of organic (e.g. acetate, propionate, butyrate, lactate, ethanol) and inorganic fermentation products (e.g. H2) is driven by different guilds of microorganisms and have different trophic categories among these microbes, respectively. However, the low in situ concentrations of fermentation products in sediments are difficult to mimic in incubations because such low concentrations challenge the sensitivity of detecting and identifying active SRM without enrichment. To test the hypothesis and overcome the technical limitation, we used the highly sensitive RNA-based stable isotope probing (RNA-SIP) [41] approach with low, close to in situ concentrations of multiple organic fermentation products (max. 30 μM) in sediment incubations. RNA-SIP is an ultra-sensitive technique with a threshold below 0.001% of fully 13C-labeled nucleic acids [41, 42]. In combination with metagenomic analysis, our findings reveal novel features of fermentation product degradation in the SRZ regarding to carbon cycling in marine sediments.
Materials and methods
Sampling and incubation setup for stable isotope probing
Sediments used in this study were sampled from Helgoland mud area (North Sea; 54°05.23′N, 007°58.04′E; RV HEINCKE cruise in 2017; water depth: 27.9 m), Cumberland Bay (South Georgia; 54°15.899′S, 36°26.248′W; M134 cruise in 2017; water depth: 253 m), and Hornsund fjord (Arctic Svalbard; 76°59.325′N, 16°18.320′E; R/V Helmer Hanssen cruise in 2019; water depth: 115 m). Sediment gravity cores were kept at 4°C on board, then the cores were cut in 25 cm sections and stored at 4°C in 2.6-l jars with anoxic artificial sea water and headspace of N2. The information of sediment sampling and geochemical profiles were described in the previous studies [43-45]. Slurry incubations were set up with sulfate-rich sediments from the top layers of Helgoland mud area (16–41 cm), Cumberland Bay (14–39 cm), and Hornsund fjord (0–15 cm). Sediments were homogenized with artificial water (w: v = 1: 4, 50 ml; 26.4 g l−1 NaCl, 11.2 g l−1 MgCl2·6H2O, 1.5 g l−1 CaCl2·2H2O, and 0.7 g l−1 KCl) and filled in sterile 120-ml serum bottles, which were sealed with butyl rubber stoppers. The slurry was vacuumed three times for 3 min in order to remove O2 introduced during incubation setup, and headspace of culture was flushed with N2 at 1.5 atm as described previously [46]. For improved isotope labelling, slurries were preincubated for 10 days at 10°C to deplete organic substrates, O2 and nitrate remaining in the original slurry [15]. Thus, O2 was not introduced into incubations in order to mimic the anoxic condition of the sediment used for incubations [44, 47, 48]. After preincubation, all the slurries were amended with low concentrations (60 μM carbon) of fully 13C-labeled (99%) organic fermentation products (acetate: 30 μM; propionate: 20 μM; lactate: 20 μM; butyrate: 15 μM and ethanol: 30 μM; provided by Cambridge Isotope Laboratories, Tewksbury, MA). For the inorganic fermentation products, i.e. H2/CO2, ~100 μM H2 in slurry was transferred (15% of H2 in the headspace gas given its very low solubility [49]), and 10 mM 13C dissolved inorganic carbon was added. All incubations were amended with 18 mM sulfate. Since without amendment of substrate during preincubation will not trigger strong bacterial community shift [50], microbial activity will be identified when 13C-labeled substrates were utilized. In order to prove that SRM were present and had ability to degrade organic fermentation products, five antibiotics with the concentration of 50 mg l−1 (streptomycin, ampicillin, kanamycin, vancomycin and D-Cycloserine) for each were amended to potentially inhibit the activity of metal-reducing bacteria in incubations using one temperate (Helgoland mud) and one permanently cold (Cumberland Bay) sediments. SRMs such as Desulfovibrionaceae and Desulfobacteraceae are able to resist antibiotics [51-56]. A parallel set of controls containing unlabeled substrates was also conducted. All slurries were incubated at 10°C in order to have a better comparison among different sediments. After 6–15 days, incubations were stopped based on the development of δ13C values of CO2 in headspace, which was measured as described previously [57] (see Fig. 1 for the details of the incubation time). For incubations with inorganic fermentation product, i.e. H2/CO2, samples were harvested after 17 days. To identify the ability of glucose degradation by SRM, 10 μM 13C-glucose (i.e. 60 μM carbon) was amended into the SIP incubations, a same setup with the study for organic fermentation products degradation. In order to reveal metal reduction and avoid effect of sulfate, deep sediment from methanic zone (Helgoland mud area: 95–120 cm [58]) was used for SIP incubation setup using organic fermentation products.
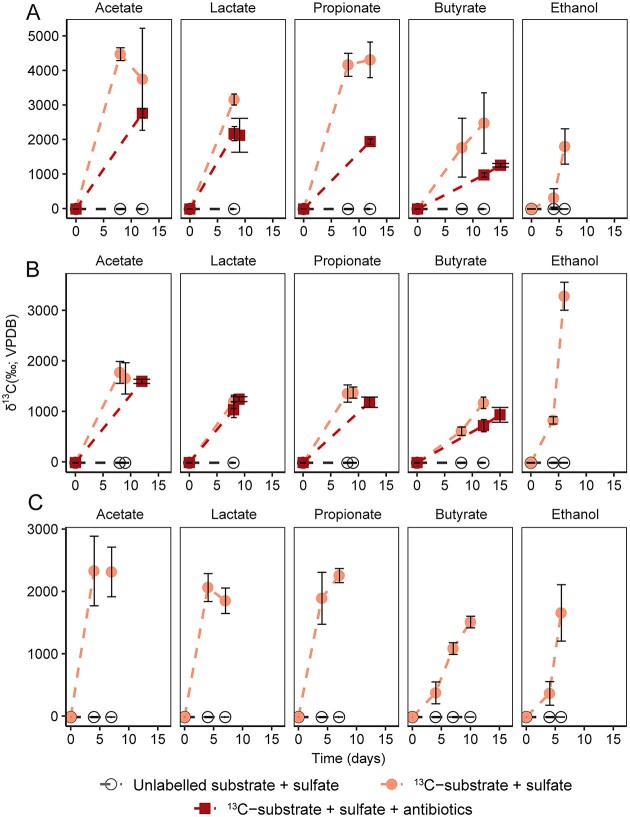
Figure 1.
Turnover of fermentation products in SIP incubations; development of δ13C-values of headspace CO2 in incubations amended with low concentration of fermentation products using Helgoland mud (A), Cumberland Bay (B), and Hornsund fjord sediment (C) (n = 3, error bar = SD); VPDB: The Vienna Peedee belemnite; samples for SIP analysis were harvested after the last time point.
Isopycnic centrifugation, gradient fractionation
For RNA-SIP analysis, RNA was extracted from slurries as described previously [59, 60]. Isopycnic centrifugation and gradient fractionation were employed to separate 13C-labeled from unlabeled RNA. Briefly, in order to obtain enough RNA for SIP, we combined RNA from biological replicates (n = 3). About 500–1000 ng of RNA was loaded with formamide (240 μl), cesium trifluoroacetate solution (6 ml, CsTFA, GE Healthcare, Buckinghamshire, UK), and gradient buffer solution. RNA was density separated by an Optima L-90 XP ultracentrifuge (Beckman Coulter, Brea, CA). At the same time, a mixture of fully 13C-labeled and unlabeled RNA from Escherichia coli was used as standard during density separation for defining heavy and light gradient fraction density ranges. After centrifugation at 124 000g at 20°C for 65 h, a total of 14 fractions (~ 410 μl) were collected from each sample. Complementary DNA (cDNA) was then obtained from reverse transcription of RNA using GoScript reverse transcription kit (Promega, Madison, WI). Combination of cDNA from fraction 4 and 5 (heavy), 6 and 7 (middle), 8 and 9 (light) and 10 and 11 (ultra-light) was performed for 16S rRNA sequencing, respectively. RNA quantification was conducted using Quanti-iT RiboGreen (Applied Biosystems, Foster City, CA). SIP fractions including 13C-labeled RNA were defined by standardization with RNA of fully labeled and unlabeled RNA standards from E. coli.
16S rRNA gene sequencing
Polymerase chain reaction (PCR) was performed with barcoded bacterial primer pair (Bac515F: 5′-GTGYCAGCMGCCGCGGTAA-3′; Bac805R: 5′-GACTACHVGGGTATCTAATCC-3′) [61] using KAPA HiFi HotStart PCR kit (KAPA Biosystems, Cape Town, South Africa). Thermocycling was set as follows: 95°C for 3 min; 35 cycles at 98°C for 20 s, 61°C for 15 s, and 72°C for 15 s; 72°C for 1 min. PCR products were then purified and quantified for library preparation [62]. Amplicons were sequenced through NovaSeq 6000 platform (Illumina, San Diego, USA; 2× 250 bp) at Novogene (Cambridge, UK). The raw reads were analysed according to Hassenrück 2022 [63]. Briefly, barcodes were extracted followed by de-multiplexing and primer clipping using cutadapt (version 2.1). The de-multiplexed reads were then analysed using dada2 (version 1.16.0). In detail, the quality of sequencing reads was checked and then the reads were trimmed, followed by the correction of error estimates and error learning in order to retrieve the final clean reads. The clean reads were then dereplicated and denoised, which were further merged for both forward and reverse reads to obtain the long sequences. The chimera reads were then filtered and the unusual reads below 248 bp or above 256 bp were removed. Taxonomy was assigned using the final reads based on the database SILVA 138 database [64] For each sample, 8000 to 60 000 reads were retrieved for abundance analysis.
Quantitative polymerase chain reaction
cDNA from the heavy, middle, light and ultra-light fractions was used for qPCR in order to quantify dsrA transcripts from RNA-SIP fractions as described previously [25]. Each 20 μl reaction mixture consisted of 10 μl of MESA Blue qPCR Master Mix (Eurogentec, Seraing, Belgium), 400 nM primers, 0.2 mg/ml bovine serum albumin (Roche, Mannheim, Germany), 1 ng DNA templates or 2 μl of cDNA samples. The primers DSR1-F+ (5′-ACSCACTGGAAGCACGGCGG-3′ [65]) and DSR-R (5′-GTGGMRCCGTGCAKRTTGG-3′ [66]) were used for qPCR, which are well-designed and have been widely used to identify marine SRM [25, 67, 68]. The qPCR protocol comprised an initial denaturation for 5 min at 95°C and 40 cycles amplification (95°C for 30 s, 60°C for 30 s and 72°C for 40 s). The detection thresholds were above 100 gene copies with an efficiency of 90%–110%.
Metagenomic analysis
Metagenomic sequencing on the Hiseq 4000 platform (2 × 150 bp) at Novogene (Cambridge, UK) was performed using DNA extracts from the original samples collected from Helgoland mud area (16–41 cm, 50–75 cm, and 222–238 cm) and Cumberland Bay sediments (15 cm, 225 cm, and 975 cm) with different depths, as well as a variety of enrichments using the sediments from the three sites (see Supplementary Table S1). Fourteen samples were used for metagenomic sequencing, with 440 million final clean reads. For metagenomic analysis, the raw reads were analyzed based on the Metawrap package (1.2.1) [69]. Briefly, quality checked reads were trimmed and then assembled using Megahit (1.1.3) with the default settings [70]. Scaffolds (>1000 bps) were binned using a combination of MaxBin2 (2.2.6), CONCOCT (1.0.0), and metaBAT2 (2.12.1). The quality of the bins was improved by remapping the raw reads using short-read mapper BWA (0.7.17) and re-assembled using SPAdes (3.13.0). The completeness and contamination of MAGs were estimated by CheckM2 (0.1.3). Taxonomic classifications of archaeal MAGs were based on GTDB database (0.3.3) (Supplementary Table S1) [71]. The MAGs with middle (≥50% and <10% contamination) and high (>90% complete with <5% contamination) quality according to MIMAG standards [72] were selected for annotation (Supplementary Table S1). Protein-coding regions were predicted using Prodigal (version 2.6.3) [73]. The Kyoto Encyclopedia of Genes and Genomes (KEGG) server (BlastKOALA) [74] (E-value cutoff ≤1e-5), eggNOG-mapper (5.0.0) [75] (E-value cutoff ≤1e-5), InterProScan tool (5.44–79.0) [76] (E-value cutoff ≤1e-10), and mmseq2 (10.6d92c) versus NCBI-nr database searched on April 2020 (E-value cutoff ≤1e-5) were used to annotate the protein-coding regions.
Phylogenetic analyses
The concatenated set of 71 ribosomal protein genes based on a previously published hidden Markov Model profile [77] were used for phylogenetic analyses in Anvi’o (6.1) [78]. Maximum-likelihood trees were built using IQ-TREE (1.6.12) [79] with the best-fit model (LG + F + R7) and 1000 times ultrafast bootstrapping. The tree files were edited using the online tool iTOL [80].
Because of the different names among GTDB and Silva databases for Sva1033 (Desulfuromonadales), 16S rRNA genes in the MAGs for all analysed Desulfuromonadales were extracted by Barrnap (version 0.3, http://www.vicbioinformatics.com/software.barrnap.shtml). The 16S rRNA genes together with references were aligned with SINA Aligner [81]. Maximum-likelihood tree was inferred using RAxML (8.2.11) with rapid bootstrapping and the GTRGAMMA model [82]. In our previous work, we have not found evidence for sulfate-reducing archaeal taxa [83], and thus archaeal analysis was not included in this study.
Amino acid sequences of reductive dehalogenase were used for orthology analysis. Reference sequences with 100 hits were retrieved from NCBI nonredundant protein database by blasting sequences of reductive dehalogenase of SRM obtained from this study. The combined sequences of each protein were filtered and clustered using cd-hit (Version 4.6.8) [84] with cut-off of 70%, which was followed by MAFFT-LINSI (Version 7.455) alignment with default parameters [85] and trimming by BMGE with flags “-t AA -m BLOSUM30” [86]. Un-rooted phylogenetic trees for protein sequence were built with 1000 times ultrafast bootstrapping using IQ-TREE with the best-fit models (LG + R8).
Results
Turnover of fermentation products in sediment incubations
RNA-SIP incubations with 13C labeled substrates and sulfate were set up in order to investigate the turnover of different fermentation products and the activity of the associated microorganisms. Three different sediments were compared including the temperate site Helgoland mud area (North Sea) and two permanently cold sites from Cumberland Bay (South Georgia, sub-Antarctic) and Hornsund fjord (Svalbard, Arctic). The degradation of 13C labeled substrate was monitored by the formation of 13CO2 in the headspace over time to determine incubation stopping time by avoiding cross-feeding when samples were incubated for too long time (Fig. 1). Delta 13C values of CO2 increased rapidly to ~1030–4470‰ within 15 days for sediments from all three sites. In Helgoland mud sediment incubations, the addition of antibiotics (to suppress SRM competitors [52-56]) to incubations slowed down the formation of 13CO2 (~1250–2760‰) compared to those without antibiotics (~2470–4470‰) within 15 days (Fig. 1A). In contrast, antibiotics had a smaller inhibitory effect on the turnover of fermentation products in incubations using Cumberland Bay sediment (Fig. 1B). The chosen incubation times were similar for incubations with and without antibiotics (Fig. 1).
Differential activities of sulfate-reducing microorganisms and other active microorganisms
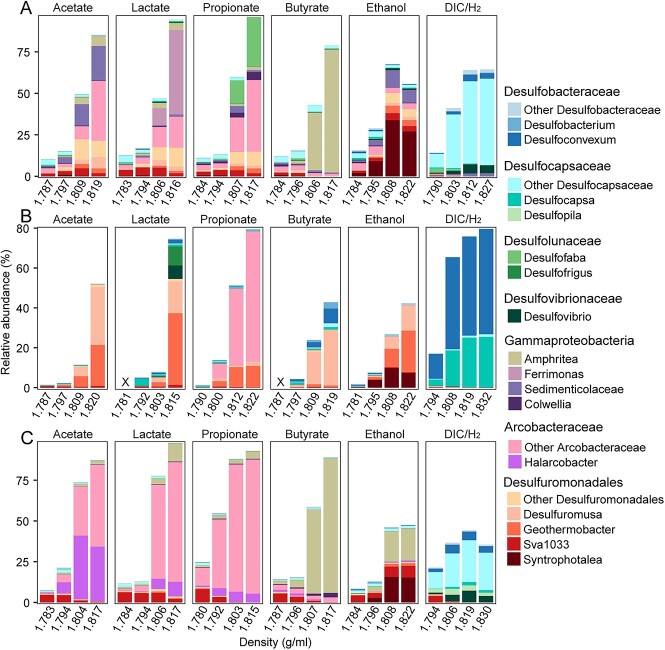
Acetate, lactate, propionate, butyrate, and ethanol were used at a concentration of 60 μM carbon, similar in situ (15–30 μM dissolved organics carbon), in RNA-SIP incubations to identify those microorganisms actively assimilating labeled substrate during fermentation product degradation. In contrast to the isotopically “light” RNA and unlabeled controls, we recovered several bacterial groups, which were highly abundant in the isotopically “heavy” RNA fractions in their respective incubations (Fig. 2, Supplementary Figs S1 and S2). Most notably, Desulfuromonadales members (~15%–50%) were highly active in Helgoland mud and Cumberland Bay sediments (Fig. 2A and B). In addition, members of Arcobacteraceae (~18%–65%) were also active when amended with acetate, lactate, and propionate (Helgoland mud) or propionate only (Cumberland Bay) (Fig. 2A and B). We found that those Desulfuromonadales groups were likely using iron oxides (lepidocrocite) as electron acceptors in methanic, sulfate-depleted sediments from Helgoland (Supplementary Fig. S3). Arcobacteraceae (>50%) also dominated in Hornsund fjord sediment incubations with 13C-labeled acetate, lactate, and propionate, whereas active Desulfuromonadales (~25%) were found in 13C-butyrate incubations of Cumberland Bay sediments. Other notable findings include large active populations of Sedimenticolaceae (up to 20%), Ferrimonas (up to 50%), and Amphritea (up to 74%), which are known as metal reducers as well [40, 87-90], respectively, in specific incubations (Fig. 2A and B). A rather uniform picture emerged from ethanol-amended incubations, in which Desulfuromonadales dominated fermentation product degradation in incubations from all three sites. However, canonical SRM were not very active in the incubations with organic fermentation products (acetate, lactate, propionate, butyrate, and ethanol), only showing minor activity in a few incubations (with propionate [Helgoland mud] and lactate[Cumberland Bay]). Instead, Desulfobacteraceae, Desulfocapsaceae, and Desulfovibrionaceae were strongly stimulated (~37–80% in the heavy fraction) when H2 and 13CO2 were amended in the incubations for all the three sites (Fig. 2).
Figure 2.
Identification of the active fermentation products degraders using RNA-SIP in incubations amended with 13C-labeled substrates and sulfate; relative abundance of 16S rRNA gene sequences of active bacteria fermentation product degraders in total bacteria from RNA-SIP gradient fractions in the Helgoland mud (A), Cumberland Bay (B), and Hornsund fjord sediment (C) incubations; × indicates that cDNA synthesis failed because of insufficient amount of RNA in these fractions; density was indicated as the average density of combined fractions for RNA-SIP samples; for sampling time points, see Figure 1.
Sulfate-reducing microorganisms were only utilizing organic fermentation products under inhibition of other microbes
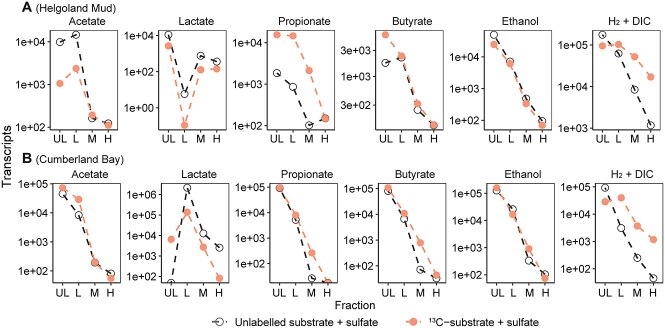
Since SRM unexpectedly had a minor contribution to organic fermentation product degradation, transcripts of dsrA (alpha subunit of the dissimilatory sulfite reductase, a marker gene for sulfate reduction [91]) were quantified within SIP fractions in order to understand the participation of sulfate reduction in incubations with representative cold (Cumberland Bay) and temperate (Helgoland) sediments (Fig. 3). We found that transcripts of dsrA from the light fraction were most abundant in any incubation, regardless if 13C-labeled or unlabeled substrates were used, while the heaviest fractions always had the lowest amount of dsrA transcripts. In contrast to the organic fermentation products, dsrA abundances in the heavy fractions from inorganic H2/DIC were comparatively high, in line with the activity of H2/DIC utilization by SRM (Figs 2 and 3).
Figure 3.
Copies of dsrA transcript in different fractions from the RNA-SIP samples; number of transcripts from the heavy (H: 1.815–1.830 g/ml), middle (M: 1.803–1.819 g/ml), light (L: 1.792–1.808 g/ml), and ultra-light (UL: 1.781–1.794 g/ml) fractions of RNA-SIP samples from incubations using Helgoland mud (A) and Cumberland Bay (B) sediment; note that values below 100 copies might be not accurate because the detection threshold was above 100 (see Method).
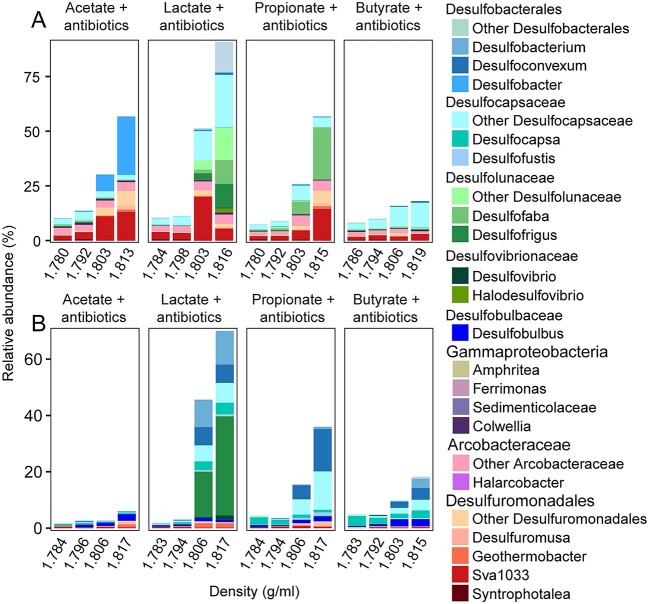
We further checked whether SRM could use these compounds if potential microbial competitors were inhibited. Typical SRM such as Desulfovibrionaceae and Desulfobacteraceae can resist antibiotics [51-56]. Therefore, multiple antibiotics were added in order to inhibit those organisms which readily used fermentation products in our incubations. In these cases, multiple SRM were actually capable of degrading various fermentation products. In detail, Desulfobacter (~27% in heavy RNA fractions) metabolized acetate in Helgoland mud sediment incubations (Fig. 4A). Desulfobacterales, Desulfocapsaceae, Desulfolunaceae, and Desulfovibrionaceae (68%–76% of total bacteria) were able to utilize lactate in both sediment types (Fig. 4). Desulfoconvexum, Desulfofaba, and Desulfocapsaceae (29%–34% of total bacteria) could degrade propionate, while Desulfobacterales, Desulfocapsaceae, and Desulfobulbus (14%–18% of total bacteria) participated in the turnover of butyrate, however, on a much lower scale compared to the other treatments (Fig. 4).
Figure 4.
Identification of the active fermentation product degraders using RNA-SIP in incubations amended with 13C-labeled substrates, sulfate, and antibiotics; relative abundance of 16S rRNA gene sequences of active bacterial fermentation product degraders in total bacteria from RNA-SIP gradient fractions in the Helgoland mud (A) and Cumberland Bay (B) sediment incubations; Helgoland mud and Cumberland Bay sediments were used as representatives for temperate and permanently cold sediment, respectively; density was indicated as the average density of combined fractions for RNA-SIP samples.
Versatility of carbon metabolic pathways for active fermentation product utilizers and noncanonical sulfate-reducing microorganisms
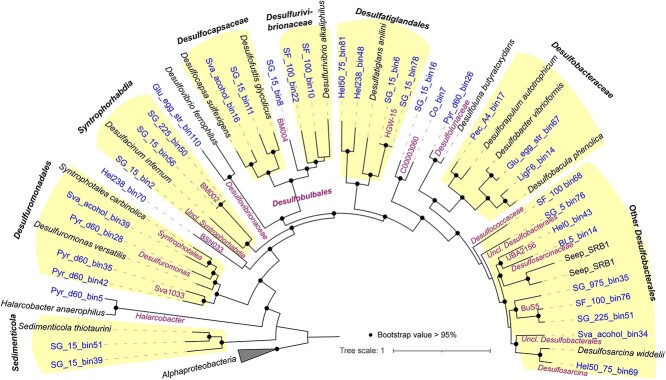
In SIP incubations, we identified the activity of fermentation product degraders and some SRM. In order to have a deeper insight into the genomes of SRM and other fermentation product degraders in sediments, we screened 36 metagenome-assembled genomes (MAGs) with middle to high quality from the original sediments and sediment enrichments (Fig. 5, Supplementary Table S1). These MAGs were affiliated to the active fermentation product degraders including Sedimenticolaceae, Halarcobacter, Sva1033, Desulfuromonadaceae Desulfovibrionaceae, Desulfocapsaceae, and Desulfobacteraceae, and noncanonical SRM such as BSN033 (class level of Desulfobacterota), Syntrophorhabdia, Desulfurivibrionaceae, Desulfatiglandales, C00003060 (order level of Desulfobacteria), and other Desulfobacterales (Fig. 5, Supplementary Fig. S4, Supplementary Table S1).
Figure 5.
Maximum likelihood tree of 71 concatenated bacterial genes; Uncl.: unclassified; the tree was built using IQ-TREE (1.6.12) with the best-fit model (LG + F + R7) and 1000 times ultrafast bootstrapping; see Supplementary Table S1 for the details of MAG information.
Based on metagenomic analysis, pathways involved in the degradation of fermentation products were widely identified in most MAGs of active microbes. In detail, MAGs of Desulfuromonadales such as Sva1033 and Desulfuromonas included the pathways for acetate, lactate, propionate, and butyrate dissimilation to the corresponding acyl-CoA, which can be oxidized to CO2 via the acetyl-CoA pathway or citric acid cycle coupled to iron reduction, but they did not feature genes for H2 oxidation (Fig. 6, Supplementary Tables S2, S3). Halarcobacter were equipped with pathways for acetate, lactate, and propionate degradation similarly to Desulfuromonadales (Fig. 6, Supplementary Table S2). In addition, Pelobacteraceae had gene sets for ethanol oxidation to acetate, a known feature of Pelobacter spp. (now partly known as Syntrophotalea spp.; [92]) such as Pelobacter acetylenicus (now Syntrophotalea acetylenica [92]) (Fig. 6, Supplementary Table S2). For active SRM, the metagenomic analysis indeed reflected that H2/CO2 utilization via Wood–Ljungdahl (WL) is a common feature for SRM indicated by the presence of complete WL pathway and hydrogenases including group 1a, 1b, or 1c (hydrogenotrophic respiration using sulfate [93, 94]) in the MAGs of Desulfobacterales and Desulfobulbales (Supplementary Table S3).
Figure 6.
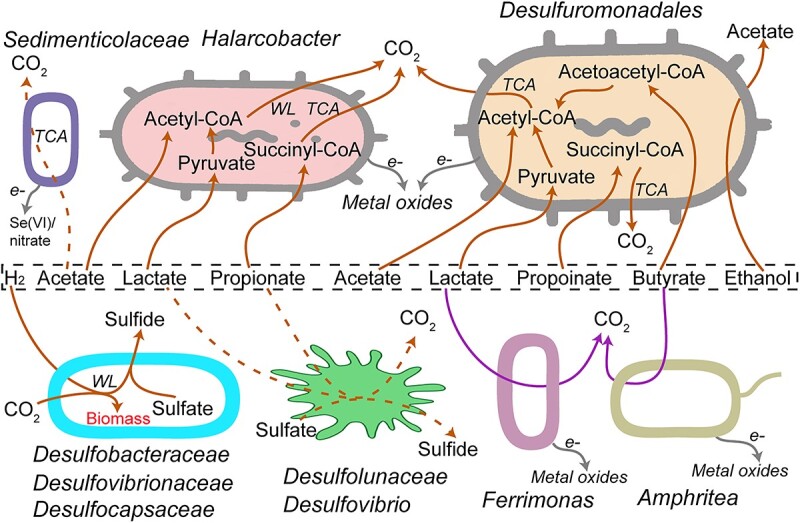
Patterns of fermentation products utilization in SRZ of temperate and permanently cold marine sediments based on SIP and metagenomics; solid and dashed brown lines indicate the strong and weak activity for fermentation product utilization, respectively; see Table S2 for the details of annotated genes; TCA: Citric acid cycle.
We further checked the metagenomic pathways of noncanonical SRM to predict their potentials for alternative organic carbon degradation pathways. We found that SRM groups including Desulfatiglandales and other Desulfobacterales harbored the pathways for reductive dehalogenesis and potentials involving in aromatic compound degradation (Supplementary Figs S5 and S6, Supplementary Table S2). Furthermore, multiple MAGs have pathways for glycolysis and fatty acid degradation in SRM including Syntrophorhabdia, Desulfobulbales, C00003060, and other Desulfobacterales (Supplementary Fig. S4). Such sugar utilization was also indicated from the SIP incubations in which the RNA of Desulfobacterales (Desulfocapsaceae) was specifically labeled by 13C-glucose in the presence of antibiotics (Supplementary Figs S7 and S8).
Discussion
Partitioning of fermentation product degradation among canonical fermentation product degraders
Using ultra-high sensitivity RNA-SIP in combination with relevant 13C-labeled fermentation products, we found a consistent partitioning pattern in temperate and permanently cold sediments: organic fermentation products were mostly used by known and novel metal-reducing bacteria, whereas SRM were strongly active in using H2 autotrophically.
In general, the concentration of organic fermentation product in coastal sediments is low, ranging from nanomolar to few-hundred micromolar [9-14]. In incubations, we used only 60 μM carbon (15–30 μM fermentation products) for SIP and thus, matched closely in situ concentrations of organic fermentation products. This in turn is likely avoiding enrichment artifacts originating from irrelevant carbon compound concentrations in incubations and thus, better reflects microbial activities in the studied sediments. We did not detect a strong enrichment of SRM on H2/CO2 since the abundance of unlabeled dsrA was still higher in the light fractions than that of the heavy fractions. Based on this, we found that Desulfuromonadales (Desulfuromusa, Geothermobacter, Syntrophotalea, and Sva1033), Arcobacteraceae, Ferrimonas, Sedimenticolaceae, Amphritea, and Syntrophotalea were the main organic fermentation product utilizers while SRM were incorporating 13CO2 with H2 as electron donor. The former microorganisms are not known as SRM [25, 38, 92], which is corroborated by the absence of genes encoding the dissimilatory sulfate reduction pathway in their MAGs (Supplementary Table S2). As revealed in our studies, Desulfuromonadales were iron-reducing bacteria (Supplementary Fig. S3, Fig. 6). Certainly, dissolved Fe(II) and Mn(II) were not detectable due to low concentration of amended substrates in SIP incubations, and abiotic reactions resulting in the formation of insoluble minerals (e.g. siderite) [95]. However, the identified active non-SRM (Desulfuromonadales – Desulfuromusa, Geothermobacter, and Sva1033) here were identified previously in Helgoland mud and Cumberland Bay sediment incubations as iron-reducing microbes (Supplementary Fig. S3) [25, 58]. It is also feasible that Arcobacteraceae reduce Fe(III) or Mn(IV) as electron acceptor linked to the oxidation of organic carbon compounds [25, 40, 96, 97]. The other active 13C-labeled bacteria, such as Sedimenticolaceae, Ferrimonas, and Amphritea, can utilize metal oxides such as manganese, iron, and selenium oxides or have been identified in incubations amended with metal oxides in several studies [25, 40, 87-90, 98]. Gas exchange in headspace and preincubation of sediments (see Method) ensured that alternative electron acceptors (e.g. traces of oxygen, nitrate) were depleted. In our study, we have used marine sediment from one temperate and two permanently cold sites. The temperate sediment from the Helgoland mud area is characterized by high sedimentation rate resulting in deeply buried iron oxides, which can fuel microbial activity in anoxic sediment layers [58, 99]. In cold sediment from Cumberland Bay (South Georgia, sub-Antarctic) and Hornsund fjord (Svalbard, Arctic), metal oxides are originating from glacier-associated erosion and meltwater, and thus metal oxides such as iron and manganese oxides are present [100, 101]. Thus, the identification of active degraders of fermentation product as known metal reducing microorganisms suggests that sediments contained sufficient amounts of metal oxides as electron acceptors to support their metal-reducing activity, even in deep sediment from the methanic zone (Supplementary Fig. S3) [25, 40].
In our SIP incubations, SRM were not actively degrading and incorporating label from organic fermentation products; this is surprising as sulfate is present at high concentration (15 to 28 mM in situ; 18 mM in our incubations) in the upper sediment layer of the studied sediments, and SRM have been identified in this and previous studies at high abundance (Supplementary Fig. S9) [25, 43, 99, 102]. Albeit their limited activity, SRM were still active in incubations but rather using other substrates since dsrA transcripts were much more abundant in the light fraction than those from heavy fraction, and thus, were not labeled from 13C-organic fermentation products (Figs 2 and 3). Only in the presence of antibiotics, SRM were found to incorporate 13C-label from added organic substrates (Fig. 4), ruling out the possibility that amended low concentrations of organic fermentation products were limiting the activity of SRM, and corroborating that the turnover of organic fermentation products is apparently not under thermodynamic control in marine sediments [14-16]. The low in situ temperature might trigger the observed partitioning of organic fermentation product degradation among SRM and other microbes: low activity of SRM at temperatures below 10°C; iron reduction in temperate marine sediment was apparently favored at low temperatures (4–10°C) [62], while the optimal temperature for sulfate reduction was found to be above 15°C for cold sediments [15], hinting to a potentially better adapted metabolism of iron-reducing microorganisms at lower temperature. With antibiotics, most metal-reducing bacteria were inhibited and therefore SRM were identified, which further indicates that there was no O2 contamination that might inhibit the activity of SRM. In this study, we used temperate and permanently cold sediment and found that these two types of sediments have very similar features for the activity of SRM at relatively low temperate condition. It is still very interesting for the future study to test the ability of SRM for organic fermentation product utilization in high temperature sediments.
Unlike organic fermentation products, the H2 concentration is typically under thermodynamic control in marine sediments [15, 49, 103, 104], thus, the hydrogen partial pressure is determined by the free energy available of the energetically most favorable electron accepting process. In sulfate reduction-dominated coastal sediment, hydrogen partial pressures were on similar levels regardless whether metal oxides were added or sulfate was present [103], suggesting that out-competition of SRM by metal reducing microorganisms based on terminal electron acceptor thermodynamics [105] was not operative. Albeit high quality of MAGs, we found a lack of respiratory H2-uptake [NiFe]-hydrogenase and incomplete WL pathways in MAGs of Sva1033 and Desulfuromonas spp. (Supplementary Tables S1–S3), suggesting that these iron-reducing microorganisms cannot oxidize hydrogen and fix CO2. In contrast, SRM were strongly stimulated in SIP incubations with H2/13CO2. Many SRM species in our marine sediment incubations are actually autotrophs capable of fixing inorganic carbon and using H2 as electron donor (Fig. 6). Besides primary fermentation, secondary, syntrophic oxidations of organic fermentation products are important sources of H2 in marine sediments [2], and H2 can contribute up to 75% in electron flow [106]. Based on our study, SRM can indirectly participate in organic matter degradation by interspecies hydrogen transfer interactions during fermentation of macromolecules such as protein, carbohydrates, and cell biomass [33, 37]. Thermodynamically, hydrogen is also a sufficiently strong reductant for CO2 fixation in the relevant reactions (oxidation of H2: H2 = 2e− + 2H+; E°′ = −414 mV; reduction of CO2 to formate, E°′ = −430 mV; CO2 to CO, E°′ = −520 mV, acetyl-CoA and CO2 to pyruvate (E°′ = −500 mV) [107]). In fact, some genera affiliated with the family Desulfobacteraceae remain lithoautotrophic in the presence of H2 when acetate is amended [108], indicating H2-based preference for lithoautotrophy in some SRM. In addition, other than H2, direct interspecies electron transfer might be another mechanism supporting autotrophy of SRM in environments. For example, Desulfosarcina/Desulfococcus utilize electrons transferred from their methanotrophic ANME partners for autotrophic growth [109], a syntrophic consortium mediating anaerobic methane oxidation [110]. Although ~100 μM H2 in slurry was amended, given that SRM were not active for almost all the common organic fermentation products (acetate, lactate, propionate, butyrate, ethanol) and the substantial overpressure in sediments [111], utilization of H2/CO2 or interspecies electron most likely reflected the activity of SRM in situ. Overall, the SIP results suggested that SRM were of minor importance during organic fermentation product degradation (Fig. 6).
Beyond fermentation products: non-canonical organic carbon utilization by sulfate-reducing microorganisms
Beyond active SRM, marine sediments inhabit diverse uncultivated SRM (Supplementary Fig. S9), and thereby this leads to the question of what their potential role in carbon degradation in these sediments is. Our SIP incubations amended with glucose suggested that SRM were able to degrade glucose in the presence of antibiotics (Supplementary Fig. S7), and thus they had the ability for glucose uptake into cells and also harbor the complete pathway for glucose degradation (Supplementary Fig. S5). Although the SRM were outcompeted by sugar fermenters when antibiotics were not present (Supplementary Fig. S8), it is still notable that some other carbohydrates might be the substrates for SRM in marine sediments, which is in line with the observation of carbohydrate degradation by SRM in a few studies [112-115]. Apart from glucose utilization, our enrichment incubations and metagenomic analysis also indicated a wider spectrum of substrate utilization by noncanonical SRM than expected, such as halogens (see supplemental discussion), and thus SIP experiments should focus on the carbon utilization versatility of SRM in the future study.
Our study has new implications for the role of SRM on biogeochemical cycling in marine sediment: (i) canonical SRM have limited contribution on the degradation of organic fermentation products at low concentrations in marine sediments, (ii) canonical SRM appear to prefer autotrophic lifestyle using H2 oxidation instead of heterotrophy, (iii) many SRM have potentials for utilizing noncanonical carbon compounds. Canonical SRM may actually have an autotrophic lifestyle in environments. In the SRZ, CO2 assimilation has been identified in some archaeal groups such as Lokiarchaeota, Bathyarchaeota, and ANMEs [116-118], while their activities are quite low. It has been recognized that autotrophy is an important lifestyle for sulfur oxidizers with a relatively high activity of bacteria [119]. Based on our findings, we propose that SRM are additional CO2 assimilators that have to be considered to regulate carbon fluxes in marine sediments.
Supplementary Material
Acknowledgements
We thank the captain, crew, and scientists of R/V HEINCKE expeditions HE443 and RV METEOR 134 H19-891-BC. We thank Dr S.I. Nam at the Korean Polar Research Institute, and crew members of RV Helmer Hanssen who assisted with Hornsund sediment collection under the project of “Research on environmental changes in fjords and coastal geomorphology: Toward a better understanding of the erosion and redeposition processes of the Svalbard archipelago in the Arctic.”
Contributor Information
Xiuran Yin, State Key Laboratory of Marine Resource Utilization in South China Sea, Hainan University, 58 Renmin Avenue, Haikou 570228, China; Faculty of Biology/Chemistry, University of Bremen, Leobener Strasse 3, Bremen D-28359, Germany; MARUM, Center for Marine Environmental Sciences, University of Bremen, Leobener Strasse 8, Bremen D-28359, Germany; Max Planck Institute for Marine Microbiology, Celsiusstrasse 1, Bremen D-28359, Germany.
Guowei Zhou, State Key Laboratory of Marine Resource Utilization in South China Sea, Hainan University, 58 Renmin Avenue, Haikou 570228, China; School of Resources and Environmental Engineering, Anhui University, 111 Jiulong Road, Hefei, Anhui 230601, China.
Haihua Wang, State Key Laboratory of Marine Resource Utilization in South China Sea, Hainan University, 58 Renmin Avenue, Haikou 570228, China; Faculty of Biology/Chemistry, University of Bremen, Leobener Strasse 3, Bremen D-28359, Germany; College of Urban and Environmental Sciences, Peking University, No. 5 Yiheyuan Road, Beijing 100871, China.
Dukki Han, Department of Marine Bioscience, Gangneung-Wonju National University, 7 Jukheon-gil, Gangneung-si 25457, Republic of Korea.
Mara Maeke, Faculty of Biology/Chemistry, University of Bremen, Leobener Strasse 3, Bremen D-28359, Germany; Max Planck Institute for Marine Microbiology, Celsiusstrasse 1, Bremen D-28359, Germany.
Tim Richter-Heitmann, Faculty of Biology/Chemistry, University of Bremen, Leobener Strasse 3, Bremen D-28359, Germany.
Lea C Wunder, Faculty of Biology/Chemistry, University of Bremen, Leobener Strasse 3, Bremen D-28359, Germany; Max Planck Institute for Marine Microbiology, Celsiusstrasse 1, Bremen D-28359, Germany.
David A Aromokeye, Faculty of Biology/Chemistry, University of Bremen, Leobener Strasse 3, Bremen D-28359, Germany.
Qing-Zeng Zhu, MARUM, Center for Marine Environmental Sciences, University of Bremen, Leobener Strasse 8, Bremen D-28359, Germany.
Rolf Nimzyk, Faculty of Biology/Chemistry, University of Bremen, Leobener Strasse 3, Bremen D-28359, Germany.
Marcus Elvert, MARUM, Center for Marine Environmental Sciences, University of Bremen, Leobener Strasse 8, Bremen D-28359, Germany; Faculty of Geosciences, University of Bremen, Klagenfurter Strasse 2-4, Bremen D-28359, Germany.
Michael W Friedrich, Faculty of Biology/Chemistry, University of Bremen, Leobener Strasse 3, Bremen D-28359, Germany; MARUM, Center for Marine Environmental Sciences, University of Bremen, Leobener Strasse 8, Bremen D-28359, Germany.
Author contributions
Xiuran Yin, Guowei Zhou, Haihua Wang, and Michael W. Friedrich designed the research; Xiuran Yin, Guowei Zhou, Haihua Wang, and Dukki Han, performed RNA-SIP experiments. Xiuran Yin, Guowei Zhou, Haihua Wang, Mara Maeke, and Rolf Nimzyk, analyzed metagenomic data; Qing-Zeng Zhu and Marcus Elvert, analyzed 13C-CO2; Xiuran Yin, Guowei Zhou, Haihua Wang, Dukki Han, Michael W. Friedrich, Tim Richter-Heitmann, Mara Maeke, David A. Aromkeye, Lea C. Wunder, Qing-Zeng Zhu, and Marcus Elvert contributed to the discussion of the results and wrote the paper.
Conflicts of interest
The authors declare that they have no conflict of interest.
Funding
This research was supported by the Research Center/Cluster of Excellence EXC 309 (project-ID 49926684) “The Ocean in the Earth System” and the Cluster of Excellence EXC 2077 (project-ID 390741601) “The Ocean Floor – Earth’s Uncharted Interface” funded by the Deutsche Forschungsgemeinschaft (DFG) and the University of Bremen, and the start-up research fund (project-ID XJ2300006031) of Hainan University.
Data availability
The bacterial MAGs data are available in NCBI database under the project PRJNA678468 with the accession numbers of SAMN32874205 to SAMN32874239. Sequencing data of RNA-SIP samples have been deposited in the Short Reads Archive under the project PRJNA505997 with accession numbers from SAMN32873837 to SAMN32874024. The metagenomic reads sequenced from original sediments have been deposited under the project PRJNA1023477 with accession numbers from SAMN37668301 to SAMN37668306.
References
- 1. Hedges JI, Keil RG. Sedimentary organic matter preservation an assessment and speculative synthesis. Mar Chem 1995;49:81–115. 10.1016/0304-4203(95)00008-F [DOI] [Google Scholar]
- 2. Jørgensen BB, Findlay AJ, Pellerin A. The biogeochemical sulfur cycle of marine sediments. Front Microbiol 2019;10:10. 10.3389/fmicb.2019.00849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arnosti C. Speed bumps and barricades in the carbon cycle: substrate structural effects on carbon cycling. Mar Chem 2004;92:263–73. 10.1016/j.marchem.2004.06.030 [DOI] [Google Scholar]
- 4. Middelburg JJ, Vlug T, Jaco Fet al. Organic matter mineralization in marine systems. Glob Planet Change 1993;8:47–58. 10.1016/0921-8181(93)90062-S [DOI] [Google Scholar]
- 5. Orcutt BN, Sylvan JB, Knab NJet al. Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol Mol Biol Rev 2011;75:361–422. 10.1128/MMBR.00039-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jørgensen BB. Mineralization of organic matter in the sea bed—the role of sulphate reduction. Nature 1982;296:643–5. 10.1038/296643a0 [DOI] [Google Scholar]
- 7. Jørgensen BB, Beulig F, Egger Met al. Organoclastic sulfate reduction in the sulfate-methane transition of marine sediments. Geochim Cosmochim Acta 2019;254:231–45. 10.1016/j.gca.2019.03.016 [DOI] [Google Scholar]
- 8. Jochum LM, Schreiber L, Marshall IPGet al. Single-cell genomics reveals a diverse metabolic potential of uncultivated Desulfatiglans-related Deltaproteobacteria widely distributed in marine sediment. Front Microbiol 2018;9:2038. 10.3389/fmicb.2018.02038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Finke N, Vandieken V, Jørgensen BB. Acetate, lactate, propionate, and isobutyrate as electron donors for iron and sulfate reduction in Arctic marine sediments, Svalbard. FEMS Microbiol Ecol 2007;59:10–22. 10.1111/j.1574-6941.2006.00214.x [DOI] [PubMed] [Google Scholar]
- 10. Glombitza C, Egger M, Røy Het al. Controls on volatile fatty acid concentrations in marine sediments (Baltic Sea). Geochim Cosmochim Acta 2019;258:226–41. 10.1016/j.gca.2019.05.038 [DOI] [Google Scholar]
- 11. Sansone FJ, Martens CS. Volatile fatty acid cycling in organic-rich marine sediments. Geochim Cosmochim Acta 1982;46:1575–89. 10.1016/0016-7037(82)90315-5 [DOI] [Google Scholar]
- 12. Glombitza C, Pedersen J, Røy Het al. Direct analysis of volatile fatty acids in marine sediment porewater by two-dimensional ion chromatography-mass spectrometry. Limnol Oceanogr 2014;12:455–68. 10.4319/lom.2014.12.455 [DOI] [Google Scholar]
- 13. Zhuang G-C, Lin Y-S, Elvert Met al. Gas chromatographic analysis of methanol and ethanol in marine sediment pore waters: validation and implementation of three pretreatment techniques. Mar Chem 2014;160:82–90. 10.1016/j.marchem.2014.01.011 [DOI] [Google Scholar]
- 14. Glombitza C, Jaussi M, Røy Het al. Formate, acetate, and propionate as substrates for sulfate reduction in sub-Arctic sediments of Southwest Greenland. Front Microbiol 2015;6:6. 10.3389/fmicb.2015.00846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Finke N, Jørgensen BB. Response of fermentation and sulfate reduction to experimental temperature changes in temperate and Arctic marine sediments. ISME J 2008;2:815–29. 10.1038/ismej.2008.20 [DOI] [PubMed] [Google Scholar]
- 16. Wellsbury P, Parkes RJ. Acetate bioavailability and turnover in an estuarine sediment. FEMS Microbiol Ecol 1995;17:85–94. 10.1111/j.1574-6941.1995.tb00133.x [DOI] [Google Scholar]
- 17. Widdel F, Kohring GW, Mayer F. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids III. Characterization of the filamentous gliding Desulfonema limicola gen. nov. sp. nov., and Desulfonema magnum sp. nov. Arch Microbiol 1983;134:286–94. 10.1007/BF00407804 [DOI] [Google Scholar]
- 18. Kuever J. The family Desulfobacteraceae. In: Rosenberg E, et al. (eds.), The Prokaryotes – Deltaproteobacteria and Epsilonproteobacteria. Springer-Verlag: Berlin Heidelberg, 2014, 45–73, 10.1007/978-3-642-39044-9_266. [DOI] [Google Scholar]
- 19. Watanabe M, Galushko A, Fukui Met al. Desulfosarcinaceae. Bergey's Manual of Systematics of Archaea and Bacteria. Hoboken, New Jersey: John Wiley & Sons, 2020, 1–4 [Google Scholar]
- 20. The KJ, Desulfovibrionaceae F. In: Rosenberg E., DeLong E.F., Lory S.et al. (eds.), The Prokaryotes: Deltaproteobacteria and Epsilonproteobacteria. Berlin, Heidelberg: Springer Berlin Heidelberg, 2014, 107–33 [Google Scholar]
- 21. Lovley DR. Dissimilatory Fe(III) and Mn(IV) reduction. Microbiol Rev 1991;55:259–87. 10.1128/mr.55.2.259-287.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Buongiorno J, Herbert LC, Wehrmann LMet al. Complex microbial communities drive iron and sulfur cycling in Arctic fjord sediments. Appl Environ Microbiol 2019;85:e00949–19. 10.1128/AEM.00949-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Canfield DE, Thamdrup B, Hansen JW. The anaerobic degradation of organic matter in Danish coastal sediments iron reduction, manganese reduction, and sulfate reduction. Geochim Cosmochim Acta 1993;57:3867–83. 10.1016/0016-7037(93)90340-3 [DOI] [PubMed] [Google Scholar]
- 24. Thamdrup B, Fossing H, Jørgensen BB. Manganese, iron and sulfur cycling in a coastal marine sediment, Aarhus bay, Denmark. Geochim Cosmochim Acta 1994;58:5115–29. 10.1016/0016-7037(94)90298-4 [DOI] [Google Scholar]
- 25. Wunder LC, Aromokeye DA, Yin Xet al. Iron and sulfate reduction structure microbial communities in (sub-)Antarctic sediments. ISME J 2021;15:3587–604. 10.1038/s41396-021-01014-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ozuolmez D, Moore EK, Hopmans ECet al. Butyrate conversion by sulfate-reducing and methanogenic communities from anoxic sediments of Aarhus Bay, Denmark. Microorganisms 2020;8:606. 10.3390/microorganisms8040606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ozuolmez D, Stams AJM, Plugge CM. Propionate converting anaerobic microbial communities enriched from distinct biogeochemical zones of Aarhus Bay, Denmark under sulfidogenic and methanogenic conditions. Microorganisms 2020;8:394. 10.3390/microorganisms8030394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kendall MM, Liu Y, Boone DR. Butyrate-and propionate-degrading syntrophs from permanently cold marine sediments in Skan Bay, Alaska, and description of Algorimarina butyrica gen. nov et al, sp. nov. FEMS Microbiol Lett 2006;262:107–14. 10.1111/j.1574-6968.2006.00380.x [DOI] [PubMed] [Google Scholar]
- 29. Fukui M, Takii S. Microdistribution of sulfate-reducing bacteria in sediments of a hypertrophic lake and their response to the addition of organic matter. Ecol Res 1996;11:257–67. 10.1007/BF02347783 [DOI] [Google Scholar]
- 30. Meier J, Piva A, Fortin D. Enrichment of sulfate-reducing bacteria and resulting mineral formation in media mimicking pore water metal ion concentrations and pH conditions of acidic pit lakes. FEMS Microbiol Ecol 2012;79:69–84. 10.1111/j.1574-6941.2011.01199.x [DOI] [PubMed] [Google Scholar]
- 31. Sánchez-Andrea I, Stams AJ, Amils Ret al. Enrichment and isolation of acidophilic sulfate-reducing bacteria from Tinto River sediments. Environ Microbiol Rep 2013;5:672–8. 10.1111/1758-2229.12066 [DOI] [PubMed] [Google Scholar]
- 32. Dyksma S, Lenk S, Sawicka JEet al. Uncultured Gammaproteobacteria and Desulfobacteraceae account for major acetate assimilation in a coastal marine sediment. Front Microbiol 2018;9:9. 10.3389/fmicb.2018.03124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dyksma S, Pjevac P, Ovanesov Ket al. Evidence for H2 consumption by uncultured Desulfobacterales in coastal sediments. Environ Microbiol 2018;20:450–61. 10.1111/1462-2920.13880 [DOI] [PubMed] [Google Scholar]
- 34. Webster G, Watt LC, Rinna Jet al. A comparison of stable-isotope probing of DNA and phospholipid fatty acids to study prokaryotic functional diversity in sulfate-reducing marine sediment enrichment slurries. Environ Microbiol 2006;8:1575–89. 10.1111/j.1462-2920.2006.01048.x [DOI] [PubMed] [Google Scholar]
- 35. Ansbaek J, Blackburn TH. A method for the analysis of acetate turnover in a coastal marine sediment. Microb Ecol 1980;5:253–64. 10.1007/BF02020333 [DOI] [PubMed] [Google Scholar]
- 36. BB JØ, BB JØ, Parkes RJ. Role of sulfate reduction and methane production by organic carbon degradation in eutrophic fjord sediments (Limfjorden, Denmark). Limnol Oceanogr 2010;55:1338–52. 10.4319/lo.2010.55.3.1338 [DOI] [Google Scholar]
- 37. Beulig F, Roy H, Glombitza Cet al. Control on rate and pathway of anaerobic organic carbon degradation in the seabed. Proc Natl Acad Sci U S A 2018;115:367–72. 10.1073/pnas.1715789115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Greene AC. The family Desulfuromonadaceae. In: Rosenberg E., DeLong E.F., Lory S.et al. (eds.), The Prokaryotes: Deltaproteobacteria and Epsilonproteobacteria. Berlin, Heidelberg: Springer Berlin Heidelberg, 2014, 143–55 [Google Scholar]
- 39. Vandieken V, Thamdrup B. Identification of acetate-oxidizing bacteria in a coastal marine surface sediment by RNA-stable isotope probing in anoxic slurries and intact cores. FEMS Microbiol Ecol 2013;84:373–86. 10.1111/1574-6941.12069 [DOI] [PubMed] [Google Scholar]
- 40. Vandieken V, Pester M, Finke Net al. Three manganese oxide-rich marine sediments harbor similar communities of acetate-oxidizing manganese-reducing bacteria. ISME J 2012;6:2078–90. 10.1038/ismej.2012.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Aoyagi T, Hanada S, Itoh Het al. Ultra-high-sensitivity stable-isotope probing of rRNA by high-throughput sequencing of isopycnic centrifugation gradients. Environ Microbiol Rep 2015;7:282–7. 10.1111/1758-2229.12243 [DOI] [PubMed] [Google Scholar]
- 42. Aoyagi T, Morishita F, Sugiyama Yet al. Identification of active and taxonomically diverse 1,4-dioxane degraders in a full-scale activated sludge system by high-sensitivity stable isotope probing. ISME J 2018;12:2376–88. 10.1038/s41396-018-0201-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Oni OE, Schmidt F, Miyatake Tet al. Microbial communities and organic matter composition in surface and subsurface sediments of the Helgoland Mud Area, North Sea. Front Microbiol 2015;6:1290. 10.3389/fmicb.2015.01290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bohrmann G, Aromokeye AD, Bihler Vet al. R/V METEOR cruise report M134, emissions of free gas from cross-shelf troughs of South Georgia: distribution, quantification, and sources for methane ebullition sites in sub-Antarctic waters, port Stanley (Falkland Islands) - Punta Arenas (Chile). Berichte aus dem MARUM und dem Fachbereich Geowissenschaften der Universität Bremen. Staats- und Universitätsbibliothek Bremen 2017;317:1–220 [Google Scholar]
- 45. Kostka JE, Thamdrup B, Glud RNet al. Rates and pathways of carbon oxidation in permanently cold Arctic sediments. Mar Ecol Prog Ser 1999;180:7–21. 10.3354/meps180007 [DOI] [Google Scholar]
- 46. Yin X, Kulkarni AC, Friedrich MW. DNA and RNA stable isotope probing of methylotrophic methanogenic archaea. In: Dumont M, Hernández García M (eds), Stable Isotope Probing, Methods in Molecular Biology, Spring Street, NY, Humana Press, 2019, 189–206, 10.1007/978-1-4939-9721-3_15. [DOI] [PubMed] [Google Scholar]
- 47. Jørgensen B, Glud R, Holby O. Oxygen distribution and bioirrigation in Arctic fjord sediments (Svalbard, Barents Sea). Mar Ecol Prog Ser 2005;292:85–95. 10.3354/meps292085 [DOI] [Google Scholar]
- 48. Lohse L, Epping EHG, Helder Wet al. Oxygen pore water profiles in continental shelf sediments of the North Sea: turbulent versus molecular diffusion. Mar Ecol Prog Ser 1996;145:63–75. 10.3354/meps145063 [DOI] [Google Scholar]
- 49. Lin Y-S, Heuer VB, Goldhammer Tet al. Towards constraining H2 concentration in subseafloor sediment: a proposal for combined analysis by two distinct approaches. Geochim Cosmochim Acta 2012;77:186–201. 10.1016/j.gca.2011.11.008 [DOI] [Google Scholar]
- 50. Yin X, Zhou G, Cai Met al. Catabolic protein degradation in marine sediments confined to distinct archaea. ISME J 2022;16:1617–26. 10.1038/s41396-022-01210-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Deng Y, Zhang Y, Gao Yet al. Microbial community compositional analysis for series reactors treating high level antibiotic wastewater. Environ Sci Technol 2012;46:795–801. 10.1021/es2025998 [DOI] [PubMed] [Google Scholar]
- 52. Dzierzewicz Z, Cwalina B, Jaworska-Kik Met al. Susceptibility to antibiotics and biochemical properties of Desulfovibrio desulfuricans strains. Acta Pol Pharm 2001;58:439–45 [PubMed] [Google Scholar]
- 53. Ohge H, Furne JK, Springfield Jet al. The effect of antibiotics and bismuth on fecal hydrogen sulfide and sulfate-reducing bacteria in the rat. FEMS Microbiol Lett 2003;228:137–42. 10.1016/S0378-1097(03)00748-1 [DOI] [PubMed] [Google Scholar]
- 54. Karnachuk OV, Rusanov II, Panova IAet al. Microbial sulfate reduction by Desulfovibrio is an important source of hydrogen sulfide from a large swine finishing facility. Sci Rep 2021;11:10720. 10.1038/s41598-021-90256-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jia Y, Zhang H, Khanal SKet al. Insights into pharmaceuticals removal in an anaerobic sulfate-reducing bacteria sludge system. Water Res 2019;161:191–201. 10.1016/j.watres.2019.06.010 [DOI] [PubMed] [Google Scholar]
- 56. Ye M-Q, Chen G-J, Du Z-J. Effects of antibiotics on the bacterial community, metabolic functions and antibiotic resistance genes in mariculture sediments during enrichment culturing. J Mar Sci Eng 2020;8:604. 10.3390/jmse8080604 [DOI] [Google Scholar]
- 57. Ertefai TF, Heuer VB, Prieto-Mollar Xet al. The biogeochemistry of sorbed methane in marine sediments. Geochim Cosmochim Acta 2010;74:6033–48. 10.1016/j.gca.2010.08.006 [DOI] [Google Scholar]
- 58. Oni O, Miyatake T, Kasten Set al. Distinct microbial populations are tightly linked to the profile of dissolved iron in the methanic sediments of the Helgoland mud area, North Sea. Front Microbiol 2015;06:365. 10.3389/fmicb.2015.00365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yin X, Wu W, Maeke Met al. CO2 conversion to methane and biomass in obligate methylotrophic methanogens in marine sediments. ISME J 2019;13:2107–19. 10.1038/s41396-019-0425-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lueders T, Manefield M, Friedrich MW. Enhanced sensitivity of DNA-and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ Microbiol 2004;6:73–8. 10.1046/j.1462-2920.2003.00536.x [DOI] [PubMed] [Google Scholar]
- 61. Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 2016;18:1403–14. 10.1111/1462-2920.13023 [DOI] [PubMed] [Google Scholar]
- 62. Aromokeye DA, Richter-Heitmann T, Oni OEet al. Temperature controls crystalline iron oxide utilization by microbial communities in methanic ferruginous marine sediment incubations. Front Microbiol 2018;9:2574. 10.3389/fmicb.2018.02574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hassenrück C. Paired-end amplicon sequence processing workflow configurable for mixed-orientation libraries and highly variable insert sizes. 2022. https://gitio-warnemuendede/bio_inf/workflow_templates/src/branch/master/Amplicon_dada2_MiSeq. http://doi.io-warnemuende.de/10.12754/misc-2022-0002
- 64. McLaren MR, Callahan BJ. SSU taxonomic training data formatted for DADA2 (Silva version 138) [data set]. Zenodo. 2020. 10.5281/zenodo.4587955 [DOI]
- 65. Kondo R, Nedwell DB, Purdy KJet al. Detection and enumeration of sulphate-reducing bacteria in estuarine sediments by competitive PCR. Geomicrobiol J 2004;21:145–57. 10.1080/01490450490275307 [DOI] [Google Scholar]
- 66. Leloup J, Loy A, Knab NJet al. Diversity and abundance of sulfate-reducing microorganisms in the sulfate and methane zones of a marine sediment, Black Sea. Environ Microbiol 2007;9:131–42. 10.1111/j.1462-2920.2006.01122.x [DOI] [PubMed] [Google Scholar]
- 67. Bourne DG, Muirhead A, Sato Y. Changes in sulfate-reducing bacterial populations during the onset of black band disease. ISME J 2011;5:559–64. 10.1038/ismej.2010.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Emilson EJS, Carson MA, Yakimovich KMet al. Climate-driven shifts in sediment chemistry enhance methane production in northern lakes. Nat Commun 2018;9:1801. 10.1038/s41467-018-04236-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Uritskiy GV, DiRuggiero J, Taylor J. MetaWRAP-a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 2018;6:158. 10.1186/s40168-018-0541-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li D, Luo R, Liu CMet al. MEGAHIT v1.0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 2016;102:3–11. 10.1016/j.ymeth.2016.02.020 [DOI] [PubMed] [Google Scholar]
- 71. Chaumeil PA, Mussig AJ, Hugenholtz Pet al. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. J Bioinform 2019;36:1925–7. 10.1093/bioinformatics/btz848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bowers RM, Kyrpides NC, Stepanauskas Ret al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol 2017;35:725–31. 10.1038/nbt.3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hyatt D, Chen GL, LoCascio PFet al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010;11:119. 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 2016;428:726–31. 10.1016/j.jmb.2015.11.006 [DOI] [PubMed] [Google Scholar]
- 75. Huerta-Cepas J, Forslund K, Coelho LPet al. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol 2017;34:2115–22. 10.1093/molbev/msx148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Jones P, Binns D, Chang H-Yet al. InterProScan 5: genome-scale protein function classification. Bioinformatics 2014;30:1236–40. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lee MD. GToTree: a user-friendly workflow for phylogenomics. Bioinformatics 2019;35:4162–4. 10.1093/bioinformatics/btz188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Eren AM, Esen OC, Quince Cet al. Anvi'o: an advanced analysis and visualization platform for 'omics data. PeerJ 2015;3:e1319. 10.7717/peerj.1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Nguyen LT, Schmidt HA, von Haeseler Aet al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 2015;32:268–74. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 2006;23:127–8. 10.1093/bioinformatics/btl529 [DOI] [PubMed] [Google Scholar]
- 81. Pruesse E, Peplies J, Glöckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 2012;28:1823–9. 10.1093/bioinformatics/bts252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014;30:1312–3. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Baker BJ, De Anda V, Seitz KWet al. Diversity, ecology and evolution of Archaea. Nat Microbiol 2021;5:887–900. 10.1038/s41564-020-0715-z [DOI] [PubMed] [Google Scholar]
- 84. Fu L, Niu B, Zhu Zet al. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 2012;28:3150–2. 10.1093/bioinformatics/bts565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013;30:772–80. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Criscuolo A, Gribaldo S. BMGE (block mapping and gathering with entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol 2010;10:210. 10.1186/1471-2148-10-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Balzano S, Statham PJ, Pancost RD, Lloyd J. Role of microbial populations in the release of reduced iron to the water column from marine aggregates. Aquat Microb Ecol 2009;54:291–303. 10.3354/ame01278. [DOI] [Google Scholar]
- 88. Narasingarao P, Häggblom M. Sedimenticola selenatireducens, gen. nov., sp. nov., an anaerobic selenate-respiring bacterium isolated from estuarine sediment. Syst Appl Microbiol 2006;29:382–8. 10.1016/j.syapm.2005.12.011 [DOI] [PubMed] [Google Scholar]
- 89. Nakagawa T, Iino T, Suzuki KIet al. Ferrimonas futtsuensis sp. nov. and Ferrimonas kyonanensis sp. nov., selenate-reducing bacteria belonging to the Gammaproteobacteria isolated from Tokyo Bay. Int J Syst Evol Microbiol 2006;56:2639–45. 10.1099/ijs.0.64399-0 [DOI] [PubMed] [Google Scholar]
- 90. Rosselló-Mora RA, Ludwig W, Kämpfer Pet al. Ferrimonas balearica gen. nov., spec. nov., a new marine facultative Fe(III)-reducing bacterium. Syst Appl Microbiol 1995;18:196–202. 10.1016/S0723-2020(11)80390-5 [DOI] [Google Scholar]
- 91. Klein M, Friedrich M, Roger AJet al. Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. J Bacteriol 2001;183:6028–35. 10.1128/JB.183.20.6028-6035.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Waite DW, Chuvochina M, Pelikan Cet al. Proposal to reclassify the proteobacterial classes Deltaproteobacteria and Oligoflexia, and the phylum Thermodesulfobacteria into four phyla reflecting major functional capabilities. Int J Syst Evol Microbiol 2020;70:5972–6016. 10.1099/ijsem.0.004213 [DOI] [PubMed] [Google Scholar]
- 93. Greening C, Biswas A, Carere CRet al. Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J 2016;10:761–77. 10.1038/ismej.2015.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Sondergaard D, Pedersen CN, Greening C. HydDB: a web tool for hydrogenase classification and analysis. Sci Rep 2016;6:34212. 10.1038/srep34212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Poulton SW, Canfield DE. Development of a sequential extraction procedure for iron: implications for iron partitioning in continentally derived particulates. Chem Geo 2005;214:209–21. 10.1016/j.chemgeo.2004.09.003 [DOI] [Google Scholar]
- 96. Cho H, Kim B, Mok JSet al. Acetate-utilizing microbial communities revealed by stable-isotope probing in sediment underlying the upwelling system of the Ulleung Basin, East Sea. Mar Ecol Prog Ser 2019;634:45–61. 10.3354/meps13182 [DOI] [Google Scholar]
- 97. Berg C, Beckmann S, Jost Get al. Acetate-utilizing bacteria at an oxic-anoxic interface in the Baltic Sea. FEMS Microbiol Ecol 2013;85:251–61. 10.1111/1574-6941.12114 [DOI] [PubMed] [Google Scholar]
- 98. Henkel JV, Schulz-Vogt HN, Dellwig Oet al. Biological manganese-dependent sulfide oxidation impacts elemental gradients in redox-stratified systems: indications from the Black Sea water column. ISME J 2022;16:1523–33. 10.1038/s41396-022-01200-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Aromokeye DA, Kulkarni AC, Elvert Met al. Rates and microbial players of iron-driven anaerobic oxidation of methane in methanic marine sediments. Front Microbiol 2020;10:10. 10.3389/fmicb.2019.03041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Löffler B. Geochemische Prozesse und Stoffkreisläufe in Sedimenten innerhalb und außerhalb des Cumberland-Bay Fjords, Süd Georgien. Bachelor Thesis, University of Bremen, 2013. [Google Scholar]
- 101. Wehrmann LM, Formolo MJ, Owens JDet al. Iron and manganese speciation and cycling in glacially influenced high-latitude fjord sediments (West Spitsbergen, Svalbard): evidence for a benthic recycling-transport mechanism. Geochim Cosmochim Acta 2014;141:628–55. 10.1016/j.gca.2014.06.007 [DOI] [Google Scholar]
- 102. Ravenschlag K, Sahm K, Pernthaler Jet al. High bacterial diversity in permanently cold marine sediments. Appl Environ Microbiol 1999;65:3982–9. 10.1128/AEM.65.9.3982-3989.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Hoehler TM, Alperin MJ, Albert DBet al. Thermodynamic control on hydrogen concentrations in anoxic sediments. Geochim Cosmochim Acta 1998;62:1745–56. 10.1016/S0016-7037(98)00106-9 [DOI] [Google Scholar]
- 104. Hoehler TM, Alperin MJ, Albert DBet al. Apparent minimum free energy requirements for methanogenic Archaea and sulfate-reducing bacteria in an anoxic marine sediment. FEMS Microbiol Eco 2001;38:33–41. 10.1111/j.1574-6941.2001.tb00879.x [DOI] [Google Scholar]
- 105. Lovley DR, Goodwin S. Hydrogen concentrations as an indicator of the predominant terminal electron-accepting reactions in aquatic sediments. Geochim Cosmochim Acta 1988;52:2993–3003. 10.1016/0016-7037(88)90163-9 [DOI] [Google Scholar]
- 106. Vandieken V, Finke N, Thamdrup B. Hydrogen, acetate, and lactate as electron donors for microbial manganese reduction in a manganese-rich coastal marine sediment. FEMS Microbiol Eco 2014;87:733–45. 10.1111/1574-6941.12259 [DOI] [PubMed] [Google Scholar]
- 107. Thauer RK, Kaster AK, Goenrich Met al. Hydrogenases from methanogenic archaea, nickel, a novel cofactor, and H2 storage. Annu Rev Biochem 2010;79:507–36. 10.1146/annurev.biochem.030508.152103 [DOI] [PubMed] [Google Scholar]
- 108. Brysch K, Schneider C, Fuchs Get al. Lithoautotrophic growth of sulfate-reducing bacteria, and description of Desulfobacterium autotrophicum gen. nov., sp. nov. Arch Microbiol 1987;148:264–74. 10.1007/BF00456703 [DOI] [Google Scholar]
- 109. Wegener G, Krukenberg V, Riedel Det al. Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature 2015;526:587–90. 10.1038/nature15733 [DOI] [PubMed] [Google Scholar]
- 110. Hinrichs KU, Boetius A. The anaerobic oxidation of methane: new insights in microbial ecology and biogeochemistry. In: Wefer G., Billett D., Hebbeln D.et al. (eds.), Ocean Margin Systems. Berlin, Heidelberg: Springer Berlin Heidelberg, 2003, 457–77 [Google Scholar]
- 111. Li C, Zhan L, Lu H. Mechanisms for overpressure development in marine sediments. J Mar Sci Eng 2022;10:490. 10.3390/jmse10040490 [DOI] [Google Scholar]
- 112. Sass A, Rütters H, Cypionka Het al. Desulfobulbus mediterraneus sp. nov., a sulfate-reducing bacterium growing on mono- and disaccharides. Arch Microbiol 2002;177:468–74. 10.1007/s00203-002-0415-5 [DOI] [PubMed] [Google Scholar]
- 113. Casalot L, Hatchikian CE, Forget Net al. Molecular study and partial characterization of iron-only hydrogenase in Desulfovibrio fructosovorans. Anaerobe 1998;4:45–55. 10.1006/anae.1997.0137 [DOI] [PubMed] [Google Scholar]
- 114. Boopathy R, Robichaux M, LaFont Det al. Activity of sulfate-reducing bacteria in human periodontal pocket. Can J Microbiol 2002;48:1099–103. 10.1139/w02-104 [DOI] [PubMed] [Google Scholar]
- 115. Dicker HJ, Smith DW. Effects of organic amendments on sulfate reduction activity, H2 consumption, and H2 production in salt marsh sediments. Microb Ecol 1985;11:299–315. 10.1007/BF02016814 [DOI] [PubMed] [Google Scholar]
- 116. Wegener G, Niemann H, Elvert Met al. Assimilation of methane and inorganic carbon by microbial communities mediating the anaerobic oxidation of methane. Environ Microbiol 2008;10:2287–98. 10.1111/j.1462-2920.2008.01653.x [DOI] [PubMed] [Google Scholar]
- 117. Yin X, Cai M, Liu Yet al. Subgroup level differences of physiological activities in marine Lokiarchaeota. ISME J 2020;15:848–61. 10.1038/s41396-020-00818-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Yu T, Wu W, Liang Wet al. Growth of sedimentary Bathyarchaeota on lignin as an energy source. Proc Natl Acad Sci U S A 2018;115:6022–7. 10.1073/pnas.1718854115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Dyksma S, Bischof K, Fuchs BMet al. Ubiquitous Gammaproteobacteria dominate dark carbon fixation in coastal sediments. ISME J 2016;10:1939–53. 10.1038/ismej.2015.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The bacterial MAGs data are available in NCBI database under the project PRJNA678468 with the accession numbers of SAMN32874205 to SAMN32874239. Sequencing data of RNA-SIP samples have been deposited in the Short Reads Archive under the project PRJNA505997 with accession numbers from SAMN32873837 to SAMN32874024. The metagenomic reads sequenced from original sediments have been deposited under the project PRJNA1023477 with accession numbers from SAMN37668301 to SAMN37668306.