Abstract
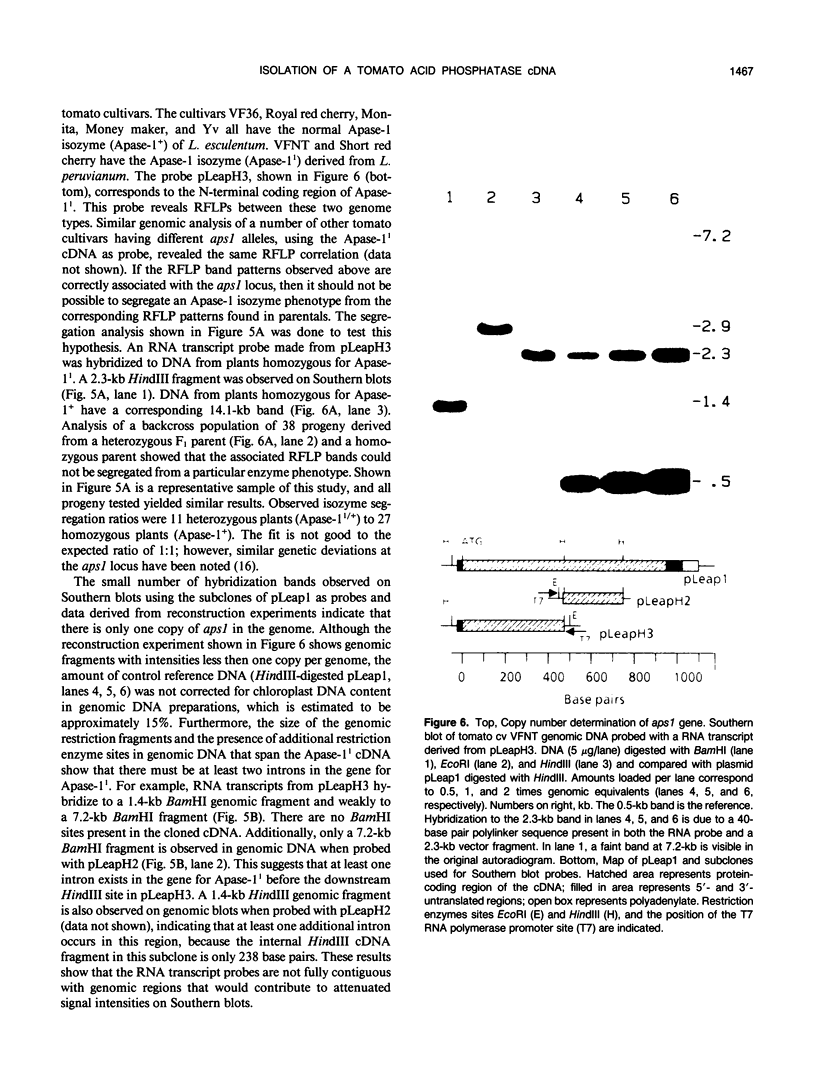
The tomato (Lycopersicon esculentum) acid phosphatase-1 (Apase-11, EC 3.1.3.2) isozyme variant, genetically linked to the root-knot nematode resistance locus (Mi) on chromosome 6, has been purified by a rapid procedure from tomato cell suspension cultures. Peptide fragments of the purified enzyme were generated from trypsin and Lys-C endoprotease digests and separated by reverse-phase high-performance liquid chromatography. Amino acid sequences derived from the purified peptide fragments represented >50% of the total amino acid content of the protein and enabled the construction of degenerate oligonucleotide probes that were used to screen a tomato cell culture complementary DNA library. Clones corresponding to full-length coding sequences for Apase-1 have been isolated and sequenced. Southern blot analysis of DNA isolated from a number of tomato cultivars shows that the Apase-11 gene (aps1) is present at one copy per genome and that genotypes containing the aps11 allele have restriction fragment length polymorphisms that distinguish them from cultivars having the aps1+ allele. Segregation analysis demonstrates that the restriction fragment length polymorphisms are associated with the aps1 locus. Tomato Apase-11 is also found to have significant homology at the amino acid sequence level to a class of vegetative storage proteins characterized in soybean.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aarts J. M., Hontelez J. G., Fischer P., Verkerk R., van Kammen A., Zabel P. Acid phosphatase-1(1), a tightly linked molecular marker for root-knot nematode resistance in tomato: from protein to gene, using PCR and degenerate primers containing deoxyinosine. Plant Mol Biol. 1991 Apr;16(4):647–661. doi: 10.1007/BF00023429. [DOI] [PubMed] [Google Scholar]
- Alexander D. C., McKnight T. D., Williams B. G. A simplified and efficient vector-primer cDNA cloning system. Gene. 1984 Nov;31(1-3):79–89. doi: 10.1016/0378-1119(84)90197-5. [DOI] [PubMed] [Google Scholar]
- Bernatzky R., Tanksley S. D. Toward a saturated linkage map in tomato based on isozymes and random cDNA sequences. Genetics. 1986 Apr;112(4):887–898. doi: 10.1093/genetics/112.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D. T., Carle G. F., Olson M. V. Cloning of large segments of exogenous DNA into yeast by means of artificial chromosome vectors. Science. 1987 May 15;236(4803):806–812. doi: 10.1126/science.3033825. [DOI] [PubMed] [Google Scholar]
- Capecchi J. T., Loudon G. M. Substrate specificity of pyroglutamylaminopeptidase. J Med Chem. 1985 Jan;28(1):140–143. doi: 10.1021/jm00379a024. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Collins F. S., Weissman S. M. Directional cloning of DNA fragments at a large distance from an initial probe: a circularization method. Proc Natl Acad Sci U S A. 1984 Nov;81(21):6812–6816. doi: 10.1073/pnas.81.21.6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A. H., Danon A., Baertlein D. A., McDaniel R. G. Phosphate Starvation Inducible Metabolism in Lycopersicon esculentum: II. Characterization of the Phosphate Starvation Inducible-Excreted Acid Phosphatase. Plant Physiol. 1988 Jul;87(3):716–720. doi: 10.1104/pp.87.3.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld R., Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- Paul E. M., Williamson V. M. Purification and properties of Acid phosphatase-1 from a nematode resistant tomato cultivar. Plant Physiol. 1987 Jun;84(2):399–403. doi: 10.1104/pp.84.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D., Halvorson H. O. A putative signal peptidase recognition site and sequence in eukaryotic and prokaryotic signal peptides. J Mol Biol. 1983 Jun 25;167(2):391–409. doi: 10.1016/s0022-2836(83)80341-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P. E. Novel Regulation of Vegetative Storage Protein Genes. Plant Cell. 1990 Jan;2(1):1–6. doi: 10.1105/tpc.2.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P. E. Soybean vegetative storage protein structure and gene expression. Plant Physiol. 1988 May;87(1):250–254. doi: 10.1104/pp.87.1.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M., Minard K., Horvath S., McNicholas J., Srelinger J., Wake C., Long E., Mach B., Hood L. A molecular map of the immune response region from the major histocompatibility complex of the mouse. Nature. 1982 Nov 4;300(5887):35–42. doi: 10.1038/300035a0. [DOI] [PubMed] [Google Scholar]
- Tanaka H., Hoshi J. I., Nakata K., Takagi M., Yano K. Isolation and some properties of acid phosphatase-1(1) from tomato leaves. Agric Biol Chem. 1990 Aug;54(8):1947–1952. [PubMed] [Google Scholar]
- Wittenbach V. A. Purification and characterization of a soybean leaf storage glycoprotein. Plant Physiol. 1983 Sep;73(1):125–129. doi: 10.1104/pp.73.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983 Jun 1;133(1):17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]





