Abstract
Objectives
The present study aimed to determine the inheritance pattern and genetic cause of congenital radial hemimelia (RH) in cats.
Methods
Clinical and genetic analyses were conducted on a Siamese cat family (n = 18), including two siblings with RH. Radiographs were obtained for the affected kittens and echocardiograms of an affected kitten and sire. Whole genome sequencing was completed on the two cases and the parents. Genomic data were compared with the 99 Lives Cat Genome data set of 420 additional domestic cats with whole genome and whole exome sequencing data. Variants were considered as homozygous in the two cases of the siblings with RH and heterozygous in the parents. Candidate variants were genotyped by Sanger sequencing in the extended pedigree.
Results
Radiographs of the female kitten revealed bilateral absence of the radii and bowing of the humeri, while the male kitten showed a dysplastic right radius. Echocardiography suggested the female kitten had restrictive cardiomyopathy with a positive left atrial-to-aortic root ratio (LA:Ao = 1.83 cm), whereas hypertrophic cardiomyopathy was more likely in the sire, showing diastolic dysfunction using tissue Doppler imaging (59.06 cm/s). Twenty-two DNA variants were unique and homozygous in the affected kittens and heterozygous in the parents. Seven variants clustered in one chromosomal region, including two frameshift variants in cardiomyopathy associated 5 (CMYA5) and five variants in junction mediating and regulatory protein, P53 cofactor (JMY ), including a missense and an in-frame deletion.
Conclusions and relevance
The present study suggested an autosomal recessive mode of inheritance with variable expression for RH in the Siamese cat family. Candidate variants for the phenotype were identified, implicating their roles in bone development. These genes should be considered as potentially causal for other cats with RH. Siamese cat breeders should consider genetically testing their cats for these variants to prevent further dissemination of the suspected variants within the breed.
Keywords: Domestic cat, Felis catus, radial dysplasia, precision medicine, whole genome sequencing
Introduction
Radial hemimelia (RH) (intercalary longitudinal hemimelia of the radius; OMIA 002225-9685) is a congenital absence of the radius characterised by partial or total absence of the bone. RH has been documented in various domesticated species;1 –7 however, opportunities for genetic studies have been limited in diverse and non-traditional biomedical models. Limb patterning and development is highly conserved across vertebrate species 8 and animals with rare diseases are important resources for assigning new functions to genes and deciphering syndromic conditions.
RH is suggested as one of the most common dysostoses affecting the forelimbs in cats 9 and several sporadic, rare cases of RH are reported in randomly bred, domestic shorthairs10 –16 where cases are often combined with other anomalies, including vertebral abnormalities, anal atresia and cardiovascular defects. 17 The RH phenotype is also known as ‘Squitten’, ‘Kangaroo cats’ or ‘Twisty cats’ by the lay public; however, scientific evaluations and documentation are not available in peer-reviewed literature for most presentations described on social media. 18
In humans, RH is rare, occurring in 1 in 5000–30,000 live births, but is slightly more common in males than in females (sex ratio of 3:2), and is observed in more than 200 genetic syndromes as isolated cases, as part of chromosomal aneuploidies and owing to exposure to teratogenic chemicals during embryonal development. 19 In accordance with the phenotypic severity, RH is classified into four categories: type I – mild shortening of the distal radius; type II – more severe radial shortening, where ulna shortening and bowing may also be present; type III – severe radial shortening and radial deviation of the hand and wrist; and type IV – complete absence of the radius with severe radial deviation of the hand and wrist. 20 More severe phenotypes (II, III, IV) generally are accompanied with absence of the first metacarpal bones.
Presentations of RH are heterogeneous and therefore likely to be genetically heterogeneous as well. Heritable forms of RH are caused by different gene variants in humans, including X-linked (OMIM: 312190), autosomal recessive as part of Baller–Gerold syndrome (BGS; OMIM: 218600) and Rothmund–Thomson syndrome type 2 (OMIM: 268400) and an autosomal dominant form as part of Duane-radial ray syndrome (OMIM: 607323). In cattle, tetra-amelia has been associated with a r-spondin 2 (RSPO2) disruption. 2
The present study aimed to identify the genetic variants responsible for the rare RH malformation observed in a Siamese cat family. The researchers conducted whole genome sequencing (WGS) on the parents and two affected kittens to identify potential candidate variants that may be linked to the RH phenotype. To validate the findings, the genomic data obtained were compared with the 99 Lives Cat Genome data set, which included 420 other domestic cats with both whole genome and whole exome sequencing data.
Materials and methods
Cat ascertainment
The study was approved by the Animal Ethics Committee of Ankara University 2018/4/33. A nuclear family of Siamese cats (n = 5) was identified, including the two normal parents, one male and one female kitten with RH, and a normal female sibling. Shortly after birth, the queen stopped nursing the affected female and male kittens. The owner noticed forelimb anomalies in two of the three kittens. The kittens were submitted to a private clinic (Kuki Veterinary Clinic, Ankara, Türkiye) for radiological examination. Lateral and ventrodorsal radiographs (70 kV, 0.3 s) were taken of the forelimbs and upper body, respectively. Oral swabs and EDTA-anticoagulated whole blood were collected by venipuncture. A complete blood count was performed using a BC-2800Vet Auto Hematology Analyzer (Mindray). An additional 13 normal cats representing the extended family were available for sampling using oral swabs.
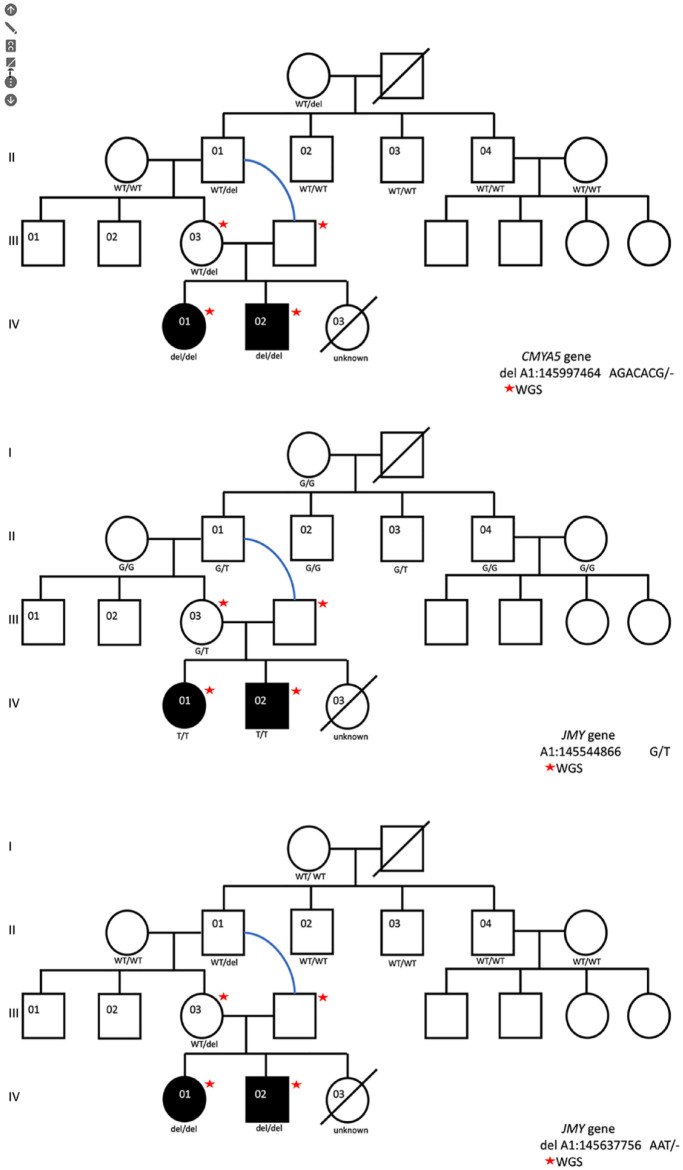
A cardiological examination of only the affected female kitten and sire was completed at a private clinic (Vet Vital VM, Ankara, Türkiye) using a Z6 ultrasound system (Mindray). CT was conducted on the female kitten for a better visualisation of the bones, using a paediatric dose (80 kV/50 mAs, 0.5 s/0.5 mm, HP 65.0, 512 × 512 pixel resolution) at the Radiology Department of Ankara University, Faculty of Medicine. A whole forearm video of the female kitten was constructed using CT images with an image-processing package (see the video in the supplementary material). 21 Cross-sectional CT images were uploaded to the 3D slicer software (3D slicer, version 4.11.20; GitHub) for semi-automatic segmentation. 22 During the segmentation process, the ‘segment editor’ function was used to separate bone from surrounding tissue. Intersection angle of lines drawn from the middle of the proximal and distal endpoints of the ulna were measured. Angular measurements of the ulnar bow was calculated with guidance of 3D reconstructed models as described by Ekblom et al. 23 Cross-sectional CT images of the female kitten were used to measure the width of the ulna; the width of ulnar midshaft was measured at midshaft from medial-lateral and from craniocaudal.
WGS and bioinformatics analysis
DNA extractions were performed using a commercial kit (Qiagen) in accordance with the manufacturer’s recommendations and approximately 3 µg was submitted for WGS. Sequencing libraries and WGS was performed by BGI Sequencing Center (BGI, Hong Kong, China) using HiSeq instrumentation (Illumina). The genomic sequence data, Fastq files, were provided to the 99 Lives Consortium for variant analyses. The WGS 12 workflow for processing the sequences, alignment to the Felis catus V9.0 reference sequence24,25 and variant calling was conducted as previously described. 24 DNA variants were viewed, filtered and annotated using VarSeq (Golden Helix) with the Ensembl annotation 101.26,27 Exons and 10 bp of flanking sequences were imported for analyses for the WGS and whole exome sequencing (WES) data. Because both parents had normal phenotypes, an autosomal recessive mode of inheritance was assumed for the RH. Candidate variants were identified as any variants that were homozygous in the two affected cats, heterozygous in the parents and absent in 358 cats with WGS data and 62 cats with WES data included in the 99 Lives Consortium. Sequencing data for the 99 Lives project are available at NCBI BioProject PRJNA987775. The specific cats for this study were accessions: felCat.Minti.Minti SAMN36026287; felCat.Pasa.Pasa SAMN36026288; felCat.Zivziv.Zivziv SAMN36026308; felCat.Minnos.Minnos SRX20808889.
Pedigree genotyping
Oligonucleotides were designed using the web-based software Primer3Plus 28 for the cardiomyopathy associated 5 (CMYA5) gene by ENSFCAG00000000223 reference. A 239 bp region in exon 1 of the CMYA5 gene harbouring the candidate causative mutation was amplified by using Forward: TGTGGAAGATGCCCATGTAA and Reverse: CGCAGCCTTCCGAAATAATA primers. A 540 bp region in exon 1 and 690 bp region in exon 9 of the JMY gene harbouring the missense and in-frame deletion mutations were amplified using forward: TGTCTTCATTGTGGCCTGGA and reverse: CTCCTGCCCAGAATCGCA and forward: AATTTGACCAGTTACAATCACTTCT and reverse: GGGCCTTTGATTATGACTCAC primers, respectively. PCR had 1 pmol of each oligonucleotide, 200 μmol/l dNTP, 1 × buffer, 1.5 μmol/l MgCl2, 3 IU of Taq polymerase (MBI Fermentas) and approximately 40 ng of genomic DNA added as the template in a volume of 25 μl. PCR conditions included an initial denaturation at 95°C for 4 mins, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, extension at 72°C for 30 s and 5 mins at 72°C for the final extension. Obtained PCR products were purified, and dideoxy chain termination (Sanger) sequencing was performed using the same forward and reverse primers on an ABI310 DNA Sequencer (Applied Biosystems). Bidirectional sequence results were analysed using Molecular Evolutionary Genetics Analysis (MEGA11) software. 29
In silico protein sequence comparison analysis was conducted for the CMYA5 and JMY proteins using Ensembl. 30 Gene ORGANizer was used to determine gene-target organ/system associations (http://geneorganizer.huji.ac.il). Also, Sorting Intolerant From Tolerant (SIFT) analysis was conducted to assess the effects of amino acid changes in the proteins in Ensembl, Variant Effect Prediction (VEP). 31
Results
An extended pedigree was ascertained consisting of 18 Siamese cats (Figure 1). One normal (Figure 1, IV-01) and one affected female kitten (Figure 1, IV-03) and an affected male kitten (Figure 1, IV-02) were born from an unaffected queen (Figure 1, III-03) that was backcrossed to her unaffected sire to produce the litter (Figure 1, II-02), thereby suggesting an autosomal recessive mode of inheritance.
Figure 1.
The Siamese cat family pedigree with radial hemimelia. Circles represent females and squares represent males. Black, filled shapes are the female and male kittens with radial hemimelia. Blue connector expresses the father mating with its daughter. Genotypes are presented below each symbol. Only cats III-01, III-02 and IV-03 were unavailable. Cats with asterisks were submitted for whole genome sequencing (WGS)
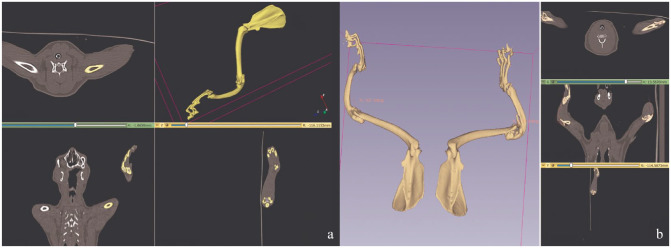
Radiographs of the 1-year-old female kitten showed bilateral absence of the radii (Figure 2) with the left radius more severely affected than the right radius. The left humerus showed bowing, and the first metacarpal was dysplastic. The right radius exhibited hypoplasia of the proximal end. Additionally, the ulna appeared to be thicker than normal. The width of the left ulnar midshaft cranial-caudal was 7.56 mm, and the medial-lateral width was 3.97 mm, while the right ulnar midshaft cranial-caudal was 8.63 mm, and the medial-lateral width was 3.91 mm. The angular distortion of the ulna was measured at 170.1° on the left forearm and 87.3° on the right forearm (Figures 2 and 3; see also the video in the supplementary material). Despite these abnormalities, the female kitten was able to ambulate by supporting its contact with the ground by using its forelimbs, although it often adopted a ‘kangaroo’ stance posture. The female kitten was consistent with a type III dysplasia on the left and type IV dysplasia on the right.
Figure 2.
Forelimb radiography of the affected female kitten. Hemimelia of radius on the left side. Hypoplasia of the proximal end of the radius on the right side. Types III and IV dysplasia in female kitten
Figure 3.
Three-dimensional reconstruction models of the forearm bones using CT images. (a) Sagittal, transversal and dorsal plane images of the left forearm extremity with a three-dimensional image of the left forearm bones. (b) Angular measurements of the left and right ulna with a three-dimensional image and cross-sectional images
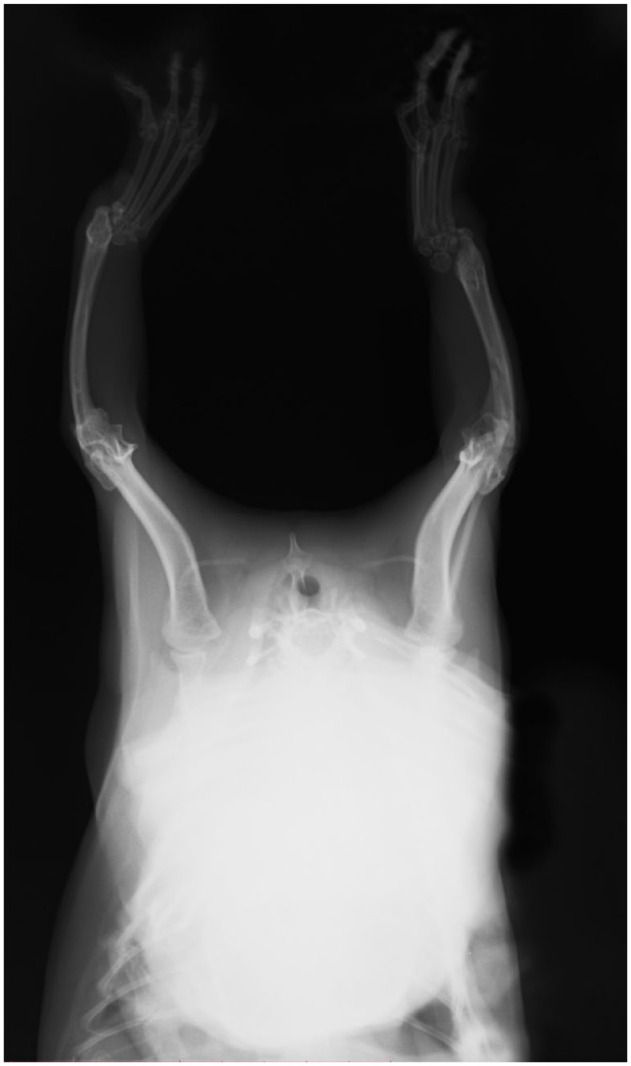
Radiographs of the 1-year-old male kitten also showed a more severe presentation on the left than on the right side with hemimelia of the left radius and a partially developed right radius; presentation in the male kitten was less severe than the female (Figure 4). The left antebrachium was dysplastic and the first metacarpal bones were absent in this cat as well. The male kitten had near-normal ambulation but was more weightbearing on the right forelimb and did not exhibit the ‘kangaroo’ stance. The male kitten’s dysplasia is consistent with a type type II on the left and type III dysplasia on the right. Both kittens had an extra pair of ribs, with 14 pairs each (Figure 5).
Figure 4.
Forelimb radiography of the affected male kitten. Hemimelia of radius on the left side (white dot indicates left). Hypoplasia of proximal end of the radius on the right side. Types II and III dysplasia in the male kitten
Figure 5.
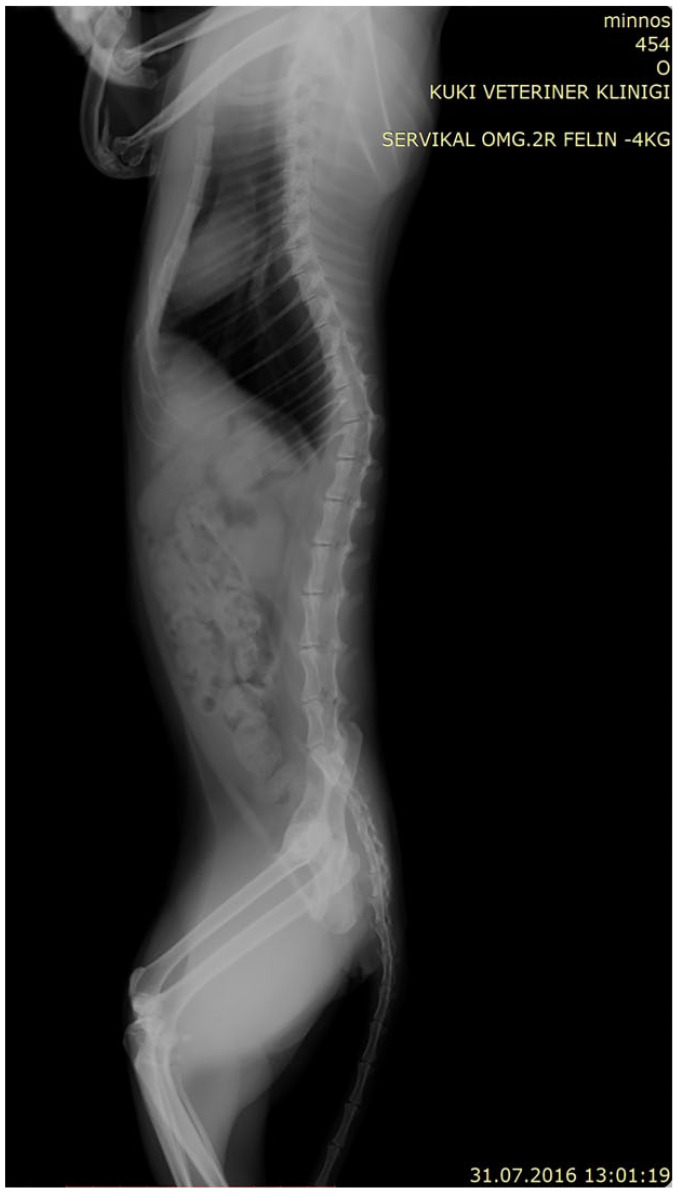
Lateral radiography of the affected female kitten. Fourteen sets of ribs and hypoplasia of the proximal end of the radius on the right side are evident
The queen had a kinked half tail and the sire and male kitten had kinked tails, but the female kitten with RH did not have a tail malformation. Radiography did not show any other bone malformation in the parents.
Echocardiography of the female kitten showed signs of congenital cardiac abnormality by having a large left atrium and a normal left ventricle (LA:Ao = 1.83 cm) 32 (Table 1).
Table 1.
Echocardiography results of female kitten and sire
| Measurement dimensions LA:Ao (M) |
Ventricle volume Teichholz (M) |
Doppler measurements | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LA Diam (cm) | LA:Ao | Ao Diam (cm) | IVSd (cm) | LVPWd | LVIDs (cm) | HR (bpm) | ESV (Teich) (ml) |
EF (Teich) (%) |
FS (Teich) (%) |
LVIDd (cm) | IVSs (cm) | LVPWs (cm) | EDV (Teich) (ml) |
SV (Teich) (ml) |
CO (Teich) (l/min) |
Mitral valve MV E Vel (cm/s) |
|
| Sire | 1.40 | 1.46 | 0.95 | 0.31 | 0.43 | 0.51 | 168 | 0.33 | 89.98 | 57.14 | 1.20 | 0.46 | 0.54 | 3.34 | 3.00 | 0.497 | 59.06 |
| Female kitten | 1.01 | 1.83 | 0.55 | 0.38 | 0.34 | 0.58 | 178 | 0.47 | 76.73 | 42.02 | 1.01 | 0.56 | 0.40 | 2.11 | 1.65 | 0.298 | NA |
LA Diam = left atrium diameter; LA:Ao = left atrium-to-aortic ratio (normal measurements: 1.21 ± 0.14); Ao Diam = aortic diameter; IVSd = interventricular septum wall diameters at end-diastole (normal measurements: 0.40 ± 0.03); LVPWd = left ventricular posterior wall (normal measurements: 0.40 ± 0.04); LVIDs = left ventricular internal diameter in end-systole (normal measurements: 1.60 ± 0.10); HR = heart rate (normal measurements: 165 ± 39); bpm = beats per minute; ESV = end systolic volume; EF = ejection fraction; FS = fractional shortening; LVIDd = left ventricular internal dimension in diastole; IVSs = interventricular septal thickness in systole; LVPWs = left ventricular posterior wall thickness in systole; EDV = end-diastolic volume; SV = stroke volume; CO = cardiac output; MV E Vel = mitral valve E velocity; NA = not available
WGS variant analyses
The 99 Lives Cat Genome Sequencing data set (n = 424) included 358 additional domestic cats with approximately 15–30 × whole genome coverage and 62 cats with >50 × whole exome sequence data.26,27 The complete variant data set included 313,3925 variants considering the exonic regions and 10 bp of flanking intronic sequence. An autosomal recessive inheritance pattern was suggested for radial hemimelia disease in cats, which is consistent with the established pedigree. 9 After filtering based on inheritance and the condition being specific to the four cats, only 34 variants were identified as candidates for RH (Table 2; see also the video in the supplementary material). A majority of the variants (n = 22) were within nine genes and three putative transcripts clustered between 119279957 and 146045193 on cat chromosome A1. Two frameshifts were identified cardiomyopathy associated 5 (CMYA5; myospryn) with suggested loss of function effects. Eight variants were identified in junction mediating and regulatory protein, P53 cofactor (JMY), including a missense, two synonymous, an in-frame deletion and four 3′ UTR variants. However, 11 variants had lower quality scores (VQSRTranche<100), including the in-frame deletion, the missense (SIFT score = 0.14) and two 3′ UTR variants in JMY. The missense mutation in Desmoglein 1 (DSG1) resulted in a deleterious SIFT score of 0 and coagulation factor II thrombin receptor (F2R) was 0.17 and not suggested as deleterious.
Table 2.
Candidate DNA variants for radial hemimelia: homozygous in affected kittens and heterozygous in parents with high quality scores
| Chromosome: position (bp) |
Reference alternative |
Gene name | Sequence ontology | Effect | Transcript change | Protein effect |
|---|---|---|---|---|---|---|
| A1:119279957 | T/– | LOC111560918 | Non-coding_exon | Other | XR_002743256.1:n.1601delA | |
| A1:142086375 | C/T | HMGCR | Upstream_gene | Unknown | XM_003981075.5:c.-139C>T | |
| A1:143261775 | A/G | F2R | Missense | Missense | XM_003981083.5:c.1196A>G | p.Tyr399Cys |
| A1:143349502 | C/T | F2RL1 | 3_prime_UTR | Other | XM_006927801.4:c.*823C>T | |
| A1:143378565 | A/C | S100Z | 5_prime_UTR | Other | XM_023257685.1:c.-257A>C | |
| A1:143383727 | G/T | S100Z | 3_prime_UTR | Other | XM_023257685.1:c.*592G>T | |
| A1:143384113 | T/C | S100Z | 3_prime_UTR | Other | XM_023257685.1:c.*978T>C | |
| A1:145626270 | A/G | JMY | Synonymous | Other | XM_011284222.3:c.1599A>G | p.Gln533= |
| A1:145645611 | A/G | JMY | 3_prime_UTR | Other | XM_011284222.3:c.*973A>G | |
| A1:145646719 | TGA/- | JMY | 3_prime_UTR | Other |
XM_011284222.3: c.*2087_*2089delTGA |
|
| A1:145659708 | T/A | LOC111561510 | Non-coding_exon | Other | XR_002744166.1:n.2516A>T | |
| A1:145997464 | AGACACG/- | CMYA5 | Frameshift | LoF |
XM_011284243.3: c.9084_9090delAGACACG |
p.Glu3028Aspfs*3 |
| A1:145997473 | TG/- | CMYA5 | Frameshift | LoF | XM_011284243.3:c.9094_9095delGT | p.Val3032Lysfs*3 |
| A1:146045193 | G/C | CMYA5 | 3_prime_UTR | Other | XM_011284243.3:c.*270G>C | |
| B2:11934569 | G/A | FAM8A1 | 3_prime_UTR | Other | XM_019831563.2:c.*3212C>T | |
| B4:71658965 | C/T | GXYLT1 | 3_prime_UTR | Other | XM_023256949.1:c.*2939G>A | |
| B4:74289208 | T/C | NELL2 | Intron | Other | XM_023256973.1:c.-32+10A>G | |
| B4:78207512 | C/G | LOC109501567 | Non-coding_exon | Other | XR_002159713.2:n.930G>C | |
| C1:175836247 | G/A | FSIP2 | Missense | Missense | XM_023259535.1:c.2586G>A | p.Met862Ile |
| D3:55279658 | C/T | DSC1,DSG1 | Missense | Missense |
XM_019815118.1:c.111+27104G>A XM_011288059.3:c.3218C>T |
p.Thr1073Met |
| D3:55969070 | C/T | LOC111557318 | Non-coding_exon | Other | XR_002737317.1:n.351C>T | |
| D3:55970578 | G/A | MEP1B | 5_prime_UTR | Other | XM_023241859.1:c.-375G>A | |
| D3:55970715 | G/A | MEP1B | 5_prime_UTR | Other | XM_023241859.1:c.-238G>A |
LoF = loss of function
Considering information provided by GeneCards (https://www.genecards.org), CMYA5 has been shown to be involved with limb-girdle muscular dystrophy and several cardiomyopathies. JMY is stated as a cofactor of p53 and JMY may have an important role in p53/TP53 stress response in DNA damage. The function of the CMYA5 gene in cats was defined by its similarity in humans. 33 CMYA5 has two transcripts in cats: CMYA5-201 (ENSFCAT00000082556.1), which is 13,193 bp and consists of 3716 amino acid residues; and CMYA5-202 (ENSFCAT00000000223.6), which is 12,394 bp and consists of 4108 amino acids. The identified mutations (c.8856_8862delAGACACG at A1:145997464 and c.8866_8867delGT at A1:145997473) are suggested to cause a loss of function (LoF) in both transcripts, leading to p.Glu2952Aspfs*3 and p.Val2956Lysfs*3, respectively, therefore likely disrupting the terminal 20–25% of the protein. The proximity of the two variants suggested that the annotation defined two variants based on only one deletion. Bidirectional Sanger sequencing confirmed the deletions (Figure 6). Bidirectional Sanger sequencing also confirmed the missense and in-frame deletion mutations in JMY.
Figure 6.
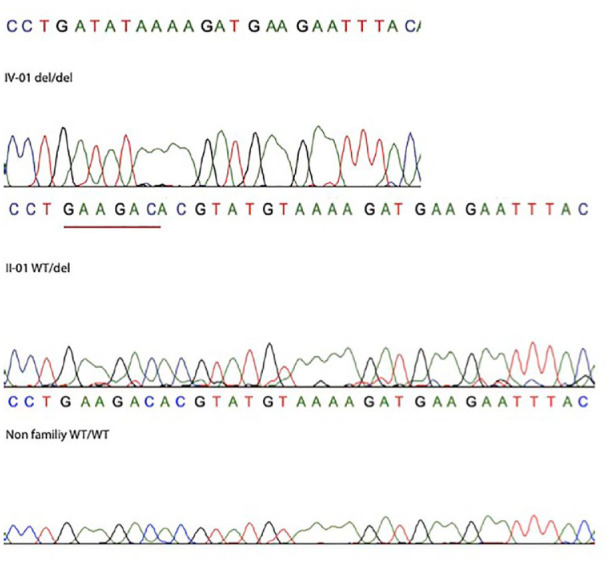
Sanger sequencing of cats with radial hemimelia (RH). Homozygous female kitten (IV-01 del/del), heterozygous sire (II-01 WT/del) and unrelated Siamese control cat (WT/WT). Del = presence of the 7 bp deletion in CMYA5. WT = wild-type variant (ie, variant of the reference sequence)
Variant screening in extended pedigree
In the Siamese pedigree consisting of four generations, the 7 bp deletion frameshift CMYA5 variant at position A1:145997464 was determined in the sire (II-01) (also known as the maternal sire) and the paternal queen (I-01) and in the two affected kittens (Figure 1). Thus, the CMYA5 variant was concordant with phenotype and an autosomal recessive mode of inheritance. No other cats in the extended pedigree had the variant. The missense mutation in the JMY gene was determined only in the sire (II-01) and therefore did not segregate with disease or the mode of inheritance.
Discussion
Radial development failures owing to genetic or non-genetic factors are classified as embryological formation defects. 34 Classifications for radial dysplasia in humans are defined according to the severity of the phenotype. 20 Despite the similarities of the phenotype, owing to rare occurrences of radial dysplasia in cats, RH has never been classified in cats. Although bowing of the long bones has been reported in humans with RH, only the bowing of the ulna is evaluated 35 for classification. Because the findings on the humerus in cats are also of clinical importance, the status of the humeri in cats should be considered to determine overall severity and classification type. The radiological findings of the two affected kittens according to the classification by Heikel 36 suggests the female kitten as having types III and IV dysplasia, whereas the male kitten is suggested as having types II and III for the right and left antebrachium, respectively.
RH has been suggested as a hereditary trait in cats by Swalley and Swalley 14 after observation of affected parents of one cat that produced a litter of eight kittens, and later on in Siamese and domestic shorthair cats; 37 sporadic cases in random bred cats from various regions of the world have been noted and popularised on social media also. However, neither the inheritance pattern, nor the genetic background of the congenital defect has been determined. In addition to the antebrachium disruptions, each of the parental cats and the male offspring had vertebral abnormalities, presenting as kinked tails and an extra set of ribs. In Asian domestic cats and the Japanese and Kurilian Bobtail breeds, the HES7 gene is associated with a short and kinked tail formation due to hemivertebrae, as well as extra ribs and occasionally an absent thoracic or lumbar vertebrae. 38 However, no variant was detected in HES7 in the parents and male kitten genome (data not shown). Therefore, the presence of the abnormal rib count and the kinked tails in the parental cats and one kitten may be incidental or potentially part of the skeletal abnormalities within these cats. If the tail and rib abnormalities are incidental, the mode of inheritance is highly suggestive of an autosomal recessive trait with variable expression, as supported by the genotypic data, the close inbreeding and absence of the antebrachium dysplasia in the parents. However, because the maternal queen and sire also had kinked tails, an autosomal dominant mode of inheritance could be possible if the tail presentation is a result of a heterozygous variant, and the more severe presentation is found in the homozygous kittens.
According to the Gene Organizer online tool, the function of CMYA5 is related to cardiac muscles. 39 To check the possible effect of the variant in the heart, the RH female kitten’s heart was examined by cardiac ultrasound. In comparison with other cats at same age, this kitten showed a restrictive cardiomyopathy. The sire was 11 years old, suggesting that the hypertrophic cardiomyopathy (HCM) was probably age-related or the reduced penetrance of gene. No known variants causing HCM in cats were detected within the four cats (data not shown).
It is likely that other transcription factors or induced expression of other factors are required for hind formation. 40 Although in silico protein function analysis of CMYA5 did not suggest bone formation association and a strong interaction with RH major causative genes, such as SALL4 or TBX5, CMY5A gene expression could be one of the transcription factors that is necessary for a proper forelimb formation. Like CMYA5, SALL4 and TBX5 genes are both important in heart development and a recent study confirmed TBX5 and CMYA5 interaction during this process. 41 CMYA5 is an under-studied striated muscle protein. 42 To date, around 40 papers have been published associating CMYA5 with cardiac dyad architecture, 42 tibial and limb-girdle muscle distrophies, 43 schizophrenia 44 and even with different types of cancer.45,46 Furthermore a recent study demonstrated CMYA5 as a novel interaction partner of FHL2 in the cardiac myocytes, 47 which supports the HCM findings of the present study. The position of CMYA5 is adjacent to Z-lines, which precede junctional sarcoplasmic reticulum positioning or transverse tubule formation during cardiac development, 42 whereas TBX5 is required for patterning of the cardiac conduction system and maintenance of mature cardiomyocyte function. 48
RH cases are associated with more than 30 genes in humans and mice (https://monarchinitiative.org/phenotype/HP:0003974#gene). Fanconi anaemia is especially genetically and clinically heterogenous and RH, together with other congenital abnormalities, is one of the clinical findings. Fanconi anaemia cases are caused by genomic instability based on deficiencies in the DNA repair. 49 Upon DNA damage, JMY triggers the p53 response. Via this path, p53-dependent transcription-induced apoptosis begins. 50 Considering the importance of DNA repair and apoptosis mechanisms, JMY cannot be neglected as a second candidate gene. The protein conservation determination for the first 63 amino acids strengthens the importance for the Gln61His missense mutation. Moreover, variations found on this region reported with deleterious effect on Ensembl VEP. 31 The A1:145544866 G/T and the A1:154637756 AAT/- in-frame deletion variations resulted in changes in the protein, but did not segregate concordantly with disease in the pedigree. However, because there are several variations found on this gene, sequencing analysis were performed to confirm variations.
The variants in both genes have not been identified previously, confirming the importance of these cats and other such natural knockdown animal models in supporting gene functions. CMYA5 has no documented single gene, pathogenic variants in the clinical variants database (ClinVar; https://www.ncbi.nlm.nih.gov/clinvar). Recently, several causative variants have been determined in diseases such as dwarfism, 51 cardiomyopathy 52 and autosomal dominant ciliopathy 53 that represent novel genes for these phenotypes. The identified variant in CMYA5 is suggested to cause a loss of function of the protein, segregates in the pedigree with phenotype, although a limited pedigree, and is a rare variant. This variant is likely pathogenic; however, functional data to support the role of CMYA5 in RH is warranted because its role would be a novel and a new contribution to developmental biology. 54
Conclusions
The findings of the present study suggest an autosomal recessive mode of inheritance with variable expression for radial hemimelia in the Siamese cat family. One deletion and several base substitutions in CMYA5 and JMY are proposed as tentative candidate variants for the phenotype, implicating their role in bone development. These genes should be investigated in other cats with RH and evaluated in a larger population to determine allele frequencies in various breeds and populations. Cat breeders, especially those of Siamese cats, could consider genetically testing their cats for these variants to prevent further dissemination within the breed if RH presentations persist.
Footnotes
Accepted: 20 July 2023
Supplementary material: The following file is available as supplementary material:
Video 1: Three-dimensional image of the affected female kitten using CT.
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article. Dr Lyons has long-standing interactions and periodic funding from several commercial genetic testing groups; however, those interactions do not influence the research and interpretations of the investigations.
Funding: This study was supported by Ankara University Scientific Research Projects Coordination Unit (Project no: 18B0239001). Funding was provided by the University of Missouri, College of Veterinary Medicine, Gilbreath McLorn Endowment, donations to the 99 Lives Cat Genome Sequencing project and funding from the Winn Feline Foundation (EveryCat and the George and Phyllis Miller Trust (MT–13-010, MTW15-017 MT18-009; MTW18-009; MT19-001) (LAL).
Ethical approval: The work described in this manuscript involved the use of non-experimental (owned or unowned) animals and procedures that differed from established internationally recognised high standards (‘best practice’) of veterinary clinical care for the individual patient. The study therefore had prior ethical approval from an established (or ad hoc) committee as stated in the manuscript.
Informed consent: Informed consent (verbal or written) was obtained from the owner or legal custodian of all animal(s) described in this work (experimental or non-experimental animals, including cadavers) for all procedure(s) undertaken (prospective or retrospective studies). For any animals or people individually identifiable within this publication, informed consent (verbal or written) for their use in the publication was obtained from the people involved.
ORCID iD: Nüket Bilgen  https://orcid.org/0000-0003-2324-7965
https://orcid.org/0000-0003-2324-7965
Bengi Çınar Kul  https://orcid.org/0000-0002-8955-0097
https://orcid.org/0000-0002-8955-0097
Mustafa Yenal Akkurt  https://orcid.org/0000-0002-8139-0252
https://orcid.org/0000-0002-8139-0252
Leslie A. Lyons  https://orcid.org/0000-0002-1628-7726
https://orcid.org/0000-0002-1628-7726
Özge Şebnem Çıldır  https://orcid.org/0000-0001-7070-4212
https://orcid.org/0000-0001-7070-4212
Furkan Kutlu  https://orcid.org/0000-0003-0310-2590
https://orcid.org/0000-0003-0310-2590
References
- 1. Alam MR, Heo SY, Lee HB, et al. Preaxial longitudinal intercalary radial hemimelia in a dog: a case report. Vet Med (Praha) 2006; 51: 118–123. [Google Scholar]
- 2. Becker D, Weikard R, Schulze C, et al. A 50-kb deletion disrupting the RSPO2 gene is associated with tetradysmelia in Holstein Friesian cattle. Genet Sel Evol 2020; 52: 68. DOI: 10.1186/s12711-020-00586-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Corbera JA, Pulido M, Morales M, et al. Radiological findings in three cases of paraxial radial hemimelia in goats. J Vet Med Sci 2002; 64: 843–845. [DOI] [PubMed] [Google Scholar]
- 4. Hildreth BE, III, Johnson KA. Ulnocarpal arthrodesis for the treatment of radial agenesis in a dog. Vet Comp Orthop Traumatol 2007; 20: 231–235. [DOI] [PubMed] [Google Scholar]
- 5. McKee WM, Reynolds J. Ulnocarpal arthrodesis and limb lengthening for the management of radial agenesis in a dog. J Small Anim Pract 2007; 48: 591–595. [DOI] [PubMed] [Google Scholar]
- 6. Pedersen NC. Surgical correction of a congenital defect of the radius and ulna of a dog. J Am Vet Med Assoc 1968; 153: 1328–1331. [PubMed] [Google Scholar]
- 7. Rahal SC, Volpi RS, Ciani RB, et al. Use of the Ilizarov method of distraction osteogenesis for the treatment of radial hemimelia in a dog. J Am Vet Med Assoc 2005; 226: 65–68, 52. [DOI] [PubMed] [Google Scholar]
- 8. Varga Z, Varga M. Gene expression changes during the evolution of the tetrapod limb. Biol Futur 2022; 73: 411–426. [DOI] [PubMed] [Google Scholar]
- 9. Towle HA, Breur GJ. Dysostoses of the canine and feline appendicular skeleton. J Am Vet Med Assoc 2004; 225: 1685–1692. [DOI] [PubMed] [Google Scholar]
- 10. Bezhentseva A, Singh H, Boudrieau RJ. Bilateral radial agenesis in a cat treated with bilateral ulnocarpal arthrodesis. Vet Comp Orthop Traumatol 2018; 31: 379–384. [DOI] [PubMed] [Google Scholar]
- 11. Lockwood A, Montgomery R, McEwen V. Bilateral radial hemimelia, polydactyly and cardiomegaly in two cats. Vet Comp Orthop Traumatol 2009; 22: 511–513. [DOI] [PubMed] [Google Scholar]
- 12. O’Brien CR MR, Nicoll RG. What is your diagnosis? Bilateral forelimb deformity and abnormal gait in a young Devon Rex. J Feline Med Surg 2002; 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pisoni L, Cinti F, Del Magno S, et al. Bilateral radial hemimelia and multiple malformations in a kitten. J Feline Med Surg 2012; 14: 598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Swalley J, Swalley M. Agenesis of the radius in a kitten. Feline Pract 1978; 8: 25–26. [Google Scholar]
- 15. Winterbotham EJ, Johnson KA, Francis DJ. Radial agenesis in a cat. J Small Anim Pract 1985; 26: 393–398. [Google Scholar]
- 16. Babar TK, Wadhokar OC, Deshmukh MK. A rare case of radius hemimelia: a case report. Pan Afr Med J 2022; 41. DOI: 10.11604/pamj.2022.41.304.32909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moura E, Cirio SM, Villanova JA., Jr. VACTERL association in a cat. Am J Med Genet A 2010; 152A: 777–780. [DOI] [PubMed] [Google Scholar]
- 18. Squitten. Wikipedia, 2023. Available at: https://en.wikipedia.org/wiki/Squitten (accessed 24 August 2023). [Google Scholar]
- 19. Krakow D. Radial ray deficiency. In: Copel JA, Feltovich H, Gratacós E, et al. (eds). Obstetric imaging: fetal diagnosis and care. Philapdelphia, PA: Elsevier, 2018, pp 273–277.e1. [Google Scholar]
- 20. Goldfarb CA, Wall L, Manske PR. Radial longitudinal deficiency: the incidence of associated medical and musculoskeletal conditions. J Hand Surg Am 2006; 31: 1176–1182. [DOI] [PubMed] [Google Scholar]
- 21. Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012; 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fedorov A, Beichel R, Kalpathy-Cramer J, et al. 3D slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging 2012; 30: 1323–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ekblom AG, Dahlin LB, Rosberg HE, et al. Hand function in children with radial longitudinal deficiency. BMC Musculoskelet Disord 2013; 14. DOI: 10.1186/1471-2474-14-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Buckley RM, Davis BW, Brashear WA, et al. A new domestic cat genome assembly based on long sequence reads empowers feline genomic medicine and identifies a novel gene for dwarfism. PLoS Genet 2020; 16. DOI: 10.1371/journal.pgen.1008926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Buckley RM, Gandolfi B, Creighton EK, et al. Werewolf, there wolf: variants in hairless associated with hypotrichia and roaning in the Lykoi cat breed. Genes (Basel) 2020; 11. DOI: 10.3390/genes11060682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rodney AR, Buckley RM, Fulton RS, et al. A domestic cat whole exome sequencing resource for trait discovery. Sci Rep 2021; 11: 7159. DOI: 10.1038/s41598-021-86200-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Katz ML, Buckley RM, Biegen V, et al. Neuronal ceroid lipofuscinosis in a domestic cat associated with a DNA sequence variant that creates a premature stop codon in CLN6. G3 (Bethesda) 2020; 10: 2741–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Untergasser A, Nijveen H, Rao X, et al. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res 2007; 35: W71–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stecher G, Tamura K, Kumar S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol Biol Evol 2020; 37: 1237–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yates AD, Achuthan P, Akanni W, et al. Ensembl 2020. Nucleic Acids Res 2020; 48: D682–D688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McLaren W, Gil L, Hunt SE, et al. The Ensembl Variant Effect Predictor. Genome Biol 2016; 17: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Locatelli C, Pradelli D, Campo G, et al. Survival and prognostic factors in cats with restrictive cardiomyopathy: a review of 90 cases. J Feline Med Surg 2018; 20: 1138–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rappaport N, Fishilevich S, Nudel R, et al. Rational confederation of genes and diseases: NGS interpretation via GeneCards, MalaCards and VarElect. Biomed Eng Online 2017; 16: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kozin SH. Upper-extremity congenital anomalies 2003. J Bone Joint Surg Am 2003; 85: 12. [DOI] [PubMed] [Google Scholar]
- 35. Elmakky A, Stanghellini I, Landi A, et al. Role of genetic factors in the pathogenesis of radial deficiencies in humans. Curr Genomics 2015; 16: 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Heikel HVA. Aplasia and hypoplasia of the radius: studies on 64 cases and on epiphyseal transplantation in rabbits with the imitated defect. Acta Orthop Scand 1959; 30: 3–155. [PubMed] [Google Scholar]
- 37. Hoskins JD. Congenital defects of cats. Compend Contin Educ Vet 1995; 17: 20. [Google Scholar]
- 38. Xu X, Sun X, Hu XS, et al. Whole genome sequencing identifies a missense mutation in HES7 associated with short tails in Asian domestic cats. Sci Rep 2016; 6. DOI: 10.1038/srep31583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gokhman D, Kelman G, Amartely A, et al. Gene ORGANizer: linking genes to the organs they affect. Nucleic Acids Res 2017; 45: W138–W45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tickle C. How the embryo makes a limb: determination, polarity and identity. J Anat 2015; 227: 418–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rathjens FS, Blenkle A, Iyer LM, et al. Preclinical evidence for the therapeutic value of TBX5 normalization in arrhythmia control. Cardiovasc Res 2020; 177: 1908–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lu F, Ma Q, Xie W, et al. CMYA5 establishes cardiac dyad architecture and positioning. Nat Commun 2022; 13: 2185. DOI: 10.1038/s41467-022-29902-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sarparanta J, Blandin G, Charton K, et al. Interactions with M-band titin and calpain 3 link myospryn (CMYA5) to tibial and limb-girdle muscular dystrophies. J Biol Chem 2010; 285: 30304–30315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tsoupri E, Kostavasili I, Kloukina I, et al. Myospryn deficiency leads to impaired cardiac structure and function and schizophrenia-associated symptoms. Cell Tissue Res 2021; 385: 675–696. [DOI] [PubMed] [Google Scholar]
- 45. Guo Y, Li Y, Li J, et al. DNA methylation-driven genes for developing survival nomogram for low-grade glioma. Front Oncol 2021; 11. DOI: 10.3389/fonc.2021.629521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Feng J, Jiang L, Li S, et al. Multi-omics data fusion via a joint kernel learning model for cancer subtype discovery and essential gene identification. Front Genet 2021; 12. DOI: 10.3389/fgene.2021.647141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stathopoulou K, Schnittger J, Raabe J, et al. CMYA5 is a novel interaction partner of FHL2 in cardiac myocytes. FEBS J 2022; 289: 4622–4645. [DOI] [PubMed] [Google Scholar]
- 48. Steimle JD, Moskowitz IP. TBX5: a key regulator of heart development. Curr Top Dev Biol 2017; 122: 195–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Giampietro PF, Verlander PC, Davis JG, et al. Diagnosis of Fanconi anemia in patients without congenital malformations: an International Fanconi Anemia Registry study. Am J Med Genet 1997; 68: 58–61. [PubMed] [Google Scholar]
- 50. Adighibe O, Pezzella F. The role of JMY in p53 regulation. Cancers (Basel) 2018; 10. DOI: 10.3390/cancers10060173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lyons LA, Fox DB, Chesney KL, et al. Localization of a feline autosomal dominant dwarfism locus: a novel model of chondrodysplasia. bioRxiv 2019. DOI: 10.1101/687210. [DOI] [Google Scholar]
- 52. Ontiveros ES, Ueda Y, Harris SP, et al. Precision medicine validation: identifying the MYBPC3 A31P variant with whole-genome sequencing in two Maine Coon cats with hypertrophic cardiomyopathy. J Feline Med Surg 2019; 21: 1086–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cogne B, Latypova X, Senaratne LDS, et al. Mutations in the kinesin-2 motor KIF3B cause an autosomal-dominant ciliopathy. Am J Hum Genet 2020; 106: 893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lyons LA. Genetic testing in domestic cats. Mol Cell Probes 2012; 26: 224–230. [DOI] [PMC free article] [PubMed] [Google Scholar]