Abstract
A highly reproducible and robust cell-based high-throughput screening (HTS) assay was adapted for screening of small molecules for antiviral activity against influenza virus strain A/Vietnam/1203/2004 (H5N1). The NIH Molecular Libraries Small Molecule Repository (MLSMR) Molecular Libraries Screening Centers Network (MLSCN) 100,000-compound library was screened at 50 μM. The “hit” rate (>25% inhibition of the viral cytopathic effect) from the single-dose screen was 0.32%. The hits were evaluated for their antiviral activity, cell toxicity, and selectivity in dose-response experiments. The screen yielded 5 active compounds (SI value >3). One compound showed an SI50 value of greater than 3, 3 compounds had SI values ranging from greater than 14 to 34, and the most active compound displayed an SI value of 94. The active compounds represent 2 different classes of molecules, benzoquinazolinones and thiazoloimidazoles, which have not been previously identified as having antiviral/anti-influenza activity. These molecules were also effective against influenza A/California/04/2009 virus (H1N1) and other H1N1 and H5N1 virus strains in vitro but not H3N2 strains. Real-time qRT-PCR results reveal that these chemotypes significantly reduced M1 RNA levels as compared to the no-drug influenza-infected Madin Darby canine kidney cells. (Journal of Biomolecular Screening 2011:73-81)
Keywords: 1-benzoyl-3-arylthioureas, anti-infective drugs, automation/robotics, benzothiazinones, carboxanilides, cell-based assays, HTS, influenza virus, sulfonamides
INTRODUCTION
Influenza A and B are negative-strand RNA viruses that infect the upper and lower respiratory tracts, causing substantial morbidity and mortality annually.1 Human infection by avian influenza is caused by the transmission of influenza viruses across the species barrier from birds to humans. Highly pathogenic avian influenza (HPAI) H5N1 viruses pose considerable pandemic potential since humans are immunogenically naive to these viruses. Furthermore, unlike human influenza A viruses, avian H5N1 viruses can infect the lower respiratory tract, causing hypercytokinemia and increased tissue damage.2 In 1997, the Special Administrative Region of China in Hong Kong garnered worldwide attention when an epidemic of highly pathogenic avian influenza (H5N1) virus was transmitted from poultry to humans resulting in 18 human cases, of which 6 were fatal.3 From 1997 to June 2008, there have been more than 130 out-breaks from Asia to Europe and Africa. As of December 2009, there is an approximately 60% mortality rate due to HPAI making this H5N1 virus the most lethal influenza virus to be transferred from birds to humans.4 Transmission of the H5N1 virus from birds to humans is inefficient; however, the presence of the pathogen in farm animals has resulted in the culling of hundreds of millions of birds and awareness of more human cases.5
Most recently, the World Health Organization declared the 2009 “swine” flu outbreak a pandemic in response to a new strain of influenza A virus subtype H1N1. Of the 8 viral gene segments from flu strain A/California/07/2009, 6 are reassorted from a variety of swine, avian, and human influenza viruses typically found in Asia and Europe. Two hundred eight countries and overseas territories have reported laboratory-confirmed cases with at least 13,554 deaths.
Since the 1997 H5N1 virus outbreak, several vaccine strategies have been under way for H5N1 influenza viruses.6,7 Development of vaccines for H5N1 has been hampered by antigenic differences of the hemagglutinin protein. Early attempts to create an inactivated vaccine using an antigenically similar H5N3 strain were unsuccessful. However, with the development of the reverse genetics systems,8-11 safe and effective attenuated vaccines could be produced. This approach also had challenges as development of a vaccine using a reverse genetics approach based on a H5N1 strain isolated from Vietnam in 2004 did not achieve immune protection.12,13 Finally, in 2006, a trial was conducted using the same modified 2004 vietnam strain as the others. To achieve satisfactory antibody titers, very high doses of the vaccine were required.14 Acceptable levels of protection were achieved in only 54% to 58% of subjects compared with the 70% to 90% usually achieved with seasonal vaccine.14 This H5N1 vaccine has been approved by the Food and Drug Administration (FDA); however, it is only recommended for people aged 18 to 64 who could be at risk for exposure to this strain. Most recently, an inactivated, whole-virus, adjuvant H5N1 vaccine elicited a cell-mediated immune response without apparent toxicity in adult and elderly volunteers receiving a single low dose.15
Three classes of drugs are used to treat influenza virus infections: the neuraminidase inhibitors, oseltamivir16 and zanamivir17; the M2 ion channel inhibitors (adamantanes), amantadine18 and rimantadine19; and the IMP dehydrogenase inhibitors, ribavirin and mycophenolic acid. All of these inhibitors, except ribavirin, are FDA approved, although ribavirin is approved in Europe (European Union’s Committee for Medicinal Products for Human Use [CHMP]). Mycophenolic acid is an immunosuppressive drug used for prevention of organ rejection in transplant recipients. Of these, only oseltamivir and zanamivir were recommended for treatment of seasonal influenza in 2007-2008 by the Centers for Disease Control and Prevention (CDC, Atlanta, GA). Each of these classes of drugs targets the 3 surface proteins, M2, HA, and NA. M2 inhibitors block the proton influx that is required for cleavage of the ribonucleoprotein complex from the matrix protein. The NA drugs inhibit the release of progeny virus from host cells by cleaving sialic acid residues on sialoglycans on the host cell.20
The emergence of resistant influenza viruses has been associated with continued drug use. The 2009 H1N1 viral resistance to Tamiflu has been reported at a low incidence but is expected to increase to levels similar to those observed in seasonal H1N1 resistance.21,22 The CDC estimates an H5N1 influenza pandemic could potentially kill 1 billion people worldwide. As well, the recent H1N1 pandemic illustrates the crucial gap that antivirals can fill as vaccines are manufactured. Thus, there is a critical need for new antiviral drugs to supplement vaccine development and existing chemotherapeutics. To date, other high-throughput (HT) assays have proven successful in discovering lead-candidate antiviral compounds against influenza A infection. These include a cell-based reporter assay that uses HA pseudotyped retroviral vectors that express luciferase23 and a cell-based high-throughput screening (HTS) assay that uses viral neuraminidase (NA) as a readout.24 We have adapted a cell-based HT assay25 to screen 100,000 compounds from the NIH Molecular Libraries Screening Centers Network (MLSCN) compound library at 50 μM against H5N1 virus to identify potential novel influenza inhibitors. We report the discovery of novel chemotypes with potent anti-influenza activity against several strains of influenza A viruses (H5N1 and H1N1).
MATERIALS AND METHODS
Cell growth conditions and media
Madin Darby Canine Kidney cells (ATCC CCL-34; American Type Culture Collection [ATCC], Manassas, VA) were maintained as adherent cell lines in Eagle minimum essential medium with 2 mM L-glutamine and 10% fetal bovine serum (FBS) at 37 °C in a humidified 5% CO2 atmosphere as described previously.25 Cells were passaged as needed and harvested from flasks using 0.25% trypsin-EDTA. Prior to cell plating, cells were resuspended in serum-free Dulbecco’s modified Eagle medium (DMEM) with 4 mM L-glut and 1% bovine serum albumin (BSA; Assay Media).
Influenza virus culture
Influenza A/NWS/33 (H1N1) was provided by K. Cochran of the University of Michigan (Ann Arbor, MI). A/Victoria/3/75 (H3N2) was obtained from the ATCC. A/Duck/MN/1525/81 (H5N1) and A/Gull/PA/4175/83 (H5N1) were obtained from R. Webster of the St. Jude Children’s Research Hospital (Memphis, TN). A/California/04/2009 (H1N1), A/New Caledonia/20/99 (H1N1), A/Solomon Islands/03/2006 (H1N1), A/Beijing/32/92 (H3N2), A/California/7/04 (H3N2), A/Panama/2007/99 (H3N2), A/Sydney/05/97 (H3N2), A/Wisconsin/67/2005 (H3N2), A/ Hong Kong/156/97 (H5N1), A/Vietnam/1203/2004 (H5N1), B/ Sichuan/379/99, and B/Malaysia/2506/2004 were acquired from the Influenza Branch of the CDC. A/Vietnam/1203/2004 × A/Ann Arbor/6/60 (H5N1), a hybrid virus containing the H5 and N1 genes from A/Vietnam and the internal genes from A/Ann Arbor (H1N1), was provided by Medimmune (Mountain View, CA). Most of the viruses were propagated in Madin Darby canine kidney (MDCK) cells to prepare pools for use in these experiments. Three virus strains were passaged in embryonated chicken eggs: A/California/04/2009 (H1N1), A/Hong Kong/156/97 (H5N1), and A/Vietnam/1203/2004 (H5N1). Chicken eggs were inoculated for amplification of virus, and allantoic fluid was recovered aliquoted and stored below −70 °C for use in the assay.
Compound library and controls
The positive control drug for this assay, ribavirin26 (#196066, MP Biomedicals, Solon, OH), was solubilized at 8 mg/mL in DMSO (Sigma, St. Louis, MO). The stock solution was diluted to a final concentration of 164 μM in assay media (DMEM without phenol red, 1% BSA, 4 mM L-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin; Gibco, Grand Island, NY) and added to each plate before each experiment and discarded after-wards. Final DMSO concentration in each well was 0.5%.
The MLSMR is a library of biologically relevant small organic molecules that has been used for HTS as part of the NIH Roadmap initiative, the MLSCN, and Molecular Libraries Production Center Network (MLPCN). This library has been updated and expanded since the initiation of the program in 2005, containing 95,512 compounds at the time of influenza H5N1 assay screening. Compounds were solubilized at 10 mM in DMSO. Before each experiment, all compounds were diluted to 50 μM in assay media for the screen.
Influenza high-throughput screen
The basic methods for the HTS for the identification of potential inhibitors of influenza virus have been previously described.25 Briefly, MDCK cells (3 × 105 cells/mL) were dispensed into black, clear-bottom, 384-well plates at a density of 6000 cells/well in 20 μL assay medium using a WellMate (Matrix, Hudson, NH) and incubated 24 h at 37°C, 5% CO2, with high humidity. The next day, 5 μL of each compound was added to cells using a Biomek FX liquid handler (Beckman Coulter, Fullerton, CA). This resulted in a final drug concentration of 50 μM (0.5% DMSO) for all samples. Within 30 min of compound addition, cells were infected with 5 μL of diluted virus at a concentration of 100 TCID50 doses using a WellMate (multiplicity of infection [MOI] of 0.005 PFU/cell). Virus was diluted from amplified virus stock prepared in egg allantoic fluid into assay media for a final virus stock dilution of 1:10,000. Internal controls consisted of wells containing cells only, cells infected with virus, and virus-infected cells treated with ribavirin. Plates were incubated at 37 °C, 5% CO2, for 72 h. After incubation, 30 μL of Cell Titer Glo (Promega, Madison, WI) was added to each well using a WellMate and incubated at room temperature (RT) for 10 to 30 min. Luminescence was measured using an EnVision plate reader (PerkinElmer, Wellesley, MA).
Secondary confirmatory assays
For dose-response assays, test compounds were serially diluted in serum-free media containing 0.5% DMSO final per well in a plate-to-plate matrix or “stacked plate” matrix. Briefly, all 320 compounds in a source plate were diluted together, resulting in a 10-point dose-response dilution series. Basically, this results in a serial dilution series proceeding vertically through a stack of plates with the high-dose plate on top and the low-dose plate on the bottom (final plate well concentration ranging from 60 μM to 0.117 μM and a final DMSO concentration of 0.6%).
Plate-to-plate variability is measured by normalizing the compound (cmpd) data using in-plate controls. Cell-only values are set to equal 100% inhibition of cytopathic effect (CPE), and virus values equal 0% inhibition. For compounds, the percent inhibition is calculated as follows: 100 * (Cmpd – Median Virus Ctrl)/(Median Cell Ctrl – Med Virus Ctrl). We control for positional variation during assay development and validation where methods are developed to minimize or eliminate positional artifacts such as edge effects.
Cell culture assays
Three methods were used to assess inhibition of virus-induced CPE in vitro in Tables 1 and 2: visual (microscopic) examination of the cells, increase in neutral red (NR) dye uptake into cells, and the Cell Titer Glo assay reagent (CTG; Promega) that generates a luminescent signal directly proportional to the amount of adenosine triphosphate (ATP) present, which is proportional to the number of metabolically active cells. The visual microscopic and NR CPE inhibition methods have been previously reported.27,28 Briefly, 7 concentrations of test drug were evaluated against each virus in 96-well flat-bottomed microplates. The compounds were added 5 to 10 min prior to virus, which was used at a concentration of approximately 50 cell culture 50% infection doses per well. This virus challenge dose equated to an MOI of approximately 0.001 infectious particles per cell. The tests were read after incubation at 37 °C for 72 h. In the NR uptake assay, dye (0.034% concentration in medium) was added to the same set of plates used to obtain the visual scores. After 2 h, the color intensity of the dye absorbed by and subsequently eluted from the cells was determined using a computerized EL-309 microplate autoreader (Bio-Tek Instruments, Winooski, VT). Antiviral activity was expressed as the 50% effective (virus-inhibitory) concentration (EC50) determined by plotting compound concentration versus percent inhibition.28 The CTG methodology was described earlier.
Table 1.
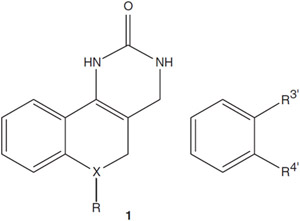
Antiviral Activity of Benzoquinazolinone Analogs (See Structure 1)
|
||||||||
|---|---|---|---|---|---|---|---|---|
| SR# | X | R | R3′ | R4′ | H1N1,aμM | H1N1 SIb | H5N1,c μM | H5N1 SIb |
| 22405 | C | H, H | Br | OCH3 | 0.425d | >118 | 0.005 | >10,000 |
| 22409 | C | H, H | Br | F | 1.4 | >36 | 0.020 | >2500 |
| 22412 | S | — | Br | OCH3 | 0.351 | >142 | 0.017 | >2941 |
| 22516 | C | H, H | CH3 | OCH3 | 10.0 | >5 | 0.051 | >980 |
| 22521 | O | — | Br | OCH3 | 4.0 | >13 | 0.006 | >8333 |
| 22700 | C | H, CH3 | Br | OH | 0.098 | >510 | 0.046 | >1087 |
| 22709 | C | H, H | Br | OH | 0.118 | >424 | 0.012 | >4167 |
Influenza A/California/04/2009 (H1N1).
Selectivity index (50% cytotoxic concentration [based on toxicity of >50 for all compounds] divided by EC50 value).
Influenza A/Vietnam/1203/2004 (H5N1).
Fifty percent virus-inhibitory concentration (EC50).
Table 2.
Antiviral Activities of SR 22700 against Influenza A and B Virus Infections in Madin Darby Canine Kidney Cells
| Virus Strain | SR 22700 | ||||
|---|---|---|---|---|---|
| HN Type | Visual EC50, μM | Selectivity Index (Vis.)a | Neutral Red EC50 μM | Selectivity Index (Neutral Red)a |
|
| A/New Caledonia/20/99 | H1N1 | 0.056 | >1800 | 0.056 | >1800 |
| A/NWS/33 | H1N1 | <0.032 | >3125 | <0.032 | >3125 |
| A/Solomon Islands/03/2006 | H1N1 | <0.032 | >3125 | <0.032 | >3125 |
| A/California/04/09 | H1N1 | 0.098 b | >487 b | ||
| Beijing/32/92 | H3N2 | >100 | 0 | >100 | 0 |
| California/7/04 | H3N2 | >100 | 0 | >100 | 0 |
| Panama/2007/99 | H3N2 | >100 | 0 | >100 | 0 |
| Sydney/05/97 | H3N2 | >100 | 0 | >100 | 0 |
| A/Victoria/3/75 | H3N2 | >100 | 0 | >100 | 0 |
| A/Wisconsin/67/2005 | H3N2 | >100 | 0 | >100 | 0 |
| A/Vietnam/1203/2004 | H5N1 | 0.046 b | >1043 | ||
| A/Hong Kong/156/97 | H5N1 | 0.009 b | >5333 | ||
| A/Duck/MN/1525/81 | H5N1 | <0.032 | >3125 | <0.032 | >3125 |
| A/Gull/PA/4175/83 | H5N1 | <0.032 | >3125 | <0.032 | >3125 |
| A/Vietnam/1203/2004 x A/Ann | H5N1 | <0.032 | >3125 | <0.032 | >3125 |
| Arbor/6/60 | |||||
| B/Sichuan/379/99 | — | >100 | 0 | >100 | 0 |
| B/Malaysia/2506/2004 | — | >100 | 0 | >100 | 0 |
The bold entry A/California/04/09 is the pandemic strain of influenza A (H1N1).
The bold H5N1 strains A/Vietnam/1203/2004 and A/Hong Kong/156/97 are avian strains of influenza.
However, both the bold H1N1 and H5N1 strains have infected humans.
Based on no toxicity seen either visually or by neutral red uptake at 100 μM.
Cell Titer Glo (CTG; Promega, Madison, WI).
Cytotoxicity of compounds was assessed in parallel with the antiviral determinations in the same microplates, except in the absence of virus. From these results, 50% cytotoxic end points (50% cell-inhibitory concentrations [IC50s]) were determined by NR assay as described above. In Table 1, EC50 values were determined by CTG assays, and the cytotoxicity cutoff was 50 μM. For Table 2, all 3 methods were used to access assay inhibition of virus-induced CPE, but the cytotoxicity cutoff for the CTG assay was 50 μM versus 100 μM for both visual and neutral red methods.
Neuraminidase inhibition assay
The assay uses amplified live virus, which has been diluted to the appropriate concentration in reaction buffer (100 mM sodium acetate, pH 6.5, 10 mM CaCl2). This diluted material is added directly to the drugged plates. Each reaction contained 5 μL of compound and 10 μL of the diluted virus stock in 1× reaction buffer. Virus and drug or compound was incubated at ambient temperature for 30 min. Substrate (15 μL) of 0.2 mM 2′-(4-methylumbelliferyl)-α-D-N-acetylneuraminic acid was added to each well. Reactions were incubated at ambient temperature for an additional 30 min and stopped with an equal volume addition of 200 mM sodium carbonate, pH 9.5. Fluorescence was measured in an EnVision plate reader using fluorescence excitation at 355 nm and emission at 460 nm.29 Oseltamivir carboxylate was provided by Jim Riordan from Southern Research (Birmingham, AL).
Hemagglutinin inhibition assay
Briefly, hemagglutinin (HA) inhibition was measured by serially diluting antiviral compounds in round-bottom 96-well plates. Forty HA units per ml or 4 HA units/well were added to the plates followed by the addition of 50 μL of turkey (0.5%) or horse (1.0%) erythrocyte suspensions. Inhibition of HA activity was demonstrated by absence of formation of an erythrocyte pellet. This was recorded using a “+” symbol. When a portion of the red blood cells (RBCs) was partially agglutinated or partially settled, a “±” symbol was used. In the absence of hemagglutination, chicken or turkey RBCs form a compact button on the bottom of the wells. A “−” symbol was used to record the absence of hemagglutination. The highest dilution of virus that causes complete hemagglutination was defined as the HA titration end point. The HA titer is the reciprocal of the dilution of virus in the last well with complete hemagglutination.
Real-time qRT-PCR analysis
MDCK cells (8 × 104 cells) were plated in 0.5 ml DMEM-ATCC, 10% FBS per well in 24-well microplates and incubated for 12 h at 37 °C, 5% CO2. The medium was removed and the cells washed 2 times with phosphate-buffered saline (PBS). The confluent monolayers were infected with influenza A/Brisbane/59/2007 (H1N1) virus on ice (4 °C) at an MOI of 0.1. After adsorption for 60 min, the monolayers were washed 2 times with PBS and incubated with DMEM-ATCC, 1% BSA, and 2.5 μg/mL TPCK trypsin. SR 22700 (50 μM) or ribavirin (82 μM) was added at 1 h or 6 h postinfection (PI). After each incubation period, the monolayers were washed 2 times with PBS and incubated with fresh medium until 9 h PI. Supernatants from 2 identically treated wells were pooled and the samples frozen at −80 °C. Total RNA was extracted using TRIZOL® LS Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Briefly, 400 μL of each sample supernatant was used for the extraction of viral genomic RNA. The RNAs were resuspended in DEPC-treated water and used in the following experiments immediately or stored at −80 °C. Generation of first-strand cDNA was accomplished using the SuperScript® VILO™ cDNA Synthesis kit (Invitrogen) according to the manufacturer’s instructions.
Conserved regions in the matrix (MA) gene from several strains of influenza A H1N1 and H5N1 viruses served as the target for the primers and TaqMan probe design. Concentrations of primers and probe were optimized to improve the sensitivity and specificity of the reactions. A plasmid containing the cDNA of influenza virus A/WSN/33 (H1N1), pHW187-M, MA gene was constructed and in vitro transcribed for a quantitative assay of copy numbers of the target gene. The linear range for detection was determined as 101 to 108 molecules in reaction.
Real-time quantitative RT-PCR (qRT-PCR) was carried out in a 20-μL mixture containing 2.5 μL RNA, 10 μL 2× TaqMan 1-step RT-PCR master mix (Applied Biosystems, Carlsbad, California), 1.0 μM MA forward primer (5′-CTT CTA ACC GAG GTC GAA ACG TA-3′), 1.0 μM MA reverse primer (5′-GGA TTG GTC TTG TCT TTA GCC A-3′), and 1.6 μM TaqMan probe (5′ FAM-CTC GGC TTT GAG GGG GCC TGA-MGB 3′) in an MJ Opticon 2 DNA Engine System (Bio-Rad, Hercules, CA). Cycling parameters were carried out for 1.5 min at 95 °C with a subsequent 38 cycles of amplification (95 °C for 8 s; 60 °C for 20 s; fluorescence was recorded at 60 °C). The samples were run in triplicate.
Time of addition compound screen
MDCK cells were plated in 96-well black tissue culture plates at 15,000 cells per well in 100 μL and incubated 24 h at 37 °C, 5% CO2. Eead candidate compounds were diluted in media to give a final concentration of 52 μM and added to plates at −1, 0, 1, 4, 6, 9, and 24 h postinfection. Cells were infected with influenza A/Vietnam/1203/2004 at an MOI of 0.1 and were incubated at 37 °C, 5% CO2. After 72 h, 20 μL (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt) (MTS) was added, and plates were incubated an additional 2 h at 37 °C, 5% CO2. Plates were read at an absorbance of 490 nm using EnVision. Ribavirin was used as a control compound at a final concentration of 82 μM.
Data analysis
HTS data were analyzed using ActivityBase software (IDBS, Inc., Guildford, UK). Percent CPE inhibition = 100 * [1 – (luminescence cmpd well – median luminescence virus ctrl)/(median luminescence cell ctrl – median luminescence virus ctrl)]. Percent viability = 100 * luminescence cmpd well/median luminescence cell ctrl. An active compound, or “hit,” was defined as a compound that exhibited a %CPE inhibition of >25% without compromising cell viability. Two dose-response curves were calculated for each substance. One assessed % CPE inhibition at each dose (EC50); the other assessed cytotoxicity at each dose (IC50). EC50 values (for % CPE inhibition) and IC50 values were calculated using the 4-parameter Levenburg-Marquardt algorithm with parameter A locked at 0 and parameter B locked at 100. Standard deviation, normalized χ2, and Hill slope were used to evaluate the curves. EC90/IC90 were also calculated to estimate the values at which 90% CPE inhibition and 10% cell viability would be achieved, respectively. Values were not extrapolated beyond the tested range of concentrations. The selective index (SI) was calculated as SI = IC50/EC50. The criteria for determining compound activity are based on its SI. Compounds with an SI value of >3 were defined as active, whereas compounds that exhibited an SI value less than 3 were defined as inactive.
Thirty-two control wells containing cells only and 24 wells containing cells and virus were included on each assay plate and used to calculate Z values for each plate and to normalize the data on a per plate basis. Eight ribavirin positive control wells were included on each plate for quality control purposes but were not used in z calculations.
The Z factor values were calculated from 1 minus [3 * standard deviation of cell ctrl (σc) plus 3* standard deviation of the virus ctrl (σv)]/[mean cell ctrl signal (μc) minus mean virus ctrl signal (μv]).30 The signal/background (S/B) was calculated from mean cell ctrl signal (μc) divided by the mean virus ctrl signal (μv). The signal/noise (S/N) was calculated from mean cell ctrl signal (μc) minus mean virus ctrl signal (μv) divided by the [standard deviation of the cell ctrl signal (σc)2 minus the standard deviation of the virus ctrl signal (σv)]1/2.
RESULTS AND DISCUSSION
Compound screening results
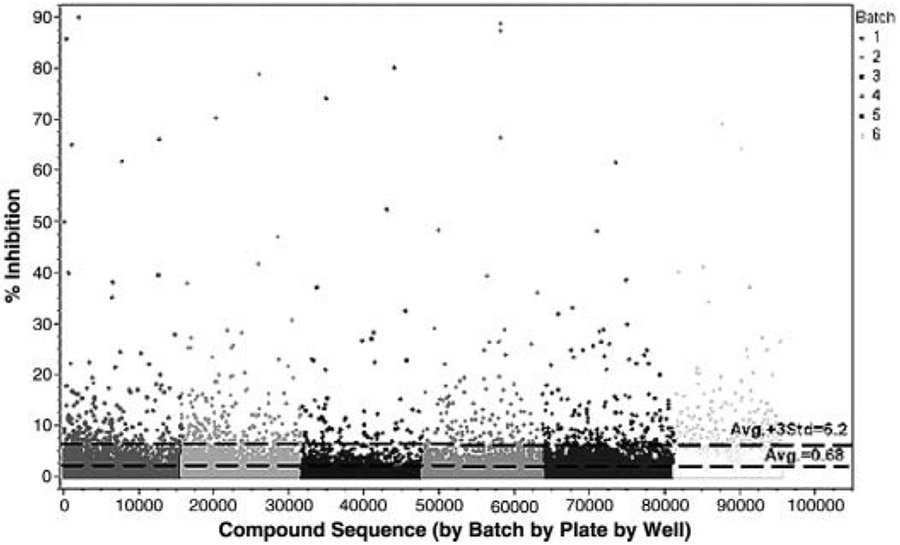
A total of 95,512 compounds were analyzed in the primary screen at a single-dose concentration of 50 μM. The overall Z score for the screening campaign was 0.86, reflecting a highly robust assay. Statistically, the average inhibition of the compound wells was 0.68% with a standard deviation of 1.84%. Inhibition values of greater than 6.2% were outside the calculated noise of the assay defined by using the average plus 3 times the standard deviation of compound well inhibition (Fig. 1).
FIG. 1.
High-throughput screen of 95,512 compounds from the NIH Molecular Libraries Small Molecule Repository (MLSMR) library. Compounds from the library, along with the influenza A Vietnam/1203/04 virus (H5N1), were added to 6 × 103 Madin Darby canine kidney (MDCK) cells per well in 384-well plates. Inhibitory effects were assessed after 72 h, as described in Materials and Methods. Reference lines were calculated from the percent (%) inhibition. Control drug used was ribavirin.
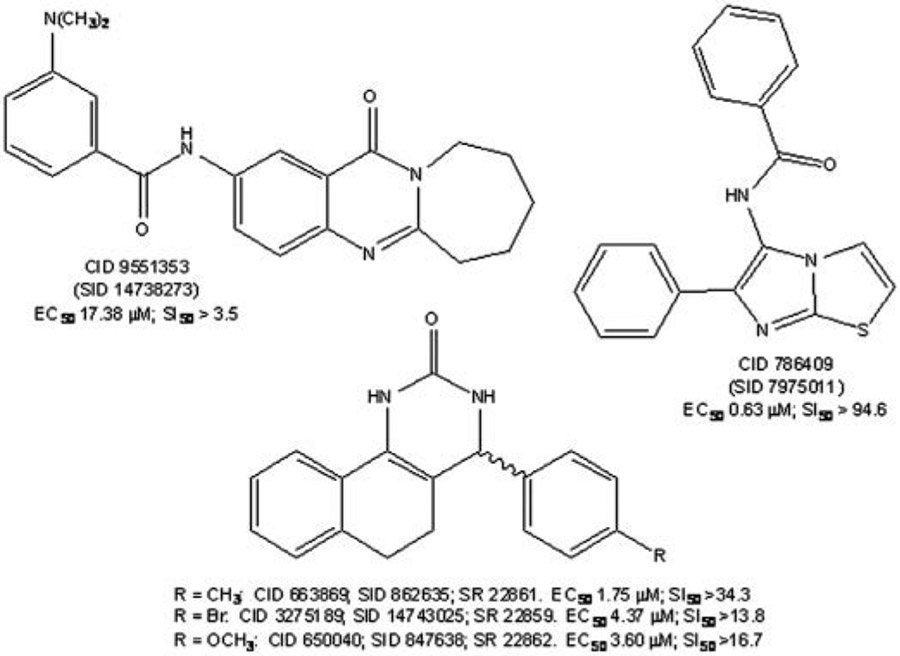
Previously, using our cell-based HTS against influenza A virus (H3N2), we determined a higher hit rate when compounds were screened at higher concentration, and more compounds were confirmed if we chose a higher percentage cutoff for dose response and cytotoxicity assays.31 Therefore, 64 compounds that showed a protective effect of >25% CPE inhibition were identified as hits. Because of the low number of hits identified in the primary single-dose screen, extensive structure-function activity analysis was performed and additional analogs to these 64 compounds were identified within the compound library for rescreening. A total of 305 compounds were ordered for secondary screening. These compounds were evaluated in more detail by measuring their dose response, cell toxicity, and selectivity. Dose-response curves were performed over a 512-fold concentration range (from 60 μM to 0.117 μM) for cytotoxicity and antiviral activity. Five of the compounds screened in dose response were confirmed as active. The EC50 (CPE inhibition) values ranged from 0.634 μM to greater than 100 μM. The IC50 (cytotoxicity) values ranged from 3.6 μM to greater than 100 μM. The selectivity index (SI = IC50/EC50) values ranged from 3.45 to 94.64, and compounds with SI values greater than 3 were considered active. Their PubChem Chemical Identifier (CID) and Substance Identifier (SID) and Southern Research identifier (SR#) are CID (SID) SR numbers, SI values: 786409 (7975011), >94.6; 663869 (862635), SR 22861, >34.3; 650040 (847638), SR 22862, >16.7; 3275189 (14743025), SR 22859 >13.8; and 9551353 (14738273), >3.5. The inhibitory activities of selected compounds are shown in Figure 2.
FIG. 2.
Structures, PubChem identifiers, and inhibitory activities of Molecular Libraries Screening Centers Network (MLSCN) assay hits against influenza A/VN/1203/2004 (H5N1). The structures, EC, and SI values are shown for PubChem Chemical Identifier (CID) and Substance Identifier (SID) and Southern Research identifier (SR#). These compounds are CID (SID) numbers, 786409 (7975011); 663869 (862635), SR 22861; 650040 (847638); 3275189 (14743025), SR 22859; and 9551353 (14738273). In parallel experiments, median inhibitory concentrations (IC50 compound concentrations that reduce cell viability by 50) and selective indices (SI = IC/EC) at 72 h postinfection were also determined and are indicated.
Dose-response data for the analogs of 2 active scaffolds, benzoquinazolinone and thiazoloimidazole structures, revealed initial structure-activity relationships (SARs). A medicinal chemistry analysis was performed on the hit compounds to identify and choose the most promising compounds for lead optimization. The declared probe reported to the MLSCN (PubChem) was used to synthesize a series of compounds from the benzoquinazolinones for secondary testing using the in vitro CPE-based HT screening assay against influenza A/Vietnam/1203/2004 (H5N1). Of these compounds, 7 met our criteria of activity: the efficacy, EC50 value of <1 μM and with toxicity to efficacy, SI of >10 (Table 1).
Although not fully developed at this point, SAR analysis of the benzoquinazolinone series suggests that the aryl substituent occupies a hydrophobic site that does not readily accommodate more polar moieties; introduction of more hydrophilic heterocyclic substituents at this position reduces or eliminates activity (data not shown). A 3,4-disubstitution pattern in the aryl substituent was quickly determined to be optimal. It is also noted that aromatization of the pyrimidinone ring, a common side reaction during analog synthesis that eliminates the stereocenter at the aryl substitution site, abolishes activity.
In contrast to aryl modifications, there is significant potential to modify the base structure in the central fused ring: replacement of an aliphatic carbon in the B-ring with either oxygen or sulfur or introduction of a substituent at this position leads to analogs retaining good activity. These latter derivatives also possess significantly improved solubility compared with the parent compounds. Especially in the oxa congener SR 22521, it is clear that the details of the SAR pattern differ between H1N1 and H5N1 and deserve further investigation.
To establish the selectivity of this chemotype, we tested SR 22409, SR 22412, SR 22521, and SR 22700 against a panel of influenza A and B viruses in cell culture (Table 2). Results showed all compounds were highly active against 4 strains of H1N1 and 4 strains of H5N1 viruses but not active against 6 strains of H3N2 and 2 strains of B viruses. Interestingly, SR 22700 was highly active against influenza A/California/04/09 virus (H1N1). For Table 2, 3 methods were used to assess inhibition of virus-induced cytopathic effect, but the cytoxicity cutoff for the Cell Titer Glo assay was 50 μM versus 100 μM for both visual and neutral red methods.
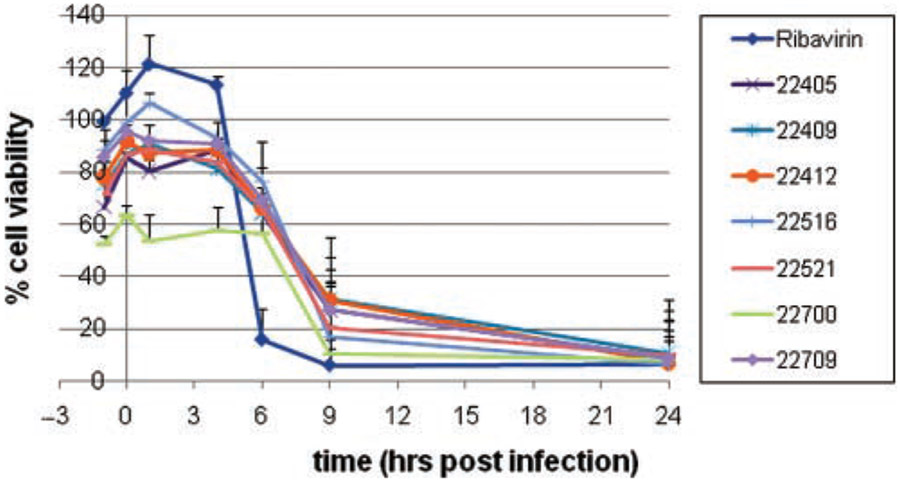
Our initial approach to probe the mechanism of action of this intriguing new class of molecules, the benzoquinazolinones, was to refine the window of inhibitory activity and complement those studies with biochemical assays for various enzyme activities. We employed a time of addition assay to ascertain if the inhibition activity of the compound was early (entry) or late (replication) in the virus life cycle. In this screen, compounds were added in triplicate to plates at time points −1, 0, 1, 4, 6, 9, and 24 h PI. Seventy-two hours later, CPE was assessed using MTS as an endpoint reagent. All 7 compounds exerted antiviral activity when they were added 1 h before infection or 1 to 6 h after infection (Fig. 3). The results suggest that these compounds may impede an early to middle stage of influenza replication, following entry but prior to release.
FIG. 3.
Time of addition compound screen against influenza A/Vietnam/1203/04 virus (H5N1). Madin Darby canine kidney (MDCK) cells were plated in 96-well black tissue plates at 15,000 cells per well and incubated 24 h at 37°C, 5% CO2. Test compounds were diluted in media to give a final concentration of 52 μM (0.5% DMSO final concentration) and added to plates at time points −1, 0, 1, 4, 6, 9, and 24 h postinfection. Cells were infected with influenza A/Vietnam/1203/04 at an multiplicity of infection (MOI) of 0.1 at time point 0 and incubated 72 h at 37°C, 5% CO2. (3-(4,5-Dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt) (MTS) was added was added, and plates were incubated an additional 2 h at 37°C, 5% CO2. Plates were read at an absorbance of 490 nm on an EnVision plate reader. Ribavirin was used as a control compound.
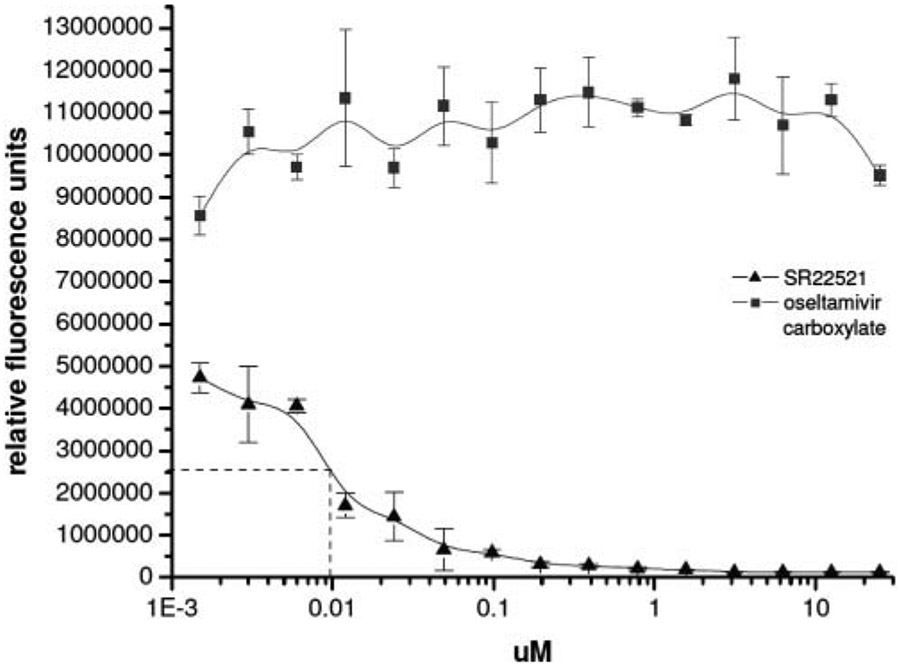
The data in Table 2 suggested neuraminidase (NA) as a possible target, specifically N1 but not N2 or the NA on B virus; hence, a neuraminidase assay was employed to test this hypothesis. Neuraminidase inhibition was measured using 2′-(4-methylumbelliferyl)-α-D-N-acetylneuraminic acid, which liberates a quantifiable fluorescent tag in response to neuraminidase activity in the presence of influenza A viruses.29 Methylumbelliferone signal can be detected with a sensitivity as low as 106 virus particles/mL (104 particles total) with a broad linear range. Because active NA is located on the surface of the virus, no purification of the protein was necessary to measure enzymatic activity. Although the control drug oseltamivir carboxylate inhibited the virus NA with an EC50 of approximately 8 nM, SR 22521 did not inhibit neuraminidase (Fig. 4).
FIG. 4.
Neuraminidase (NA) assay and EC50 curve of oseltamivir carboxylate. NA inhibition was measured using 2′-(4-methyl umbelliferyl)-α-D-N-acetylneuraminic acid (substrate), which liberates a quantifiable fluorescent tag in response to neuraminidase activity. Drug or SR 22521 was incubated with influenza A/Vietnam/1203/2004 virus (H5N1) in reaction buffer followed by the addition of substrate 30 min later. Fluorescence was measured in an EnVision plate reader using excitation at 355 nm and fluorescence detection at 460 nm. Control drug used was oseltamivir carboxylate.
Influenza infection begins when viral HA adheres to sialic acid receptors on the host cell surface. The selectivity data indicate that the compounds inhibit infectivity of H5 and H1 subtypes. Therefore, an HA inhibition assay was performed as described in Materials and Methods to examine whether SR 22700 was inhibiting the fusogenic function of the viral hemagglutinin. No hemagglutination inhibitor activity was detected in the HA inhibitor assay with SR 22700 (data not shown).
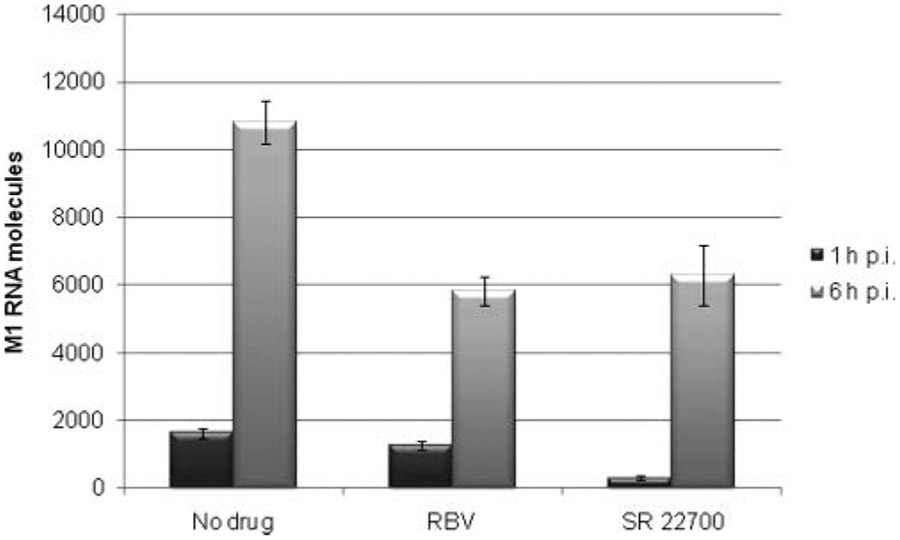
To further probe the mechanism of action of this novel compound scaffold, we established a real-time qRT-PCR method to measure the amount of viral RNA in a time of addition experiment. The assay provides direct and reliable measurements that can also suggest a mechanism of action. In this experiment, SR 22700 or ribavirin was added 1 and 6 h after virus infection and monitored by qRT-PCR as described in Materials and Methods. These time points were chosen since, typically, a window of 8 to 10 h is required to detect influenza A/Brisbane/59/2007 progeny virus. The 1-h time point showed a 5.5-fold reduction in M1 RNA levels compared to the no-drug influenza-infected MDCK cells (Fig. 5). In contrast, the 1-h time point for ribavirin showed only a 1.3-fold decrease in M1 RNA levels. In the case of the infected cells treated 6 h postinfection, there were 1.9- and 1.7-fold reductions in M1 RNA levels for ribavirin and SR 22700, respectively. This suggests that SR 22700 is targeting an early event in the viral life cycle in agreement with the time of addition assay based on CPE (Fig. 3).
FIG. 5.
Real-time quantitative RT-PCR (qRT-PCR) screen of influenza A/Brisbane/59/2007 virus (H1N1) in the presence of SR 22700 and ribavirin. Addition of compound to cells was either 1 or 6 h after virus exposure. Cell supernatant fluid was collected at 9 h for subsequent qRT-PCR experiments. The signal dynamic range was over 5 logs of virus. The linear range for detection was determined as 101 to 108 molecules in reaction. The samples were run in triplicate. Control drug was ribavirin (RBV).
In summary, primary and secondary screening has led to the discovery of 5 compounds with an SI value of greater than 3. Activity data from the confirmatory and secondary assays were analyzed in-depth to identify key scaffolds of interest, evaluate SAR, and contribute to probe optimization efforts. Subsequently, we synthesized several compounds for additional testing. Time of addition assays suggest that these compounds are most efficacious from 1 h before infection to 6 h after infection, similar to the control drug ribavirin. In addition, dose-response and toxicity assays indicate that 7 of these compounds met our activity criteria for identification of lead compounds: an efficacy EC50 value of <1 μM and toxicity to efficacy SI of >10. Furthermore, these compounds were efficacious against both H5N1 and H1N1 viruses in vitro but not against H3N2 viruses. Finally, qRT-PCR results showed the M1 RNA levels were significantly reduced for treatments starting 1 h postinfection in the presence of the benzoquinazolinones, suggesting a novel mechanism of action. Since viral resistance to H5N1 is a growing problem for the neuraminidase inhibitors32 and most clade 1 H5N1 viruses are resistant to the M2 inhibitors,33 the discovery of this chemotype that exhibits a novel mechanism of action makes them excellent candidates for further evaluation. Lead compounds will be examined for their ability to select for viral resistance strains in cell culture assays, and those that show the least virus resistance will be taken into animal models.
ACKNOWLEDGMENTS
We appreciate the technical assistance of Sara McKellip, Lakshmi Reddy, Lynn Rasmussen, and Anna Manouvakhova. The screening was performed with the support of NIMH grant R03-MH081270-01 to W.S. The compounds were made available for screening through MLSCN U54 HG003917 awarded to Gary Piazza (principal investigator).
REFERENCES
- 1.Ford SM, Grabenstein JD: Pandemics, avian influenza A (H5N1), and a strategy for pharmacists. Pharmacotherapy 2006;26:312–322. [DOI] [PubMed] [Google Scholar]
- 2.van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, et al. : H5N1 virus attachment to lower respiratory tract. Science 2006;312:399–400. [DOI] [PubMed] [Google Scholar]
- 3.Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, et al. : Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 1998;279:393–396. [DOI] [PubMed] [Google Scholar]
- 4.Melidou A: Avian influenza A (H5N1): current situation. Euro Surveill 2009;14(18):19199. [DOI] [PubMed] [Google Scholar]
- 5.Webster RG, Govorkova EA: H5N1 influenza: continuing evolution and spread. N Engl J Med 2006;355:2174–2177. [DOI] [PubMed] [Google Scholar]
- 6.Brady RC, Treanor JJ, Atmar RL, Keitel WA, Edelman R, Chen WH, et al. : Safety and immunogenicity of a subvirion inactivated influenza A/H5N1 vaccine with or without aluminum hydroxide among healthy elderly adults. Vaccine 2009;27:5091–5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noah DL, Hill H, Hines D, White EL, Wolff MC: Qualification of the hemagglutination inhibition assay in support of pandemic influenza vaccine licensure. Clin Vaccine Immunol 2009;16:558–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicholson KG, Colegate AE, Podda A, Stephenson I, Wood J, Ypma E, et al. : Safety and antigenicity of non-adjuvanted and MF59-adjuvanted influenza A/Duck/Singapore/97 (H5N3) vaccine: a randomised trial of two potential vaccines against H5N1 influenza. Lancet 2001;357:1937–1943. [DOI] [PubMed] [Google Scholar]
- 9.Fodor E, Devenish L, Engelhardt OG, Palese P, Brownlee GG, Garcia-Sastre A: Rescue of influenza A virus from recombinant DNA. J Virol 1999;73:9679–9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann E, Krauss S, Perez D, Webby R, Webster RG: Eight-plasmid system for rapid generation of influenza virus vaccines. Vaccine 2002;20:3165–3170. [DOI] [PubMed] [Google Scholar]
- 11.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, et al. : Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci U S A 1999;96:9345–9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bresson JL, Perronne C, Launay O, Gerdil C, Saville M, Wood J, et al. : Safety and immunogenicity of an inactivated split-virion influenza A/Vietnam/1194/2004 (H5N1) vaccine: phase I randomised trial. Lancet 2006;367:1657–1664. [DOI] [PubMed] [Google Scholar]
- 13.Leroux-Roels I, Borkowski A, Vanwolleghem T, Drame M, Clement F, Hons E, et al. : Antigen sparing and cross-reactive immunity with an adjuvanted rH5N1 prototype pandemic influenza vaccine: a randomised controlled trial. Lancet 2007;370:580–589. [DOI] [PubMed] [Google Scholar]
- 14.Treanor JJ, Campbell JD, Zangwill KM, Rowe T, Wolff M: Safety and immunogenicity of an inactivated subvirion influenza A (H5N1) vaccine. N Engl J Med 2006;354:1343–1351. [DOI] [PubMed] [Google Scholar]
- 15.Vajo Z, Wood J, Kosa L, Szilvasy I, Paragh G, Pauliny Z, et al. : A single-dose influenza A (H5N1) vaccine safe and immunogenic in adult and elderly patients: an approach to pandemic vaccine development. J Virol 2010;84:1237–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lew W, Chen X, Kim CU: Discovery and development of GS 4104 (oseltamivir): an orally active influenza neuraminidase inhibitor. Curr Med Chem 2000;7:663–672. [DOI] [PubMed] [Google Scholar]
- 17.von Itzstein M, Wu WY, Kok GB, Pegg MS, Dyason JC, Jin B, et al. : Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature 1993;363:418–423. [DOI] [PubMed] [Google Scholar]
- 18.Davies WL, Grunert RR, Haff RF, McGahen JW, Neumayer EM, Paulshock M, et al. : Antiviral activity of 1-adamantanamine (amantadine). Science 1964;144:862–863. [DOI] [PubMed] [Google Scholar]
- 19.Privatdegarilhe M, De Rudder J: Effect of 2 arabinose nucleosides on the multiplication of herpes virus and vaccine in cell culture [in French]. C R Hebd Seances Acad Sci 1964;259:2725–2728. [PubMed] [Google Scholar]
- 20.Palese P, Shaw ML: Orthomyxoviridae: the viruses and their replication. In Knipe DM (ed): Fields Virology. Philadelphia: Wolters Kluwer/Lippincott Williams & Wilkins; 2007:1647–1690. [Google Scholar]
- 21.Oseltamivir-resistant 2009 pandemic influenza A (H1N1) virus infection in two summer campers receiving prophylaxis—North Carolina, 2009. MMWR Morb Mortal Wkly Rep 2009;58:969–972. [PubMed] [Google Scholar]
- 22.Hurt AC, Ernest J, Deng YM, Iannello P, Besselaar TG, Birch C, et al. : Emergence and spread of oseltamivir-resistant A(H1N1) influenza viruses in Oceania, South East Asia and South Africa. Antiviral Res 2009;83:90–93. [DOI] [PubMed] [Google Scholar]
- 23.Wang SY, Su CY, Lin M, Huang SY, Huang WI, Wang CC, et al. : HA-pseudotyped retroviral vectors for influenza antagonist screening. J Biomol Screen 2009;14:294–302. [DOI] [PubMed] [Google Scholar]
- 24.Eichelberger MC, Hassantoufighi A, Wu M, Li M: Neuraminidase activity provides a practical read-out for a high throughput influenza antiviral screening assay. Virol J 2008;5:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noah JW, Severson W, Noah DL, Rasmussen L, White EL, Jonsson CB: A cell-based luminescence assay is effective for high-throughput screening of potential influenza antivirals. Antiviral Res 2007;73:50–59. [DOI] [PubMed] [Google Scholar]
- 26.Eriksson B, Helgstrand E, Johansson NG, Larsson A, Misiorny A, Noren JO, et al. : Inhibition of influenza virus ribonucleic acid polymerase by ribavirin triphosphate. Antimicrob Agents Chemother 1977;11:946–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sidwell RW, Huffman JH: Use of disposable micro tissue culture plates for antiviral and interferon induction studies. Appl Microbiol 1971;22:797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smee DF, Huffman JH, Morrison AC, Barnard DL, Sidwell RW: Cyclopentane neuraminidase inhibitors with potent in vitro anti-influenza virus activities. Antimicrob Agents Chemother 2001;45:743–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blick TJ, Tiong T, Sahasrabudhe A, Varghese JN, Colman PM, Hart GJ, et al. : Generation and characterization of an influenza virus neuraminidase variant with decreased sensitivity to the neuraminidase-specific inhibitor 4-guanidino-Neu5Ac2en. Virology 1995;214:475–484. [DOI] [PubMed] [Google Scholar]
- 30.Zhang JH, Chung TD, Oldenburg KR: A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 1999;4:67–73. [DOI] [PubMed] [Google Scholar]
- 31.Severson WE, McDowell M, Ananthan S, Chung DH, Rasmussen L, Sosa MI, et al. : High-throughput screening of a 100,000-compound library for inhibitors of influenza A virus (H3N2). J Biomol Screen 2008;13:879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta RK, Nguyen-Van-Tam JS: Oseltamivir resistance in influenza A (H5N1) infection. N Engl J Med 2006;354:1423–1424; author reply 1423-1424. [DOI] [PubMed] [Google Scholar]
- 33.Bright RA, Shay DK, Shu B, Cox NJ, Klimov AI. Adamantane resistance among influenza A viruses isolated early during the 2005-2006 influenza season in the United States. JAMA 2006;295:891–894. [DOI] [PubMed] [Google Scholar]