Abstract
Traveler's diarrhea (TD) is the most common infectious illness acquired by visitors to developing nations. The purpose of this study was to utilize molecular diagnostic techniques to determine the prevalence of norovirus (NoV) in TD occurring among visitors from the United States to Guatemala and Mexico. Stool samples (n = 54) were collected from 34 TD cases and analyzed for NoV by reverse transcription-PCR and oligoprobe confirmation. The overall prevalence of NoV was 65%. Interestingly, all NoV-positive stool samples were identified as genogroup I NoVs, and time spent at travel destinations was found to be an important factor in determining the frequency of infection (P = 0.003). Eleven NoV-positive stool samples also tested positive for enterotoxigenic Escherichia coli, indicating that dual infections with this leading bacterial cause of TD were very common. Results of this study suggest that NoV infection is a frequent occurrence among travelers to Mexico and Guatemala who experience episodes of TD. In addition, the simple molecular detection method utilized here will serve to facilitate more in-depth epidemiological studies of this emergent viral pathogen in travelers and other at-risk populations.
Each year, 50 million people travel between industrialized and developing countries (28). These travelers may be at risk for a variety of illnesses. However, diarrhea, associated with exposures to bacterial, protozoal, and viral enteropathogens, is the most common infectious illness, affecting 20 to 50% of people traveling to the tropical and subtropical areas of Latin America, the Caribbean, southern Asia, and Africa (23). Traveler's diarrhea (TD) is generally defined as the passage of three or more watery or loose stools during a 24-h period, accompanied by at least one additional symptom (i.e., nausea, vomiting, abdominal pain or cramps, urgency, or loss of appetite) (8). Generally, recovery occurs within 5 days; nevertheless, immunocompromised and elderly individuals may experience longer recovery times and more severe symptoms (6). Comprehensive bacterial and protozoal diagnostic methods have been utilized to identify agents associated with TD, including enterotoxigenic Escherichia coli (ETEC), one of the leading causes of TD among travelers from the United States (11, 27). Yet, in some studies as many as 40 to 50% of TD cases remain undiagnosed (4).
Noroviruses (NoVs) are recognized as one of the leading causes of nonbacterial gastroenteritis outbreaks in countries such as The Netherlands, the United Kingdom, and the United States (7, 9, 12). NoVs are estimated to cause over 23 million cases of illness each year in the United States (5, 15). The genus Norovirus is currently subdivided into five genogroups (GI through GV) on the basis of sequence homologies in the viral RNA, with GI and GII the most commonly described in human infections (2). Investigations of the extensive cruise ship gastroenteritis outbreaks that occurred between July and December 2002 exemplified the usefulness of molecular diagnostic techniques for determining the role of NoVs in outbreaks (5). However, since molecular detection methods for NoVs are not readily available in many laboratories in the United States or other countries, the role of NoVs in the etiology of TD remains unclear. In a previous study, the prevalence of NoV infection among travelers to Mexico was elucidated with antibodies to NoV in a solid-phase microtiter radioimmunoassay, suggesting that NoV was the etiologic agent in 15% of the TD cases studied (18). Yet, estimates of NoV prevalence based solely on serological data may be imprecise, since immunoassays may either detect persisting antibodies from a NoV infection that occurred prior to travel or fail to detect low levels of NoV-specific antibodies present during an acute infection (2). Current antigen detection enzyme-linked immunosorbent assays also are limited by inadequate cross-reactivity to the numerous NoV strains in circulation (2, 9). The purpose of this study was to utilize reverse transcription-PCR (RT-PCR) with NoV-specific primers and probes that are broadly reactive to numerous NoV strains as a sensitive molecular diagnostic technique to determine the prevalence of NoVs in TD occurring among United States visitors to Guatemala and Mexico.
MATERIALS AND METHODS
Study population.
The study population consisted of 34 travelers from the United States who experienced TD during 14- to 24-day visits to Antigua, Guatemala (n = 9), and Cuernavaca, Mexico (n = 25), from 1998 to 2002. These travelers represented a subset of a larger population of students who were visiting Guatemala or Mexico for Spanish language training and participating in ongoing field trials designed to evaluate interventions for the prevention of TD. A total of 1,445 subjects participated in these trials, and 410 (28.4%) experienced TD during their time in-country. Seventy one percent (291 cases) of the TD cases detected in these language students occurred during the April-to-September time frame (the rainy season in both Mexico and Guatemala); cases for this study were randomly selected to represent a 12% sample of the TD cases occurring during this time period. TD was characterized as the occurrence of three or more watery or loose stools in a 24-h period accompanied by one or more additional gastrointestinal symptoms, including abdominal pain or cramps, nausea, vomiting, urgency, gas, loss of appetite, or frank blood in stools.
All study participants were requested to submit stool samples upon arrival in Guatemala or Mexico (acceptable range, 0 to 4 days in-country), at 1 week into their stay (acceptable range, 5 to 10 days in-country) and at the time of departure (generally 14 to 21 days after arrival; acceptable range, 11 to 23 days). Additional stool samples were collected from participants during acute diarrheal episodes. All samples were examined for the presence of classic bacterial and protozoal enteropathogens, including E. coli, Salmonella, Shigella, Campylobacter, Vibrio, Aeromonas, Plesiomonas, Giardia, Entamoeba, Cryptosporidium, and Cyclospora species, by standard culture, antigen detection, and microscopic methods (16). Up to five E. coli colonies from each stool sample were screened for ETEC by monoclonal antibody-based antigen detection assays for heat-labile and heat-stable toxins (24, 25). Stool suspensions from the 34 TD cases studied for NoV infection in this report were also screened for rotavirus infection by a commercially available antigen detection assay (Rotoclone; Meridian Diagnostics, Cincinnati, Ohio). All stool samples (10% suspensions in phosphate-buffered saline [PBS]) were cryopreserved at −70°C for additional viral analyses.
In addition to providing stool samples, study participants completed weekly diary cards designed to assess their daily health status during their stay in-country. The presence or absence of a number of general or gastrointestinal symptoms was recorded, as well as the number, time, and characteristics of all stools passed. Subjects also were interviewed twice a week to further assess their overall health status.
Optimization of NoV molecular diagnostic techniques.
Molecular diagnostic techniques for NoVs were optimized with 20 different strains of NoV (10 GI and 10 GII) present in stool samples obtained from the Centers for Disease Control and Prevention (CDC). CDC stool samples were prepared as 10% suspensions in PBS, and diluted 10, 100, and 1,000 fold in sterilized, molecular-grade H2O (Nanopure Diamond water system; Barnstead, Dubuque, Iowa). A previously described method that employs heat to release viral RNA from the capsid protein was utilized (20). Briefly, 90-μl sample dilutions (10−2 and 10−3) were heated at 95°C for 5 min and chilled on ice for 2 min. If stool samples were inhibited at the 1,000-fold dilution, they were further processed by extracting NoV RNA from 50 μl of 10% stool by a standard guanidinium extraction, followed by phenol-chloroform purification and isopropanol precipitation (22). Viral RNA isolated by either heat release or extraction was immediately assayed for NoV in 50-μl reaction mixtures by single-enzyme, single-tube RT-PCR with an MJR Peltier Thermal Cycler (PTC-200; MJ Research, Inc., Cambridge, Mass.). The RT-PCR mixture yielded a final solution containing 1× EZ Buffer, 0.2 mM deoxynucleoside triphosphates, 0.6 μM upstream and downstream primers, 2.5 mM Mn(OAc)2, 10 U of Rnasin, 5 U of rTth (Perkin-Elmer, Foster City, Calif.), and 20 μl of each heat-released sample dilution or extracted RNA. The RT reaction mixture was incubated for 10 min at 50°C for downstream primer annealing and for 50 min at 60°C for RT. cDNA was subsequently amplified under the following conditions: initial denaturation for 2 min at 94°C; 40 cycles each consisting of template denaturation for 15 s at 92°C, primer annealing for 30 s at 50°C, and primer extension for 30 s at 60°C; and a final extension for 5 min at 60°C. Region B primers developed at the CDC were used to amplify NoV RNA with an expected PCR product size of 213 bp (Table 1) (1). All samples were processed in a dedicated sample preparation laboratory, and amplicons were analyzed in a dedicated post-PCR laboratory to reduce the risk of laboratory cross-contamination.
TABLE 1.
Primer and probe sequences for RT-PCR and Southern oligoprobe confirmation
| Name | Geno- group | Sense | Use | Sequence (5′ to 3′) |
|---|---|---|---|---|
| Mon 432 | GI | + | Primer | TGGACICGYGGICCYAAYCA |
| Mon 434 | GI | − | Primer | GAASCGCATCCARCGGAACAT |
| Mon 431 | GII | + | Primer | TGGACIAGRGGICCYAAYCA |
| Mon 433 | GII | − | Primer | GAAYCTCATCCAYCTGAACAT |
| NVp35 | NVIS | − | Primer | CTTGTTGGTTTGAGGCCATAT |
| NVp36 | NVIS | + | Primer | ATAAAAGTTGGCATGAACA |
| Mon 458 B/I | GI | + | Probe | ATGTATGTRCCAGGATGGCARGCC |
| Mon 456 B/I.1 | GI | + | Probe | ATGTATGTCCCAGGATGGCAGGCC |
| Mon 457 B/I.2 | GI | + | Probe | ATGTATGTGCCAGGATGGCAAGCC |
| Mon 453 B/I.3 | GI | + | Probe | ATCTACATHCCTGGTTGGCAGGCC |
| Mon 454 B/I.4 | GI | + | Probe | ATACAGGAGATAAAGACTGGTGGT |
| Mon 455 B/I.5 | GI | + | Probe | ATGTATGTGCCAGGCTGGCAGGCC |
| Mon 459 B/II | GII | + | Probe | ATGGATTTTTACGTGCCCAGGCAA |
| Mon 451 B/II.1&4 | GII | + | Probe | GACCCATCTGAAACAATGATWCCA |
| Mon 460 B/II.2 | GII | + | Probe | AGCAAGATCAGCAAGCTTGTGATA |
| Mon 461 B/II.3 | GII | + | Probe | GACCCCAGTGAAACCATGATACCA |
| Mon 449 B/II.5 | GII | + | Probe | AATGAAACAATGATACCTCACTCT |
| Mon 448 B/II.6&7 | GII | + | Probe | GAGAACCCATACGAGAGCATGGTC |
An NoV RNA internal standard (NVIS) control also was utilized in a separate 50-μl RT-PCR (to prevent any potential multiplex competition) with each sample dilution to detect sample inhibition (i.e., false negatives) (20). The NVIS RT-PCR mixture comprised the same components and reaction conditions as previously described, except the primer set NVp35/NVp36 (Table 1) (20) replaced region B primers.
All NoV PCR products were confirmed by Southern hybridization oligoprobes. Six probes were utilized for GI, and six probes were utilized for GII (Table 1). GI and GII probes were 5′-end labeled with digoxigenin by using terminal transferase in accordance with the manufacturer's protocols (Roche Diagnostics Corporation, Indianapolis, Ind.). The GI probes were combined into one GI hybridization reaction mixture, and the GII probes were combined into one GII hybridization reaction mixture. Southern hybridization and detection were performed as previously described (21).
Laboratory and epidemiological analysis of TD stool samples.
Fifty-four stool samples (10% PBS suspensions) from the 34 TD study participants were analyzed for NoVs utilizing the molecular techniques described above. All of the samples were coded; therefore, during the laboratory analysis the symptomatology of each individual and the status of each sample (i.e., true sample, positive control, and negative control) were not known by the laboratory personnel.
Among the 54 stool samples obtained from travelers, 15 were collected prior to the onset of TD, and the remaining 39 were collected during acute TD episodes (some participants provided more than one acute sample). Access to the 15 prediarrhea stool samples (all negative for NoVs) enabled the calculation of an overall NoV infection incidence rate for 15 TD cases (5 from Guatemala and 10 from Mexico). Analyses of the 39 acute diarrheal samples enabled the calculation of an overall NoV infection prevalence rate among all 34 study participants. In addition, incidence and prevalence rates were calculated for two categories of TD cases: (i) TD cases that met the definition of TD, in which no bacterial or parasitic pathogens were identified (n = 16 cases); and (ii) TD cases that were previously documented as ETEC infections (one of which was also rotavirus positive) (n = 18 cases).
RESULTS
Optimization of NoV molecular techniques utilizing CDC-positive controls.
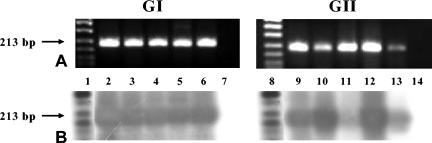
All 20 NoV-positive stool samples, previously characterized by the CDC to the genotype level, were positively confirmed to the genogroup level. Ten samples were confirmed positive for NoV GI, and 10 samples were confirmed positive for NoV GII by RT-PCR and Southern hybridization oligoprobing, demonstrating that the molecular diagnostic techniques and GI and GII region B primer sets and probes were working effectively to identify NoV-positive GI and GII stool samples (Fig. 1). All 20 CDC NoV-positive stool samples had RT-PCR amplification end points at or beyond 10−4 dilutions (data not shown). Therefore, the use of 10−2 and 10−3 sample dilutions in the heat-release viral extraction method did not fail to detect NoV-positive samples, due to dilution beyond the detection limit of the assay. Moreover, the use of the NVIS control with each sample dilution enabled the detection of potential sample inhibition that could have resulted in false negatives. Nine (45%) of 20 samples were inhibited at the 10−2 dilution, while 0 of 20 samples were inhibited at the 10−3 dilution. Thus, diluting the CDC 10% stool suspensions 1,000 fold was sufficient to remove potential PCR inhibitors; additional processing was not required for these samples.
FIG. 1.
RT-PCR (A) and Southern oligoprobe (B) results for a subset of NoV-positive stool samples obtained from the CDC. Each stool sample was analyzed separately with GI or GII primers and probes. Lanes 1 and 8, digoxigenin-labeled DNA marker; lanes 2 to 6 and 9 to 13, GI or GII NoV-positive stool samples, respectively; lanes 7 and 14, GI and GII negative controls, respectively.
RT-PCR and Southern hybridization oligoprobing of TD stool samples.
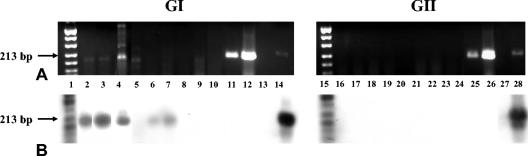
Twenty-six of 54 of the TD stool samples were positive for NoV GI. None of the samples were positive for NoV GII. A representative subset of TD samples is shown in Fig. 2. All GI- and GII-positive controls were positive, indicating that RT-PCR NoV GI and GII primers and probes worked effectively to detect NoV in TD samples. All negative controls were negative. Laboratory-generated poliovirus (PV) and hepatitis A virus (HAV) stocks (amplified with PV- and HAV-specific primers, respectively) (19) did not react with NoV oligoprobes, indicating specificity of the NoV probes for NoVs (Fig. 2). The use of an NVIS control in both dilutions of each TD sample enabled the identification of sample inhibition. Eighteen (60%) of 30 TD samples were inhibited at the 10−2 dilution, and 4 (7.4%) of 54 TD samples were inhibited at the 10−3 dilution. The four inhibited samples (formed stools) were additionally processed as described in Materials and Methods with guanidinium and phenol-chloroform and were negative for NoV.
FIG. 2.
RT-PCR (A) and Southern oligoprobe (B) results for a representative subset of TD samples. Each stool sample was analyzed separately with GI or GII primers and probes. Lanes 1 and 15, digoxigenin-labeled DNA marker; lanes 2 to 9 and 16 to 23, GI and GII 10−3 dilutions of heat- released RNA amplifications of TD stool samples, respectively; lanes 10 and 24, GI and GII negative controls, respectively; lanes 11 and 25, PV amplicons; lanes 12 and 26, HAV amplicons; lanes 13 and 27, space; lanes 14 and 28, GI and GII positive controls, respectively.
Prevalence and incidence of NoV among visitors experiencing TD.
The epidemiological analysis of the 54 TD stool samples collected from the 34 study participants indicated that NoV infection was a very common event among United States visitors to Guatemala and Mexico who experienced TD (Table 2). The overall prevalence of NoV infection was 65%, with 61% in the ETEC TD case group and 69% in the TD with no etiology case group, respectively (Table 2). As might be expected in an epidemiological setting in which travelers are at risk for a short period of time (14 to 24 days), NoV incidence rates calculated for a subset of individuals were comparable to the overall prevalence rates (73 versus 65%) (Table 2). The NoV incidence rate among students traveling to Guatemala (60%) appeared to be slightly lower than that of students traveling to Mexico (80%). However, this difference was not statistically significant (P > 0.05) (data not shown).
TABLE 2.
NoV infection among travelers from the United States experiencing an episode of traveler's diarrhea while visiting Mexico or Guatemala for 14 to 24 days
| Measure of infection | No. (%) of N.V cases detected by RT-PCR and confirmed by oligoprobing
|
||
|---|---|---|---|
| Traveler's diarrhea with no etiologya | Traveler's diarrhea with ETEC infectionb | Total | |
| Prevalence | 11/16 (69%) | 11/18 (61%) | 22/34 (65%) |
| Incidence | 6/9 (67%) | 5/6 (83%) | 11/15 (73%) |
Met the definition of traveler's diarrhea, and no bacterial or parasitic pathogens were identified.
Met the definition of traveler's diarrhea and were documented as ETEC infections based on microbiology results. One subject in this group was found to be positive for ETEC, rotavirus, and norovirus.
Influence of time spent at travel destination on NoV infection.
Further analysis of NoV infection among these TD cases indicated that time spent at their respective travel destinations was an important factor in determining the frequency of infection. NoV infections began to occur in a small percentage of travelers shortly after their arrival in Guatemala and Mexico (Table 3). Only 15% of the stools examined within the first 3 days of travel were positive for NoV (Table 3). However, as time spent at the respective travel destinations increased (≥7 days), the prevalence of NoV infections rose rapidly to over 66% (χ2 = 9.13; P = 0.003). In addition, most NoV-positive stools were from subjects with TD or a recent history of a TD episode. Of the 22 stools collected from subjects during their first 6 days in-country, 5 stools were NoV positive, and 4 of these were from subjects with TD; among the 21 NoV-positive stools collected from subjects after longer periods in-country, 20 were from subjects that were symptomatic at the time of submittal or had a history of recent TD.
TABLE 3.
NoV infection among travelers to Mexico and Guatemala: influence of time spent at travel destination on frequency of infectiona
| No. of days in countryb | No. (%) of NoV-positive stool samples/ total no. of stools samples tested
|
||
|---|---|---|---|
| Traveler's diarrhea with no etiology | Traveler's diarrhea with ETEC | Total NoV positive | |
| 0 to 3 | 1/7 (14) | 1/6 (17) | 2/13 (15) |
| 4 to 6 | 3/7 (43) | 0/2 (0) | 3/9 (33) |
| 7 to 9 | 6/6 (100) | 1/3 (33) | 7/9 (78) |
| 10+ | 4/7 (57) | 10/16 (63) | 14/23 (61) |
These traveler's diarrhea case groups were defined in the footnotes to Table 1.
RT-PCR and oligoprobing data strongly suggest that time spent at the travel destination was an important factor in contributing to NoV-positive stool samples among study participants traveling to Guatemala and Mexico. The proportion of stool samples positive for NoV was higher among those collected after subjects were in-country for ≥7 days (66%) than those collected during the first 3 days in-country (χ2 = 9.13; P ≤ 0.003).
Symptoms associated with NoV infections.
A review of incident infections acquired by travelers to both destinations indicated that NoV infection may be associated with a spectrum of TD clinical presentations that may or may not include the moderate-to-severe nausea and vomiting that is frequently associated with NoVs (as illustrated by selected cases presented in Table 4). For example, in some instances where NoV may be present as a copathogen with other etiological agents such as ETEC, infection may result in either a syndrome that is more consistent with an NoV etiology where severe abdominal pain, vomiting, and nausea are present (case 5 in Table 4) or a more traditional TD presentation where abdominal pain and vomiting are absent (case 6 in Table 4).
TABLE 4.
Clinical characteristics of traveler's diarrhea cases with NoV present as either the sole etiologic agent or a copathogen with ETEC
| Case no. | Stool samplea | NoV statusb | No. of days in country | Maximum no. of diarrheal stools/ 24 hc | Total no. of diarrheal stoolsd | Abdominal pain/crampse | Vomitinge | Nauseae | ETEC statusb |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Pre | Neg | 2 | 0 | 0 | 0 | 0 | 1 | Neg |
| Post | Pos | 13 | 12 | 30 | 1 | 1 | 2 | Neg | |
| 2 | Pre | Neg | 2 | 0 | 0 | 0 | 0 | 0 | Neg |
| Post | Pos | 8 | 3 | 7 | 1 | 2 | 3 | Neg | |
| 3 | Pref | Pos | 5 | 2 | 2 | 0 | 0 | 0 | Neg |
| Post | Pos | 7 | 8 | 11 | 0 | 3 | 2 | Neg | |
| 4 | Pre | Neg | 2 | 0 | 0 | 0 | 0 | 0 | Neg |
| Pre | Neg | 5 | 0 | 0 | 1 | 0 | 0 | Neg | |
| Post | Pos | 6 | 7 | 12 | 2 | 0 | 0 | Neg | |
| 5 | Pre | Neg | 3 | 0 | 0 | 0 | 0 | 0 | Neg |
| Post | Pos | 13 | 8 | 13 | 3 | 3 | 3 | Pos | |
| 6 | Pre | Pos | 2 | 0 | 0 | 0 | 0 | 0 | Neg |
| Post | Pos | 22 | 9 | 14 | 0 | 0 | 1 | Pos |
Pre, sample was collected before onset of traveler's diarrhea; Post, sample was collected following onset of traveler's diarrhea.
Neg, negative; Pos, positive.
Maximum number of diarrheal stools passed on a given day during the traveler's diarrhea episode.
Total number of diarrheal stools passed over the entire episode.
0, not present; 1, present, not affecting daily activity; 2, present, affecting daily activity; 3, present, preventing normal activity.
Traveler's diarrhea case definition not met at time of sample collection.
DISCUSSION
In this study, a simple molecular method was utilized to test TD stool samples for NoV, an emerging pathogen that may play a significant role in TD. The overall prevalence of NoV among the 34 cases examined was 65%, indicating that the burden of NoV infection was very high among TD cases in this study. Eleven NoV-positive stool samples also were positive for ETEC, indicating that dual infections were present. These results suggest that NoV infection is a frequent occurrence among travelers to Mexico and Guatemala who experience episodes of TD. The results also suggest that dual infections (bacterial and viral) may be more common than previously thought in individuals suffering from TD.
Although NoV has been previously implicated as a cause of TD in Mexico (18), this is the first report indicating that NoV also contributes to TD occurring among visitors to Guatemala. In both settings, the frequency of NoV infection increased sharply as subjects spent more time in-country (Table 3). While our study population was relatively small, the infection rate among travelers to Mexico also appears to be higher than previous reports (18). The difference may reflect the greater sensitivity of the RT-PCR diagnostic methods utilized in this study compared to the radioimmunoassay used in the previous study.
Another interesting finding in this study is that all NoV-positive TD samples were positive for GI; no TD samples were positive for GII. In the 1990s, GII was the predominant strain type identified in outbreaks worldwide (10, 13, 14). However, Fankhauser et al. (9) reported that the number of GI strains detected in outbreaks increased from 4% in 1996 to 1997 to 26% between 1997 to 2000. The authors proposed that this increase in GI detection may be due in part to the use of region B primers (the primers used in this study) which are more efficient in detecting GI strains previously missed with other primer sets (9).
We are confident that detecting only GI NoV-positive samples among the TD samples tested was not the result of ineffective GII primers or probes. During the optimization of our molecular techniques with CDC stools, NoV stools previously known to be positive for GII were confirmed positive with both RT-PCR gels and Southern blots (Fig. 1). Moreover, urban stream samples and outbreak samples collected from the mid-Atlantic region of the United States that are currently being analyzed for NoVs in our laboratory are predominantly positive for GII NoVs (data not shown). One potential explanation for why we detected only GI strains in this study while GII strains are believed to be more prevalent is that GI NoVs may be from different genotypes in developing versus developed countries whereas GII NoVs could be of the same genotypes, resulting in international travelers more likely to be infected with GI strains. The nature of NoVs prevalent in developing countries is an issue that is worthy of additional research.
The simple molecular detection method utilized in this study is particularly attractive in terms of furthering NoV-related research in developing countries such as Guatamala and Mexico. The heat release viral RNA extraction procedure only requires a heat block and a timer. Expensive, toxic reagents such as guanadinium-based extraction solutions and phenol are not required. Moreover, to identify potential false negatives caused by interfering substances that remain in diluted stool samples, a previously described NVIS control can be utilized in conjunction with the heat release method (20). Our results indicate that a majority of inhibitors present in stool samples can be eliminated by 100- to 1,000-fold dilutions of a 10% stool suspension, especially if the original stool sample is very loose or watery (the condition of most stool samples obtained from individuals with TD). Inhibition can occur at a higher frequency in formed stools from uninfected individuals. However, the few samples that remain inhibited can be reanalyzed following additional sample processing.
As an added precaution, separate RT-PCRs also were performed in this study for NoV GI, NoV GII, and NVIS instead of utilizing a multiplex reaction with GI, GII, and NVIS primers. In multiplex reactions, competition can occur where controls such as NVIS (which may be present in concentrations higher than that of the NoV within the environmental sample) can usurp the RT-PCR reagents, resulting in little to no amplification of NoV and false-negative results (21). Separate NVIS and NoV reaction mixtures eliminate this potential problem.
In addition to utilizing controls such as NVIS to detect sample inhibition during viral detection procedures, the results of this study reaffirm that it is crucial to confirm RT-PCR gel results by internal probing. Previous studies have shown that visual interpretation of gels alone leads to errors in interpreting the presence or absence of viral nucleic acids in environmental samples (3, 21). Figure 2 exemplifies the importance of not relying solely on gel results for the detection of viral agents. For instance, in Fig. 2, lanes 6 and 7, few to no definitive bands appeared on the GI gel; however, Southern probing indicated that the samples were NoV positive. Conversely, in Fig. 2, lanes 5 and 9, faint bands in the GI gel that are approximately the same size as a region B GI amplicon appeared. Yet, upon Southern probing, the amplicons associated with these bands did not hybridize to NoV-specific probes, indicating that the bands perceived on the gel were nonspecific.
In this study, Southern probing for GI and GII NoVs was performed using six probes in combined hybridization reactions for each genogroup; therefore, hybridization was utilized as a confirmation tool and not as a genotyping tool. For the treatment of NoV-associated gastroenteritis, physicians only need to know whether NoVs are present. They do not need to know which NoV strain caused the illness. However, reverse line blot methods of probing may be effective alternative methods that not only confirm NoV amplicons but also enable the identification of specific strains (26). Strain identification is useful in outbreak investigations where strain information can help to identify the source and/or patterns of the outbreak. The development of reverse line blot methods is ongoing in our laboratory.
In summation, NoVs are known to be a major cause of nonbacterial gastroenteritis outbreaks occurring in both domestic settings and unique travel settings such as cruise ships (5, 9, 15). The role of NoVs also has been well documented in outbreaks among small military groups going ashore for short periods of time in conjunction with naval deployment overseas (17). Yet, this study is the first of its kind to indicate that NoVs may be a major cause of illness among United States travelers who experience TD during extended stays in developing countries. The results also document concurrent NoV and bacterial infections in one-third of the cases studied, highlighting the polymicrobial nature of TD illness and suggesting that NoVs complicate both the clinical management of TD and the interpretation of vaccine and other intervention studies that may be ongoing in at-risk populations. The high frequency of NoV infection among TD cases examined in this study suggests that further investigations concerning the role of these viruses in TD are warranted. More in-depth studies regarding the role of this agent in TD will be facilitated by the application of the simplified detection methods described here in both United States and oversea laboratories.
Acknowledgments
We thank the Center for a Livable Future at the Johns Hopkins University Bloomberg School of Public Health for funding this research. A.R.C. is the recipient of a Howard Hughes Medical Institute Pre-Doctoral Fellowship in Biological Sciences.
We also extend thanks to Ann-Mari Svennerholm of the University of Göteborg, Göteborg, Sweden, for her generous efforts to determine the frequency of ETEC infections among subjects participating in this study; Robert Atmar, Baylor College of Medicine, Houston, Tex., for a constructive review and comments; and Stephan Monroe, Division of Viral and Rickettsial Diseases, Centers for Disease Control and Prevention, Atlanta, Ga., for supplying positive control NoV stool samples as well as a constructive review and comments.
REFERENCES
- 1.Anderson, A. D., V. D. Garrett, J. Sobel, S. S. Monroe, R. L. Fankhauser, K. J. Schwab, J. S. Bresee, P. S. Mead, C. Higgins, J. Campana, and R. I. Glass. 2001. Multistate outbreak of Norwalk-like virus gastroenteritis associated with a common caterer. Am. J. Epidemiol. 154:1013-1019. [DOI] [PubMed] [Google Scholar]
- 2.Atmar, R. L., and M. K. Estes. 2001. Diagnosis of noncultivatable gastroenteritis viruses, the human caliciviruses. Clin. Microbiol. Rev. 14:15-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atmar, R. L., F. H. Neill, J. L. Romalde, F. Le Guyader, C. M. Woodley, T. G. Metcalf, and M. K. Estes. 1995. Detection of Norwalk virus and hepatitis A virus in shellfish tissues with the PCR. Appl. Environ. Microbiol. 61:3014-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brewster, S. J., and D. N. Taylor. 2003. Epidemiology of traveler's diarrhea, p. 175-184. In J. S. Keystone, P. E. Kozarsky, D. O. Freedmen, H. D. Northdurft, and B. A. Connor (ed.), Travel medicine. Elsevier, Ltd., London, United Kingdom.
- 5.Centers for Disease Control and Prevention. 2002. Outbreaks of gastroenteritis associated with noroviruses on cruise ships—United States, 2002. Morb. Mortal. Wkly. Rep. 51:1112-1115. [PubMed] [Google Scholar]
- 6.Cheng, A. C., and N. M. Thielman. 2002. Update on traveler's diarrhea. Curr. Infect. Dis. Rep. 4:70-77. [DOI] [PubMed] [Google Scholar]
- 7.de Wit, M. A., M. P. Koopmans, L. M. Kortbeek, W. J. Wannet, J. Vinje, F. van Leusden, A. I. Bartelds, and Y. T. van Duynhoven. 2001. Sensor, a population-based cohort study on gastroenteritis in The Netherlands: incidence and etiology. Am. J. Epidemiol. 154:666-674. [DOI] [PubMed] [Google Scholar]
- 8.DuPont, H. L., and C. D. Ericsson. 1993. Prevention and treatment of traveler's diarrhea. N. Engl. J. Med. 328:1821-1827. [DOI] [PubMed] [Google Scholar]
- 9.Fankhauser, R. L., S. S. Monroe, J. S. Noel, C. D. Humphrey, J. S. Bresee, U. D. Parashar, T. Ando, and R. I. Glass. 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis. 186:1-7. [DOI] [PubMed] [Google Scholar]
- 10.Fankhauser, R. L., J. S. Noel, S. S. Monroe, T. Ando, and R. I. Glass. 1998. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J. Infect. Dis. 178:1571-1578. [DOI] [PubMed] [Google Scholar]
- 11.Gorbach, S. L., B. H. Kean, D. G. Evans, D. J. Evans, Jr., and D. Bessudo. 1975. Travelers' diarrhea and toxigenic Escherichia coli. N. Engl. J. Med. 292:933-936. [DOI] [PubMed] [Google Scholar]
- 12.Hale, A., K. Mattick, D. Lewis, M. Estes, X. Jiang, J. Green, R. Eglin, and D. Brown. 2000. Distinct epidemiological patterns of Norwalk-like virus infection. J. Med. Virol. 62:99-103. [DOI] [PubMed] [Google Scholar]
- 13.Lopman, B. A., D. W. Brown, and M. Koopmans. 2002. Human caliciviruses in Europe. J. Clin. Virol. 24:137-160. [DOI] [PubMed] [Google Scholar]
- 14.Maguire, A. J., J. Green, D. W. G. Brown, U. Desselberger, and J. J. Gray. 1999. Molecular epidemiology of outbreaks of gastroenteritis associated with small round-structured viruses in East Anglia, United Kingdom, during the 1996-1997 season. J. Clin. Microbiol. 37:81-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray, P. R., E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.). 1999. Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.
- 17.Oyofo, B. A., R. Soderquist, M. Lesmana, D. Subekti, P. Tjaniadi, D. J. Fryauff, A. L. Corwin, E. Richie, and C. Lebron. 1999. Norwalk-like virus and bacterial pathogens associated with cases of gastroenteritis onboard a US Navy ship. Am. J. Trop. Med. Hyg. 61:904-908. [DOI] [PubMed] [Google Scholar]
- 18.Ryder, R. W., C. A. Oquist, H. Greenberg, D. N. Taylor, F. Orskov, I. Orskov, A. Z. Kapikian, and R. B. Sack. 1981. Travelers' diarrhea in Panamanian tourists in Mexico. J. Infect. Dis. 144:442-448. [DOI] [PubMed] [Google Scholar]
- 19.Schwab, K. J., R. De Leon, and M. D. Sobsey. 1996. Immunoaffinity concentration and purification of waterborne enteric viruses for detection by reverse transcriptase PCR. Appl. Environ. Microbiol. 62:2086-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwab, K. J., M. K. Estes, F. H. Neill, and R. L. Atmar. 1997. Use of heat release and an internal RNA standard control in reverse transcription-PCR detection of Norwalk virus from stool samples. J. Clin. Microbiol. 35:511-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwab, K. J., F. H. Neill, M. K. Estes, T. G. Metcalf, and R. L. Atmar. 1998. Distribution of Norwalk virus within shellfish following bioaccumulation and subsequent depuration by detection using RT-PCR. J. Food Prot. 61:1674-1680. [DOI] [PubMed] [Google Scholar]
- 22.Schwab, K. J., F. H. Neill, R. L. Fankhauser, N. A. Daniels, S. S. Monroe, D. A. Bergmire-Sweat, M. K. Estes, and R. L. Atmar. 2000. Development of methods to detect “Norwalk-like viruses” (NLVs) and hepatitis A virus in delicatessen foods: application to a food-borne NLV outbreak. Appl. Environ. Microbiol. 66:213-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steffen, R., M. Rickenbach, U. Wilhelm, A. Helminger, and M. Schar. 1987. Health problems after travel to developing countries. J. Infect. Dis. 156:84-91. [DOI] [PubMed] [Google Scholar]
- 24.Svennerholm, A. M., M. Wikstrom, M. Lindblad, and J. Holmgren. 1986. Monoclonal antibodies against Escherichia coli heat-stable toxin (STa) and their use in a diagnostic ST ganglioside GM1-enzyme-linked immunosorbent assay. J. Clin. Microbiol. 24:585-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Svennerholm, A.-M., and J. Holmgren. 1978. Identification of Escherichia coli heat-labile enterotoxin by means of a ganglioside immunosorbent assay (GM1-ELISA) procedure. Curr. Microbiol. 1:19-23. [Google Scholar]
- 26.Vinje, J., and M. P. Koopmans. 2000. Simultaneous detection and genotyping of “Norwalk-like viruses” by oligonucleotide array in a reverse line blot hybridization format. J. Clin. Microbiol. 38:2595-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolf, M. K., D. N. Taylor, E. C. Boedeker, K. C. Hyams, D. R. Maneval, M. M. Levine, K. Tamura, R. A. Wilson, and P. Echeverria. 1993. Characterization of enterotoxigenic Escherichia coli isolated from U.S. troops deployed to the Middle East. J. Clin. Microbiol. 31:851-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.World Health Organization. 1997. The state of world health, p. 1-62. In The world health report 1996—fighting disease, fostering development. World Health Organization, Geneva, Switzerland. [PubMed]