Abstract
The DiversiLab System, which includes microfluidics-based detection, reagent kits, and software for data processing and analysis, is an automated method using repetitive sequence-based PCR (rep-PCR) for microbial strain typing. To assess the reliability of the DiversiLab System for strain characterization of Staphylococcus aureus, we tested clinical isolates sent to ARUP Laboratories for typing and compared results to those of pulsed field electrophoresis (PFGE) aided by the cluster analysis provided by BioNumerics software. spa typing was performed when the results of these two methods for an outbreak were not concordant. The study included 89 S. aureus isolates (65 mecA positive, 24 mecA negative) from 19 outbreaks (2 to 11 isolates/outbreak). The DiversiLab and PFGE-BioNumerics results were concordant for 15 of the 19 outbreaks. For the remaining four outbreaks, there was partial concordance between the two methods. spa typing results were the same as or more similar to rep-PCR results for three of those outbreaks and were more similar to PFGE results for one. With regard to performance, the DiversiLab system was considerably less labor intensive than PFGE and provided results in less than 24 h, compared with 2 to 3 days for PFGE. Additionally, the Web-based DiversiLab software provides standardized comparisons among isolates almost instantaneously and generates user-friendly, customized reports.
Staphylococcus aureus, especially oxacillin (methicillin)-resistant strains, is an important cause of nosocomial and community-acquired infections (6, 9, 11, 19). More recently, emergence of vancomycin-intermediate and vancomycin-resistant S. aureus has caused concern (2, 5, 10). To track and help limit the spread of resistant isolates, microbiology laboratories are often asked to determine the relatedness of groups of organisms. This requires the use of a molecular strain typing system with certain performance characteristics (15). For each isolate, the system should provide an interpretable result, preferably based on objective criteria. Results should be reproducible from day to day and from laboratory to laboratory and should allow differentiation of unrelated strains. Additionally, the method should be standardized and, ideally, should be technically simple, cost effective, and rapid.
Molecular methods that have been used to study relatedness of S. aureus isolates include pulsed-field gel electrophoresis (PFGE), multilocus sequence typing, staphylococcal protein A gene typing (spa typing), repetitive sequence-based PCR (rep-PCR), and various restriction fragment length polymorphism-based methods (3, 4, 7, 12, 14, 16-18, 20-22, 24-27). Of these, PFGE is the most widely used. PFGE is accurate and reproducible; however, the method is not presently standardized, and although guidelines for interpretation exist (23), they are subjective and based on visual analysis of band patterns. Moreover, PFGE is labor intensive and time consuming and, in our experience, does not always provide an interpretable result.
Only one molecular strain typing system is commercially available as a standardized kit—the automated DiversiLab system (Spectral Genomics, Inc., Houston, Tex.), which uses rep-PCR to discriminate among bacterial and fungal isolates and to identify some fungi to the species level (8). Briefly, after DNA extraction, the rep-PCR method uses primers that target and bind to multiple noncoding, repetitive sequences (generally 30 to 500 bp) interspersed throughout the bacterial genome. The outwardly facing primers generally amplify between repetitive elements, as opposed to inwardly facing primers, which amplify the repetitive element itself (as in the variable-number tandem repeat) (28). Multiple DNA amplicons of different sizes and various quantities (intensities) are generated during PCR. The manual method is an established approach for subspecies classification and strain delineation of bacteria (29, 30). The automated rep-PCR method has three components: the Agilent 2100 bioanalyzer, which separates the amplified fragments on a microfluidic chip and detects them based on fluorescent intensity and migration time, rep-PCR reagent kits, and web-based DiversiLab software (version 2.1.66). The software analyzes results by creating a proximity matrix using the Pearson correlation to calculate pair-wise similarities between all samples tested. The report generated by the DiversiLab system includes a dendrogram (which illustrates hierarchical relationships among isolates) and scatter plot (which provides a spatial, nonhierarchical view of the relationships); gel-like images and/or an electropherogram can also be incorporated. Commercially available components, automation, technical simplicity, rapid turn-around time, and user-friendly reports are features that make the DiversiLab system attractive for a busy microbiology laboratory. However, there are few published evaluations of its discriminatory power.
The objective of this study was to assess the reliability of the DiversiLab system for determining relatedness of strains of S. aureus in a large reference laboratory setting by comparing results to those of PFGE. If results of the two methods were not concordant for all isolates in an outbreak, spa typing, which has been shown to be as useful as other molecular methods (7, 12), including PFGE, for outbreak investigation purposes, was performed.
MATERIALS AND METHODS
Bacterial isolates.
From the 60 groups (398 isolates) of S. aureus isolates representing potential outbreaks that were sent to ARUP Laboratories in 2003 for strain characterization by PFGE, 20 groups (hereafter termed outbreaks A through T) consisting of a total of 91 isolates were randomly selected for analysis by the DiversiLab system. After completion of PFGE analysis, isolates were frozen at −70°C in brain heart infusion-glycerol (9.9% [vol/wt] glycerol, 3.33% [wt/vol] brain heart infusion broth). For this study, frozen stocks were thawed and grown on sheep blood agar (BBL, Sparks, Md.). Cultures were checked for purity, and the identification provided by the client was confirmed with BACTi Staph reagent (Remel, Lenexa, Kans.). The presence or absence of the mecA gene was determined by PCR as previously described (1).
PFGE.
PFGE was performed as described previously (13) with minor modifications. Briefly, a single colony was inoculated into tryptic soy broth (BBL) and incubated overnight at 37°C with vigorous shaking. Agarose plugs were prepared using 10 μl (rather than 6 μl) of achromopeptidase (Wako BioProducts, Richmond, Va.) (60 U/μl) for bacterial cell lysis. Plugs were maintained at 4°C for 15 min (rather than 10 min), and the lysed plugs were washed in 5 ml (rather than 2 ml) of TE buffer (10 mM Tris, 1 mM EDTA, pH 7.5). A restriction endonuclease (RE) mixture was prepared using 1× NEBuffer 4 and 2 μl of SmaI restriction enzyme (20 U) (New England BioLabs, Beverly, Mass.). Plugs were cut to the desired comb size and placed into a 1.5-ml sterile microcentrifuge tube. RE mixture (200 μl) was added to the cut plugs, and they were incubated at room temperature for 2 h. The RE mixture was removed, and 1 ml of 0.5× TBE (50 mM Tris, 50 mM H3BO3, 0.5 mM EDTA) was added. After 5 min, the plugs were placed on the comb by the use of 1.2% Seakem GTG agarose gel (Cambrex BioScience Rockland, Inc., Rockland, Maine). Bacteriophage lambda DNA concatemers (Lambda Ladder PFGE Marker; New England BioLabs) were similarly embedded and used as size standards.
Restriction fragments were separated over a size range of approximately 50 to 700 kb by using a contour-clamped homogeneous electric field Chef Mapper XA pulsed-field electrophoresis system (Bio-Rad Laboratories, Hercules, Calif.). Running parameters were as follows: 200 V (6 V/cm); temperature, 14°C; initial switch, 15 s; final switch, 55 s; and time, 22 h. After the electrophoresis run was completed, the gel was stained with ethidium bromide for 15 min in a covered container and destained in fresh, distilled water for 60 min. Gels were imaged by UV transillumination using a Gel Doc 2000 apparatus and Quantity One software (Bio-Rad Laboratories) and saved as TIFF images.
The relatedness of isolates in a potential outbreak was based on visual comparison of band patterns of isolates run in the same gel by the use of criteria described by Tenover et al. (23) and BioNumerics software (version 3.0; Applied Maths, Kortrijk, Belgium) for comparisons. Comparisons based on results provided by BioNumerics software were accomplished by calculating cluster analysis using the band-based Dice method to illustrate pairwise similarities between all samples in an outbreak and the dendrogram-type unweighted-pair group method using average linkages (UPGMA).
rep-PCR.
DNA from a 10-μl loopful of a S. aureus colony was extracted using an Ultra Clean microbial DNA isolation kit (Mo Bio Laboratories, Solana Beach, Calif.), following the directions recommended by Spectral Genomics. DNA genomic integrity was visualized using agarose gel electrophoresis. The extracted DNA was frozen overnight at −20°C and then amplified using a Diversilab Staphylococcus DNA fingerprinting kit (Spectral Genomics, Inc.), which includes rep-PCR master mix 1, Staphylococcus primers, and kit-specific positive and negative controls in accordance with the manufacturer's product insert. Briefly, 50 ng of genomic DNA, 2.5 U of AmpliTaq DNA polymerase, and 1.5 μl of 10× PCR buffer (Applied Biosystems, Foster City, Calif.) were added to the rep-PCR master mix to achieve a total of 25 μl. Thermal cycling parameters for a GeneAmp PCR System 9600 (Perkin-Elmer Corporation, Norwalk, Ct.) were as follows: initial denaturation at 94°C for 2 min, 35 cycles of denaturation at 94°C for 30 sec, annealing at 45°C for 30 sec, extension at 70°C for 90 sec, and a final extension at 70°C for 3 min. The amplified product was stored at −20°C until detection. Analysis of rep-PCR products was implemented using a DiversiLab system in which the amplified fragments of various sizes and intensities are separated and detected using a microfluidics Labchip with an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, Calif.).
The relatedness was determined by cluster analysis and guidelines provided by manufacturer. Isolates were categorized as indistinguishable, similar, or different. The manufacturer compared multiple sets of characterized isolates and outbreaks in “gold standard” analysis to establish the categories. In general, “different” was defined as <95% similarity and ≥2 band differences for homogeneous organisms or ≥3 band differences for heterogeneous organisms. “Similar” was defined as <97% similarity and 1 band difference for homogeneous organisms or up to 2 band differences for heterogeneous organisms. “Indistinguishable” was defined as >95% similarity and no banding differences, including no variation in intensities of individual bands, although overall intensities may differ.
For each outbreak, the analysis using the DiversiLab System was compared to PFGE (both visually and using BioNumerics). For this analysis, we defined concordance between the PFGE and rep-PCR interpretations as follows: the PFGE pattern of isolates in a potential outbreak differed by 0 to 6 bands (isolates were considered to be indistinguishable, closely related, or possibly related) and the rep-PCR pattern differed by 0 to 2 bands (indistinguishable or similar). These parameters were chosen based on which isolates would generally be considered part of an outbreak as reported to infection control. If there was no concordance in the clustering of the isolates in an outbreak between the two methods, rep-PCR and PFGE were repeated, and the isolates were sent (in a blinded fashion) to Spectral Genomics for testing by both methods.
spa typing.
The procedure recommended by Ridom Bioinformatics was modified for use on a real-time PCR platform. Briefly, DNA prepared for rep-PCR was amplified using the primers spa 1113f (5′TAAAGACGATCCTTCGGTGAGC3′) and spa 1514r (5′CAGCAGTAGTGCCGTTTGCTT3′). A total of 4 μl of template DNA was added to 1× the contents of a Lightcycler FastStart DNA Master Hybridization Probes kit (Roche-Applied Science, Penzberg, Germany), which contains deoxynucleoside triphosphates, FastStart Taq DNA polymerase, and 1 mM MgCl2; an additional 6.4 μl of 25 mM MgCl2 (final MgCl2 concentration, 4 mM); 400 nM primers; and 1× SYBR green (Molecular Probes, Inc., Eugene, Oreg.) for a total volume of 40 μl. Thermocycling parameters for a Rotor-Gene 3000 system (Corbett Research, Sydney, Australia) were as follows: polymerase activation at 95°C for 10 min, 40 cycles of denaturation at 95°C for 10 s, annealing at 60°C for 10 s, extension at 72°C for 20 s with fluorescence acquisition during each cycle, and a final extension at 72°C for 2 min. A melt curve analysis performed at temperatures from 60 to 99°C followed the amplification. The amplified product was processed using a MicroCon spin column (Millipore Corporation, Bedford, Mass.) with a 100,000-molecular-weight cutoff.
Bidirectional DNA sequence data for each spa sample was generated using fluorescently labeled terminator sequencing chemistry and sequencing primers (a 5′ spa 1113f primer and a 3′ spa 1514r primer). Briefly, 5 μl of BigDye Terminator Ready Reaction mix v. 1.1 (Applied Biosystems, Inc.) was added to 4 μl of each primer (0.8 pmol/μl) and 3 μl of purified PCR product was added to the BigDye-primer mix. Cycle sequencing was performed using a 9700 Thermal Cycler (Applied Biosystems) and the following parameters: 25 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min. Sequencing reaction products were passed through a column of Sephadex G-50 fine to remove unincorporated dye terminators. DNA sequencing data files from the purified sequencing reaction products were generated using an ABI Prism 3730 DNA Analyzer (Applied Biosystems, Inc.). Sequencher sequencing analysis software version 4.1 (Genecodes, Ann Arbor, Mich.) was used to align and edit 5′ and 3′ sequencing files to create a consensus sequence for each spa sample.
The consensus sequences were analyzed using StaphType software (Ridom), which assigns 21-, 24-, or 27-base repeats numerical values. The spa type was deduced from the order of the specific repeats. Isolates with the same spa type were considered indistinguishable. spa types with similar repeat profiles (i.e., no more than one insertion or deletion of a repeat or ≤2 bp differences within a repeat) were grouped together as part of the same cluster.
RESULTS
Of the 20 outbreaks, one was excluded from the comparison because one of the two isolates in that outbreak did not yield an interpretable fingerprint by PFGE after two attempts. All 89 isolates in the remaining 19 outbreaks were identified as S. aureus; 65 were positive for mecA by PCR. Sixteen outbreaks (2 to 9 isolates/outbreak) consisted of mecA-positive isolates; the other three (3 to 11 isolates/outbreak) consisted of mecA-negative isolates.
rep-PCR and PFGE interpretations were concordant for 12 outbreaks. Two of these outbreaks consisted of mecA-negative isolates (outbreak Q, 11 isolates; outbreak R, 3 isolates) and 10 consisted of mecA-positive isolates (outbreak B, 3 isolates; outbreak C, 2 isolates; outbreak F, 6 isolates [Fig. 1 ]; outbreak J, 3 isolates; outbreak K, 2 isolates; outbreaks L, M, and N, 3 isolates each; outbreak S, 2 isolates; and outbreak T, 3 isolates). PFGE was slightly more discriminatory for five of these outbreaks (outbreaks J, M, N, Q, and T), although the final interpretation was the same for both methods. Each of these five outbreaks consisted of two or more strains considered indistinguishable by PFGE and one considered closely or possibly related, whereas rep-PCR called all strains indistinguishable. rep-PCR was slightly more discriminatory in one outbreak (outbreak B). In this outbreak, two strains were considered indistinguishable and one was similar by rep-PCR; all three were considered indistinguishable by PFGE.
FIG. 1.
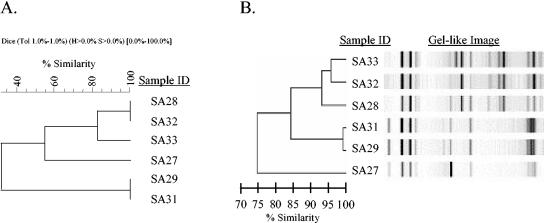
Outbreak F is an example of concordance between PFGE band cluster analysis in BioNumerics software, using dendrogram-type UPGMA and band-based Dice analysis, and rep-PCR, using a DiversiLab system. Both the BioNumerics (A) and rep-PCR (B) dendrograms show that five of the six oxacillin (methicillin)-resistant isolates clustered into two groups, one with three isolates (SA28, SA32, and SA33) and one with two (SA29 and SA31); the sixth isolate is an outlier (SA27) that is not related to the other isolates tested. The gel-like image generated by the DiversiLab software illustrates band similarities.
For four outbreaks (all mecA-positive isolates), the clustering of isolates by rep-PCR and PFGE initially was considered not concordant: outbreak A, 4 isolates; outbreak D, 6 isolates (Fig. 2); outbreak G, 9 isolates; outbreak P, 5 isolates. Therefore, all isolates in these outbreaks were retested at ARUP Laboratories and also sent to Spectral Genomics, Inc. (blinded) for testing. For outbreaks A, D, and G, the repeat PFGE fingerprint patterns and rep-PCR results of the testing done at ARUP Laboratories and the results of testing performed at Spectral Genomics were identical to initial results. The BioNumerics interpretation of PFGE testing done at ARUP Laboratories, however, changed. In retrospect, it was clear that too much weight had initially been placed on the BioNumerics cluster analysis. PFGE and rep-PCR results should have been considered concordant for outbreaks D and G and partially concordant for outbreak A. For outbreak A, interpretations by rep-PCR and PFGE were concordant for two of the four isolates, which were indistinguishable by rep-PCR and closely related by PFGE. The other two isolates in outbreak A were different by rep-PCR, whereas one was possibly related to the first two by PFGE. The results of spa typing, shown in Table 1, were in agreement with rep-PCR results. For outbreak P, the repeat PFGE fingerprint patterns and rep-PCR results of the testing done at ARUP Laboratories and the results of testing performed at Spectral Genomics were identical, and both sets of rep-PCR results were identical to each other and to the initial results. The PFGE fingerprint patterns, however, were different for two of the isolates in the outbreak. This was confirmed by testing a third time at ARUP Laboratories, suggesting that a technical error occurred when the initial isolates were frozen. Results of the repeat PFGE and initial (as well as repeat) rep-PCR testing were concordant.
FIG. 2.
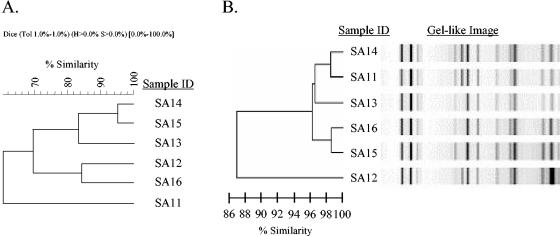
Outbreak D is an example of an outbreak initially interpreted as representing a lack of concordance between PFGE band cluster analysis in BioNumerics software, using dendrogram-type UPGMA and band-based Dice analysis, and rep-PCR, using a DiversiLab system. (A) The BioNumerics dendrogram shows two clusters, one with three isolates (SA14, SA15, and SA13) and one with two isolates (SA12 and SA16) and one outlier (SA11). However, when analyzed on the basis of the band pattern, SA12, SA13, SA14, SA15, and SA16 are shown to be related and SA11 is shown to be possibly related (data not shown). (B) The rep-PCR dendrogram has two clusters, each with three isolates that are similar to each other. Cluster 1 includes SA14 and SA11, which are indistinguishable, and SA12, which is similar. Cluster 2 includes SA13, SA15, and SA16, which are indistinguishable. The gel-like image generated by the DiversiLab software illustrates band similarities.
TABLE 1.
Spa typing results for isolates in outbreaks for which there was partial concordance between rep-PCR and PFGEa
| Outbreak | Sample(s) | Spa type |
|---|---|---|
| A | SA2, SA4 | tx032 |
| SA1 | t002 | |
| SA3 | tx033 | |
| E | SA22, SA25 | t004b |
| SA23, SA24 | t002 | |
| SA19 | tx028b | |
| SA17 | t012 | |
| SA18 | t078 | |
| SA20 | tx025 | |
| SA21 | t209 | |
| SA26 | t037 | |
| H | SA43, SA44, SA50 | t002c |
| SA47, SA48 | t242c | |
| SA45, SA49 | t018 | |
| SA46 | tx034 | |
| I | SA51 | tx027d |
| SA52 | t002d | |
| SA53 | tx030 |
The designation “t” refers to a Ridom Spa server-approved global Spa-type pattern of repeats; “tx” refers to a new local spa type, indicating that the sequence contains a spa type not found in the global Spa server database.
tx028 differed from t004 by one 24-bp insertion; therefore, these types were grouped in the same cluster.
For t242 and t002, there was a 1-bp difference in one repeat (repeat 34 of t002 and repeat 13 of t242); therefore, these types were grouped in the same cluster.
For tx027 and t002, there was a 2-bp difference in one repeat (repeat 38 of tx027 and repeat 20 of t002); therefore, these types were grouped in the same cluster.
In addition to the outbreak A results, there was partial concordance between rep-PCR and PFGE interpretations for the remaining three outbreaks. For outbreak E (mecA-negative isolates), interpretations for 7 of the 10 isolates were concordant. The other three isolates in this outbreak were considered different by PFGE, whereas rep-PCR found two of them to be indistinguishable and the third was considered similar to one of the other clusters in the outbreak. spa typing results (Table 1) for this outbreak differed from both rep-PCR and PFGE results but were more like rep-PCR results. Interpretations for seven of the eight isolates in outbreak H (mecA-positive isolates; shown in Fig. 3) were concordant; the other isolate was grouped in different clusters by PFGE and rep-PCR. spa typing results (Table 1) for this outbreak were not identical to either rep-PCR or PFGE results but were more like PFGE results. In outbreak I (mecA-positive isolates), interpretations for two of the three isolates were concordant. The remaining isolate was considered closely related to the first two by PFGE, whereas it was considered different by rep-PCR; spa typing results (Table 1) for this outbreak differed from both rep-PCR and PFGE results but more closely resembled rep-PCR results.
FIG. 3.
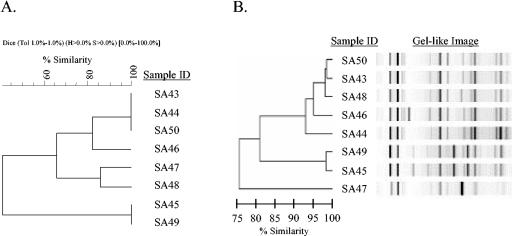
Outbreak H is an example of partial concordance between PFGE band cluster analysis in BioNumerics software, using dendrogram-type UPGMA and band-based Dice analysis, and rep-PCR, using a DiversiLab system. (A) The BioNumerics dendrogram shows three clusters: one with four isolates (SA43, SA44, SA46, and SA50) and two with two isolates each (SA47 and SA48 in one and SA45 and SA49 in the other). (B) In the rep-PCR dendrogram there are two clusters (SA43, SA44, SA46, SA48, and SA50 in one and SA45 and SA49 in the other) and one outlier (SA47). The gel-like image generated by DiversiLab software illustrates band similarities.
DISCUSSION
The primary goal of this study was to assess the reliability of the DiversiLab system for determining the relatedness of S. aureus isolates in a large reference laboratory setting. To accomplish this, results of the DiversiLab system for 19 outbreaks (89 isolates) were compared to those of PFGE (visual interpretation of band patterns) aided by the clustering provided by BioNumerics. Ideally, epidemiological data would have been incorporated into the final interpretation. Unfortunately, these data were not available. In our experience, however, it is highly unusual for clinical information to accompany isolates sent to a reference laboratory for strain typing.
We found that the correlation between rep-PCR and PFGE was quite good. For 15 of the 19 outbreaks in our study, rep-PCR and PFGE results were concordant, and for the remaining four, results of the two methods were partially concordant. For 3 of the 19 outbreaks, rep-PCR and PFGE results initially were incorrectly interpreted as not concordant. In all three cases, the clustering generated by BioNumerics was misleading, resulting in inaccurate interpretation of the PFGE fingerprints. For outbreaks in which there was partial concordance, spa typing results agreed with or were more like rep-PCR results in three of them and more closely resembled PFGE results in one. Additionally, both intra- and interlaboratory reproducibility results obtained with the DiversiLab System were excellent. Reproducibility of PFGE also was excellent; however, the clustering results provided by BioNumerics differed for three of the four outbreaks that were repeated. This likely reflects the fact that the BioNumerics software allows for parameter modifications, which in turn gives the user the freedom to influence the outcome and increases the opportunity for interuser variability.
A secondary objective of this evaluation was to assess the technical aspects of the DiversiLab system and compare them with PFGE. In our opinion, positive aspects of the DiversiLab system include a 1-day turnaround time (compared with 2 to 3 days for PFGE), standardized reagents commercially available in kit form, and the Web-based software, which we believe offers several advantages over BioNumerics. As previously mentioned, the analysis provided by BioNumerics software is less reproducible than that provided by the DiversiLab System. Transfer of the PFGE band pattern to BioNumerics is manual and, therefore, subjective and has specific gel resolution requirements. Access to BioNumerics software is local and limited by a user key, whereas DiversiLab software provides unlimited user access (although secured by password protection) for any computer with Internet connections. The DiversiLab software also generates user-friendly reports that can be customized for the testing laboratory. In addition, the DiversiLab system is considerably less technically demanding than PFGE. The most labor-intensive step is the extraction, which requires approximately 45 to 60 min of hands-on time, depending on the number of isolates being tested and the organism type (i.e., bacteria, fungus, or mycobacteria).
There are potential limitations to the DiversiLab system. Although the reagent kits include positive and negative controls, acceptable results for these controls are not indicated. Occasionally, bubbles form as DNA is loaded into the channels of the Labchip. This necessitates repeat testing of the amplified product, which increases both the turnaround time and cost of the test. Additionally, the occurrence of electrical interference when the Labchip is being analyzed within the Agilent 2100 bioanalyzer or the presence of dust in the bioanalyzer can produce an electrical spike in the electropherogram, necessitating repeat testing of the amplified DNA. Finally, for the DiversiLab system to be most cost efficient, 12 samples must be tested, because all 13 wells of the Labchip must contain DNA marker and the gel-dye matrix, even if no isolate is being tested.
In summary, the results of our evaluation showed that the performance of the DiversiLab system was comparable to that of PFGE for determining strain relatedness of isolates of S. aureus. Moreover, the DiversiLab system is considerably less labor intensive than PFGE and provides more-rapid results. Additionally, the Web-based software provides standardized comparisons among isolates almost instantaneously and generates user-friendly, customized reports.
Acknowledgments
rep-PCR reagent kits were kindly provided by Spectral Genomics, Inc.
We thank Andrea Stetler for her secretarial assistance.
REFERENCES
- 1.Carroll, K. C., R. B. Leonard, P. L. Newcomb-Gayman, and D. R. Hillyard. 1996. Rapid detection of the staphylococcal mecA gene from BACTEC blood culture bottles by the polymerase chain reaction. Am. J. Clin. Pathol. 106:600-605. [DOI] [PubMed] [Google Scholar]
- 2.Chang, S., D. M. Sievert, J. C. Hageman, M. L. Boulton, F. C. Tenover, F. P. Downes, S. Shah, J. T. Rudrik, G. R. Pupp, W. J. Brown, D. Cardo, S. K. Fridkin, and the Vancomycin-Resistant Staphylococcus aureus Investigative Team. 2003. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N. Engl. J. Med. 348:1342-1347. [DOI] [PubMed] [Google Scholar]
- 3.Del Vecchio, V. G., J. M. Petroziellom M. J. Gress, F. K. McCleskey, G. P. Melcher, H. K. Crouch, and J. R. Lupski. 1995. Molecular genotyping of methicillin-resistant Staphylococcus aureus via fluorophore-enhanced repetitive-sequence PCR. J. Clin. Microbiol. 33:2141-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deplano, A., A. Schuermans, J. Van Eldere, W. Witte, H. Meugnier, J. Etienne, H. Grundmann, D. Jonas, G. T. Noordhoek, J. Dijkstra, A. van Belkum, W. van Leeuwen, P. T. Tassios, N. J. Legakis, A. van der Zee, A Bergmans, D. S. Blanc, F. C. Tenover, B. C. Cookson, G. O'Neil, M. J. Struelens, and The European Study Group on Epidemiological Markers of the ESCMID. 2000. Multicenter evaluation of epidemiological typing of methicillin-resistant Staphylococcus aureus strains by repetitive-element PCR analysis. J. Clin. Microbiol. 38:3527-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fridkin, S. K., J. Hageman, L. K. McDougal, J. Mohammed, W. R. Jarvis, T. M. Perl, F. C. Tenover, and the Vancomycin-Intermediate Staphylococcus aureus Epidemiology Study Group. 2003. Epidemiological and microbiological characterization of infections caused by Staphylococcus aureus with reduced susceptibility to vancomycin. United States, 1997-2000. Clin. Infect. Dis. 36:429-439. [DOI] [PubMed] [Google Scholar]
- 6.Haley, R. W., N. B. Cushion, F. C. Tenover, T. L. Bannerman, D. Dryer, J. Ross, P. J. Sanchez, and J. D. Siegel. 1995. Eradication of endemic methicillin-resistant Staphylococcus aureus infections from a neonatal intensive care unit. J. Infect. Dis. 171:614-624. [DOI] [PubMed] [Google Scholar]
- 7.Harmsen, D., H. Claus, W. Witte, J. Rothganger, H. Claus, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Healy, M., K. Reece, D. Walton, J. Huong, K. Shah, and D. P. Kontoyiannis. 2004. Identification to the species level and differentiation between strains of Aspergillus clinical isolates by automated repetitive-sequence-based PCR. J. Clin. Microbiol. 42:4016-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herold, B. C., L. C. Immergluck, M. C. Maranan, D. S. Lauderdale, R. E. Gaskin, S. Boyle-Vavra, C. D. Leitch, and R. S. Daum. 1998. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 279:593-598. [DOI] [PubMed] [Google Scholar]
- 10.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135-146. [DOI] [PubMed] [Google Scholar]
- 11.Karchmer, A. W. 2000. Nosocomial bloodstream infections: organisms, risk factors, and implication. Clin. Infect. Dis. 31(Suppl. 4):S139-S143. [DOI] [PubMed] [Google Scholar]
- 12.Koreen, L., S. V. Ramaswamy, E. A. Graviss, S. Naidich, J. M. Musser, and B. N. Kreiswirth. 2004. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 42:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leonard, R. B., J. Mayer, M. Sasser, M. L. Woods, B. R. Mooney, B. G. Brinton, P. L. Newcomb-Gayman, and K. C. Carroll. 1995. Comparison of MIDI Sherlock system and pulsed-field gel electrophoresis in characterizing strains of methicillin-resistant Staphylococcus aureus from a recent hospital outbreak. J. Clin. Microbiol. 33:2723-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maslow, J. N., M. E. Mulligan, and R. D. Arbeit. 1993. Molecular epidemiology: application of contemporary techniques to the typing of microorganisms. Clin. Infect. Dis. 17:153-164. [DOI] [PubMed] [Google Scholar]
- 16.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montesinos, I., E. Salido, T. Delgado, M. Cuervo, and A. Sierra. 2002. Epidemiologic genotyping of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis at a university hospital and comparison with antibiotyping and protein A and coagulase gene polymorphisms. J. Clin. Microbiol. 40:2119-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peacock, S. J., G. D. I. de Silva, A. Justice, A. Cowland, C. E. Moore, C. G. Winearls, and N. P. J. Day. 2002. Comparison of multilocus sequence typing and pulsed-field gel electrophoresis as tools for typing Staphylococcus aureus isolates in a microepidemiological setting. J. Clin. Microbiol. 40:3764-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salgado, C. D., B. M. Farr, and D. P. Calfee. 2003. Community-acquired methicillin-resistant Staphylococcus aureus: a meta-analysis of prevalence and risk factors. Clin. Infect. Dis. 36:131-139. [DOI] [PubMed] [Google Scholar]
- 20.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shopsin, B., M. Gomez, M. Waddington, M. Riehman, and B. N. Kreiswirth. 2000. Use of coagulase gene (coa) repeat region nucleotide sequences for typing of methicillin-resistant Staphylococcus aureus strains. J. Clin. Microbiol. 38:3453-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang, Y., M. G. Waddington, D. H. Smith, J. M. Manahan, P. C. Kohner, L. M. Highsmith, H. Li, F. R. Cockerill III, R. L. Thompson, S. O. Montgomery, and D. H. Persing. 2000. Comparison of protein A gene sequencing with pulsed-field gel electrophoresis and epidemiologic data for molecular typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 38:1347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trindale, P. A., J. A. McCulloch, G. A. Oliveira, and E. M. Mamizuka. 2003. Molecular techniques for MRSA typing: current issues and perspectives. Braz. J. Infect. Dis. 7:32-43. [DOI] [PubMed] [Google Scholar]
- 25.van Belkum, A., J. Kluytmans, W. van Leeuwen, R. Bax, W. Quint, E. Peters, A. Fluit, C. Vandenbroucke-Grauls, A. van den Brule, H. Koeleman, W. Melchers, J. Meis, A. Elaichouni, M. Vaneechoutte, F. Moenens, N. Maes, M. Struelens, F. Tenover, and H. Verbrugh. 1995. Multicenter evaluation of arbitrarily primed PCR for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 33:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Zee, A., H. Verbakel, J.-C. van Zon, I. Frenay, A. van Belkum, M. Peeters, A. Buiting, and A. Bergmans. 1999. Molecular genotyping of Staphylococcus aureus strains: comparison of repetitive element sequence-based PCR with various typing methods and isolation of a novel epidemicity marker. J. Clin. Microbiol. 37:342-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Versalovic, J., F. J. de Bruijn, and J. R. Lupski. 1998. Repetitive sequence-based PCR (rep-PCR) DNA fingerprinting of bacterial genomes, p. 437-453. Bacterial genomes: physical structure and analysis, vol. 34. Chapman and Hall, New York, N.Y. [Google Scholar]
- 28.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Versalovic, J., V. Kapur, E. O. Mason, Jr., U. Shah, T. Koeuth, J. R. Lupski, and J. M. Musser. 1993. Penicillin-resistant Streptococcus pneumoniae strains recovered in Houston: identification and molecular characterization of multiple clones. J. Infect. Dis. 167:850-856. [DOI] [PubMed] [Google Scholar]
- 30.Versalovic, J., and J. R. Lupski. 2002. Molecular detection and genotyping of pathogens: more accurate and rapid answers. Trends Microbiol. 10:S15-S21. [DOI] [PubMed] [Google Scholar]