Abstract
Human metapneumovirus (hMPV) is a newly identified human respiratory virus now recognized as a major respiratory pathogen of infants and children. To define the seroepidemiology of hMPV, we developed a novel enzyme-linked immunosorbent assay (ELISA) based on expression of the fusion protein of hMPV (hMPV F) in recombinant vesicular stomatitis virus (VSV). Western blot analysis using an hMPV F-specific antiserum confirmed the expression of hMPV in recombinant VSV. The ELISA is specific for hMPV F; antibody specific for the most closely related human paramyxovirus, respiratory syncytial virus, does not bind to hMPV F. Overall, 216 serum specimens were tested. The percentages of seropositive individuals were 89.1% in children ≤5 months old, 55.0% in children 6 to 11 months old, 36.0% in children 12 to 23 months old, 45.0% in children 24 to 47 months old, 77.3% in children 48 to 59 months old, 91.3% in children 5 to 10 years old, and 95.5% for individuals 11 to 20 years old. This is the first seroepidemiological survey of hMPV in the United States and the first analysis to determine the prevalence of antibody to a specific hMPV protein. The data suggest that exposure to hMPV is common in childhood and that hMPV F is an antigenic determinant of hMPV.
In 2001, van den Hoogen et al. reported the discovery of a novel virus associated with respiratory tract disease in children. Genetic and phylogenetic analysis revealed that this new agent was the first human pathogen in the metapneumovirus genus of the Paramyxoviridae, and the virus was called human metapneumovirus (hMPV) (22). Since the initial report of the discovery of hMPV, the virus has been identified worldwide (2, 4, 6, 14, 15, 25). The data from several studies suggest that hMPV is common and accounts for a significant proportion of respiratory tract disease in infants and children (7, 22, 26). hMPV has been circulating in the human population for at least 5 decades (22, 26).
The clinical manifestations associated with hMPV infection are similar to those observed with other respiratory viruses, in particular, respiratory syncytial virus (RSV). Upper respiratory tract infection and lower respiratory tract disease have been associated with hMPV infection. Coughing, wheezing, hypoxia and abnormalities on chest radiographs occur frequently in children infected with hMPV (3, 7, 24, 26). Asymptomatic infections are rare in young children (26). hMPV has also been detected in adults with clinical evidence of respiratory tract infection (8, 18).
Based on comparisons with other paramyxoviruses, the hMPV gene order and gene sequence suggest that the virus encodes three virion envelope glycoproteins, the fusion (F), attachment (G), and short hydrophobic glycoproteins. The F protein shares 33% identity with the immunodominant F protein of the most closely related human paramyxovirus RSV (21). Like the analogous protein (RSV F) in RSV, the hMPV F glycoprotein may elicit antibodies during infection. Reports from The Netherlands, United States, Canada, and elsewhere suggest that there are at least two major genotypes or clades of hMPV (1, 7, 22, 26).
Serological evidence indicates that hMPV infection is common in childhood. Reports from The Netherlands and Japan suggest that by the age of 5, nearly all individuals tested have evidence of exposure to hMPV (5, 22). The results of these studies were based on indirect immunofluorescence assays with hMPV-infected monkey kidney cells used as antigen. While the immunofluorescence assays provide some information, they have several limitations. The interpretation of immunofluorescence assays is based on subjective criteria, and these assays are prone to batch-to-batch variation. Furthermore, this assay cannot be used to study the antibody response to specific viral glycoproteins. To further study the seroepidemiology of hMPV and to determine the presence of antibody to a specific hMPV protein, we developed an enzyme-linked immunosorbent assay (ELISA) based on recombinant expression of the hMPV F glycoprotein. We chose the hMPV F protein because the corresponding protein (F) of the closely related paramyxovirus, RSV, is the immunodominant protein of the virus. To this end, we cloned the hMPV F gene from a clinical sample into recombinant vesicular stomatitis virus (VSV). This expression system was chosen for several reasons. Foreign glycoproteins are produced in relatively high amounts (∼1% of total cellular protein) (17). Foreign viral glycoproteins are faithfully expressed, modified properly, and are biologically active (11, 12). This strongly suggests that these foreign viral glycoproteins maintain their antigenic configuration. This is essential for the development of an ELISA specific for antibodies that bind hMPV F. VSV is a virus of livestock and horses; human infection is rare. The prevalence of antibodies to the VSV vector in the human population is low (20). Therefore, when recombinant VSV is used as an antigen for an ELISA, reactivity to the VSV vector is predicted to be low. Here, we describe a novel ELISA system to detect hMPV F-specific antibodies and to define the seroepidemiology of hMPV in Connecticut.
MATERIALS AND METHODS
Cloning of the hMPV F gene.
The hMPV F gene was amplified from an hMPV-positive specimen identified previously (6). RNA from this hMPV-positive specimen was extracted by using a QiaAmp viral RNA mini kit (QIAGEN Inc., Valencia, Calif.), and cDNA was produced by using Moloney murine leukemia virus reverse transcriptase (New England Biolabs, Beverly, Mass.) and random hexamer primers. The forward primer 5′-GGACGCGTCTCGAGACCATGTCTTGGAAAGTGG and reverse primer 5′-CGGGCTAGCCTAATTATGTGGTATGAAGCCATTGTTTGTG were used for amplification of the hMPV F gene. These primers correspond to nucleotides 3052 to 3067 and 4641 to 4671 of the hMPV F gene (GenBank accession number AF371337). Restriction sites, C/G clamps, and an obligatory ACC (forward primer) required for VSV gene expression are underlined. The initiation codon ATG (forward primer) and termination codon (reverse codon, antisense strand) are shown in bold. Following digestion with the appropriate restriction endonucleases, the hMPV F gene was ligated into a plasmid containing the full-length genome of VSV (17). The ligation reactions were transformed into bacteria (Escherichia coli strain DH5α), and colonies containing the desired plasmid were confirmed by PCR and sequence analysis.
Recovery of recombinant VSV expressing hMPV F (VSV-hMPV F).
Recovery of recombinant VSV was performed as previously described (11, 17). Briefly, individual plasmids containing the VSV-N gene, the VSV-P gene, the VSV-L gene, and the full-length VSV genome containing the hMPV F gene were transfected into baby hamster kidney (BHK) cells (BHK-21; American Type Tissue Collection) infected with vaccinia virus (strain WR) expressing T7 RNA polymerase. Each of the VSV genes and the full-length VSV genome in the transfected plasmids were under T7 promoter control. Two days following the transfection, the infected cell supernatants were filtered (to remove vaccinia virus) and passaged on BHK cells. Recovered recombinant VSV was identified by the presence of cytopathic effects. The recovery of a recombinant VSV containing the hMPV F gene was confirmed by reverse transcription-PCR (RT-PCR) and sequence analysis. The preparation of working stocks of VSV and VSV recombinants has been described previously (11).
hMPV F-specific antibody.
A synthetic peptide, CQNAGSTVYYPNEKDCETRG-COOH, corresponding to amino acids 311 to 330 of the hMPV F protein (GenBank accession number AF371337), was synthesized at the W. M. Keck Biotechnology Resource Center, Yale University. To prepare antigen for animal inoculations, the synthetic peptide was first conjugated to maleimide-activated mariculture keyhole limpet hemocyanin (Pierce, Rockford, Ill.) according to the manufacturer's specifications. For primary immunization of rabbits, the conjugated peptide was emulsified in complete Freund's adjuvant, and the animal was inoculated subcutaneously with ∼400 μl (100 μl in four sites). Thereafter, the conjugated antigen was mixed with incomplete Freund's adjuvant. The animal was immunized with 400 μl at an interval of at least 4 weeks for a total of nine immunizations. Immunizations and bleeds were performed by the Yale University Veterinary Clinical Services and followed approved standard protocols.
Western blot analysis.
Cell lysates infected with wild-type VSV (VSV-wt) and VSV-hMPV F were prepared as described elsewhere (10). Proteins were separated by electrophoresis through a 7.5% polyacrylamide gel under reducing denaturing conditions. Following electrophoresis, proteins were transferred to a nitrocellulose filter as described elsewhere (16). The filters were dried, blocked in 5% BLOTTO (Carnation nonfat milk in phosphate-buffered saline [PBS]) and incubated with a rabbit anti-hMPV F peptide antiserum (described above) diluted 1:100 in 5% BLOTTO or rabbit anti-VSV antiserum (kindly provided by J. Rose, Department of Pathology, Yale University School of Medicine) diluted 1:5,000 in 5% BLOTTO. Following several washes, the filters were incubated with a horseradish peroxidase-conjugated goat anti-rabbit antibody (Jackson ImmunoResearch Laboratories, West Grove, Pa.) diluted 1:10,000 in 150 mM NaCl, 50 mM Tris-HCl (pH 7.6). Binding of the secondary antibody was detected by using an ECL Western blotting system (Amersham Pharmacia Biotech Inc., Piscataway, N.J.).
ELISA.
Antigen for ELISAs was prepared from infected cell culture material. BHK cells or HEp-2 cells (human larynx carcinoma cells; American Type Tissue Collection) were infected with VSV-wt, VSV-hMPV F (BHK), or RSV A2 (HEp-2) at a multiplicity of infection of >5. (RSV A2 was supplied by Peter Collins, National Institutes of Health.) hMPV from a clinical specimen that tested positive by RT-PCR (7) was propagated on LLC-MK-2 cells as described elsewhere (1). Monolayers were scraped when ∼75% of the cells demonstrated cytopathic effects or when viral antigen (hMPV) was detected by indirect immunofluorescence. Cells were centrifuged and resuspended in 0.5% NP-40 detergent and incubated on ice for 10 min. Cell lysate suspensions were centrifuged (Eppendorf microcentrifuge) for 1 min at 4°C to pellet cell nuclei. Supernatants were collected and stored at −20°C. ELISAs were performed as described previously (10). Briefly, 96-well plates (Costar EIA/RIA 96-well plates; Corning Inc., Corning, N.Y.) were coated with infected cell lysates (1.7 mg of protein/ml) and incubated overnight at 37°C (binding of an anti-VSV antiserum was optimal at this concentration of lysate). Lysate solutions were removed, and plates were blocked with PBS containing 0.5% gelatin and 0.05% Tween 20 for 1 h at 37°C. Wells were washed with PBS containing 0.05% Tween (PT). Serial dilutions of human serum or goat anti-RSV antibody (Fitzgerald Industries International, Inc., Concord, Mass.), in PBS containing 0.5% gelatin, 0.05% Tween 20, and 2% fetal bovine serum (PGTF), were added to the wells and allowed to bind for 1 h at 37°C. Following five washes with PT solution, a species-appropriate secondary horseradish peroxidase-conjugated antibody (Jackson ImmunoResearch Laboratories) diluted in PGTF (1:20,000) was added to each well, and plates were incubated for 1 h at 37°C. After five washes with PT solution, bound secondary antibody was detected with 3,3′,5,5′, tetramethyl benzidine (Pierce). Chromogenic reactions were stopped with 5N H3PO4. Optical densities were read at 450 nm (OD450; DynaTech MR5000). For antibody subtraction studies, serum was incubated with RSV lysate or mock-infected HEp-2 cell lysate at 37°C for 1 h prior to ELISA analysis.
Serum collection, ELISA interpretation, and statistical analysis.
Human serum specimens from individuals <20 years old were collected from the Clinical Chemistry Laboratory, Yale-New Haven Hospital. Other than the age of the individual from whom the serum was obtained, no other clinical information was available. A minimum of 20 serum samples was collected per age group. Serum samples were collected from June 2003 to March 2004. The criteria for a positive ELISA for hMPV were defined as an OD450 of >0.2 (1:32) and a VSV-hMPV F value greater than the VSV value at a fourfold-lower dilution. Confidence intervals equal the observed proportion ± 1.96 times the standard error.
RESULTS
Cloning of the hMPV F gene into recombinant VSV.
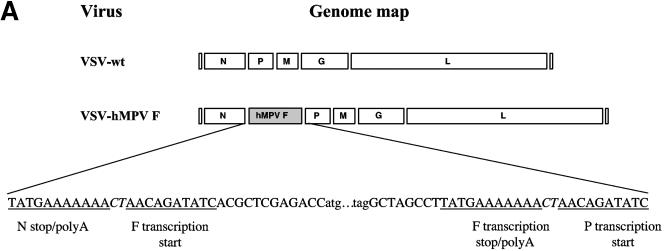
As part of an ongoing epidemiological study, hMPV-positive respiratory specimens were identified by RT-PCR and confirmed by sequence analysis. From one of these specimens, a clade B isolate (strain NH379) (7), the hMPV F gene was amplified by RT-PCR and cloned into a plasmid containing the full-length genome of VSV, and recombinant virus (VSV-hMPV F) was recovered by reverse genetics (17). The hMPV F gene was inserted into the N/P gene junction and contained the obligatory transcriptional regulatory elements required for faithful gene expression (Fig. 1A) (17). The level of gene expression in VSV is maximal at the 3′ (N) end of the genome and declines through each gene junction through the 5′ (L) end of the genome. Therefore, cloning the hMPV F gene between N and P would ensure relatively high expression in the recombinant virus (17).
FIG. 1.
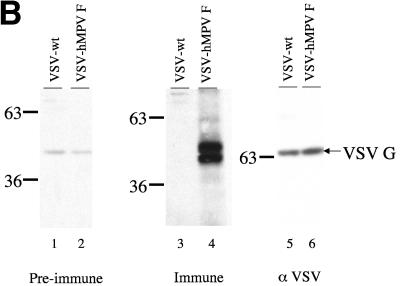
(A) Cloning and expression of the hMPV F gene in recombinant VSV. The hMPV F gene from a clinical specimen was amplified by RT-PCR and cloned into a plasmid containing the full-length genome of VSV. Recombinant VSV with the hMPV F gene inserted between the VSV N and P genes (VSV-hMPV F) was recovered by using reverse genetics. The genomic maps (3′ to 5′ orientation for the negative-strand RNA genome) of VSV-wt and VSV-hMPV are displayed in panel A. Sequences of the gene junctions (corresponding the positive-sense cDNA) are shown below the genomic maps. The transcription start and stop/poly A signals are indicated. The hMPV initiation codon (atg) and termination codon (tag) and the obligatory intergenic CT are shown. (B) Western blot of VSV-wt and VSV-hMPV F lysates with rabbit anti-hMPV F peptide serum. Proteins of infected cells were separated by polyacrylamide gel electrophoresis under reducing, denaturing conditions, transferred to nitrocellulose, and blotted with the rabbit anti-hMPV F peptide antiserum (preimmune or immune) or rabbit anti-VSV antiserum. Antibody binding was detected following incubation with a horseradish peroxidase-conjugated anti-rabbit antibody and enhanced chemiluminescence. Lane 1, VSV-wt and preimmune serum; lane 2, VSV-hMPV F and preimmune serum; lane 3, VSV-wt and immune serum; lane 4, VSV-hMPV F and immune serum; lane 5, VSV-wt and rabbit anti-VSV serum; lane 6, VSV-hMPV F and anti-VSV serum. The positions of the molecular weight markers for each panel are indicated.
The presence of the full-length, authentic copy of the hMPV F gene in the recombinant viral genome was confirmed by RT-PCR and sequence analysis of recovered virus (data not shown). To demonstrate that hMPV F was expressed in recombinant virus, a rabbit anti-hMPV F antiserum was developed. A synthetic peptide representing amino acids 311 to 330 of hMPV F was used as antigen to immunize the animal. This region of hMPV F was chosen for several reasons. This is a relatively hydrophilic domain and may be more antigenic than hydrophobic domains. This region is not conserved among the fusion glycoproteins of paramyxoviruses, and, therefore, antibodies induced by this peptide should be specific for hMPV. Lastly, this region is conserved among isolates of the two major genotypes of hMPV. Binding of the rabbit anti-hMPV F peptide antibodies to both VSV-wt and VSV-hMPV F was assayed by Western blot analysis. Figure 1B shows the detection of two bands in the VSV-hMPV F-infected cell lysates that are not present in the VSV-wt-infected cell lysates. Based on the apparent molecular weights, these two bands likely represent the cleaved and uncleaved forms of hMPV F. The 58-kDa hMPV F protein, like the fusion protein of other paramyxoviruses, is presumably cleaved to generate a ∼10-kDa N-terminal F2 and a ∼46-kDA C-terminal F1 (a 2-kDa signal peptide at the N terminus is thought to be cleaved to form the mature protein). The synthetic peptide used in the rabbit immunization corresponds to a region in F1. No specific binding was detected with preimmune serum. Western blot analysis with a VSV-specific antiserum reveals that equal amounts of the viral vector were present in the lysates of both VSV-wt and VSV-hMPV F.
hMPV F-specific ELISA based on recombinant VSV.
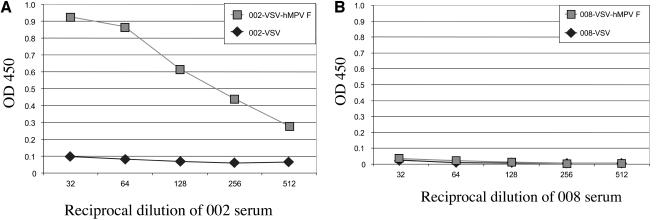
The VSV-hMPV F ELISA was used to measure hMPV-specific antibody titers in human serum. Although the prevalence of antibody to the VSV vector in the human population was considered to be very low, specific or nonspecific binding to the VSV antigens needed to be controlled for in the ELISA analysis. Therefore, each serum tested was assayed for binding to both VSV-wt and VSV-hMPV F lysates. For a vast majority (>95%) of human serum samples tested, the OD450 (at a dilution of 1:32) was ≤0.2 for binding to the VSV-wt lysate, and therefore a cutoff of 0.2 was used as one of the criteria for a positive ELISA for hMPV. For ≤5% of the serum samples tested, the OD450 was ≥0.2 for binding to the VSV-wt lysate. However, the OD450 for the VSV-hMPV F ELISA had to be greater than the VSV value at a fourfold-lower dilution for the sample to be considered positive for hMPV. The lysates used as antigen were standardized for protein concentration and binding of an anti-VSV antiserum. With this standardization, the assay is highly reproducible. The reproducibility of the assay was confirmed by using the anti-hMPV F peptide antibody (data not shown). ELISA results for both a representative seropositive and a representative seronegative individual are shown in Fig. 2A and B, respectively. Assays using serum or plasma gave identical results (data not shown).
FIG. 2.
hMPV-seropositive and hMPV-seronegative serum samples based on VSV-hMPV F recombinant ELISAs. VSV-wt lysate or VSV-hMPV F lysate was used as antigen in an ELISA of human serum. Serum specimens from subjects 002 (A) and 008 (B) were serially diluted and allowed to bind antigen. Binding was measured by optical densitometry following incubation with a conjugated secondary antibody and a chromogenic substrate.
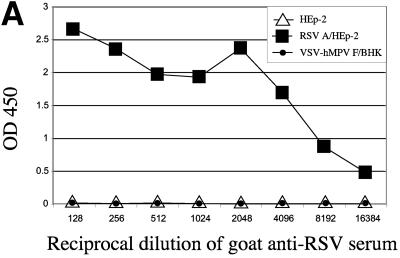
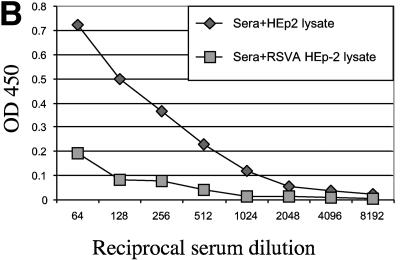
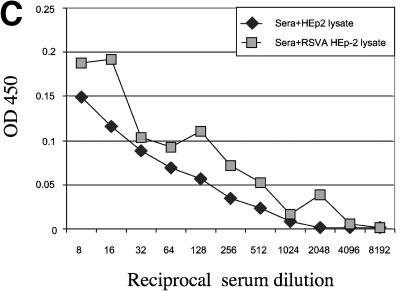
A potential shortcoming of an hMPV F ELISA is that this method may detect antibodies that bind to RSV F, thereby limiting the specificity of this assay. RSV is the most closely related human paramyxovirus, and the hMPV F and RSV F share 33% homology (21). Therefore, the potential binding of RSV-specific antibodies to the VSV-hMPV F lysates was evaluated in two ways. First, the binding of an RSV-specific antibody to VSV-hMPV F was assayed. As shown in Fig. 3A, a goat anti-RSV antiserum had a high titer of RSV-specific antibody but failed to bind VSV-hMPV F. This evidence suggested that RSV-specific antibodies do not cross-react with hMPV F. Second, serum in which RSV antibodies were subtracted was assayed for VSV-hMPV F-specific binding. The results of ELISAs in which RSV lysates were used as antigen are shown in Fig. 3B. Serum incubated with RSV lysates (Fig. 3B, sera+RSV HEp-2 lysate) had dramatically reduced binding to RSV antigens compared with serum incubated with HEp-2 lysates (Fig. 3B). However, when the same two incubated sera were assayed for VSV-hMPV F binding, no differences in the ELISA profiles were observed (Fig. 3C). These findings suggest that this ELISA is specific for hMPV antibodies.
FIG. 3.
Specificity of the hMPV ELISA based on recombinant expression of the hMPV F gene. (A) Lysates of VSV-hMPV F-infected BHK cells, RSV A-infected HEp-2 cells, and noninfected HEp-2 cells were used as antigens in ELISAs. Serial dilutions of a goat anti-RSV antiserum were incubated with the lysates, and binding was measured by optical densitometry following incubation with a conjugated secondary antibody. (B) Human serum with RSV-specific antibodies was incubated with RSV A-infected HEp-2 cell lysates or mock-infected HEp-2 cell lysates prior to ELISA analysis. RSV A lysate was used as antigen, and binding was measured following incubation with an anti-human horseradish peroxidase-conjugated antibody as described above. (C) RSV-subtracted antiserum binds to VSV-hMPV F lysates. The RSV-specific antibody-subtracted serum and mock-subtracted serum described in panel B above was used in an ELISA in which VSV-hMPV F-infected BHK cell lysates were used as antigen. Binding was measured as described above.
Seroprevalence of hMPV-specific antibodies.
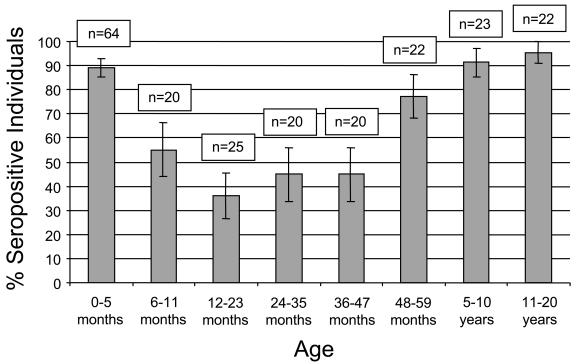
The VSV-wt and VSV-hMPV F ELISAs were used to measure the prevalence of hMPV-specific antibodies in the pediatric population. As presented in Fig. 4, the percentage of children of ≤5 months old with hMPV-specific antibody is 89.1% (95% confidence interval [CI], 85.2 to 93.0%). The percentage of seropositive individuals drops to 55.0% (95% CI, 43.9 to 66.1%) in children 6 to 11 months of age and to 36.0% (95% CI, 26.4 to 45.6%) in children 12 to 23 months of age. The percentage of seropositive children 24 to 35 months old and 36 to 47 months old is 45.0% (95% CI, 33.9 to 56.15%) in both age groups. The percentages of seropositive individuals are 77.3% (95% CI, 68.3 to 86.2%) for children 48 to 59 months old, 91.3% (95% CI, 85.4 to 97.2%) for children 5 to 10 years old, and 95.5% (95% CI, 91.0 to 99.9%) for individuals 11 to 20 years old. Phylogenetic analysis based on the F gene of hMPV isolates from our group and others suggests that at least two distinct genotypes of hMPV circulate in the community (7, 22). Whether these genotypes represent distinct serotypes is unknown. To address this issue, we cloned the F gene from a clade A isolate (strain NH539) (7) into the VSV expression vector and tested a subset of serum samples for binding antibody by ELISA. (The ELISA results presented above were based on the F gene of a clade B isolate.) Eleven sera that tested positive by ELISA for the clade B hMPV F also tested positive for the clade A hMPV F. Likewise, nine sera that tested negative for the clade B hMPV also tested negative in the clade A hMPV ELISA (data not shown). There was no quantitative difference in the ODs for binding of antibody to the VSV-hMPV F (A) lysates and the VSV-hMPV F (B) lysates for any given serum.
FIG. 4.
Rates of hMPV seropositivity based on age group. The percentage of hMPV seropositivity for each age group is indicated. The number of serum samples tested in each age group is indicated (n). Bars represent 95% CIs.
In order to confirm the sensitivity of this recombinant antigen ELISA in detecting hMPV-positive sera, we developed an ELISA based on hMPV-infected (MK2-LLC) cell lysates. Uninfected cells were used as control antigens. The hMPV-infected cell ELISA detected anti-hMPV F peptide antibodies and hMPV antibodies in sera that tested positive with the recombinant antigen ELISA. Of 20 serum specimens that tested negative for hMPV F-specific antibodies (using the recombinant VSV-hMPV F antigen ELISA), all tested negative in the hMPV-infected cell ELISA, thus establishing the sensitivity of the recombinant antigen ELISA.
DISCUSSION
This is the first report of the seroepidemiology of hMPV in the United States and the first investigation to examine the prevalence of antibody to a specific hMPV glycoprotein. Infection with hMPV is common in our population. More than 90% of individuals of ≥5 years of age had evidence of exposure to hMPV. The dynamics of the seroepidemiology of hMPV in the pediatric population is complex. The decline in the proportion of seropositive individuals during the first year of life likely represents waning maternally acquired antibody, as observed with RSV (9). However, this decline is superimposed on an increase in antibody as results of infection during the first year of life. In our previous study of hMPV infection in children <5 years old, a majority (60.4%) of hMPV-infected children were <1 year of age (7). However, this prior study was not population based but used respiratory specimens from the hospital's Clinical Virology Laboratory. Consequently, the true incidence of hMPV infection in children of <1 year of age remains to be determined.
There appear to be two periods of acquisition of hMPV infection in childhood. The first period occurs within the first 3 years of life. The percentage of individuals who are seropositive is essentially constant at 35 to 45% for children 12 to 47 months of age. The second period occurs in children of >48 months of age. The percentage of seropositive individuals increases to 77.3% (95% CI, 68.3 to 86.2%) in children 48 to 59 months old and to >90% in children who are >5 years old. This second peak likely reflects increased exposure to the virus, perhaps at day care or preschool environments.
Our findings are similar to those reported elsewhere. In the initial report of the discovery of hMPV, van den Hoogen et al., using an immunofluorescence assay and a viral neutralization assay, found that by the age of 5 years, nearly all individuals have evidence of hMPV infection (22). Of 40 Israeli children ≤24 months old, 52% had serological evidence of hMPV infection (27). The percentage of seropositive individuals in a Japanese cohort was >90% in those ≥5 years old (5). However, only 17% (3 of 17) of children 6 months to 1 year of age had serological evidence of hMPV infection, whereas in our study the percentage of seropositive children 6 to 11 months old was 55.0% (95% CI, 43.9 to 66.1%). This difference may be explained by variation in the circulation of hMPV or by the sensitivities of the methodologies used to detect hMPV antibodies. In the Japanese study, as well as The Netherlands study, hMPV-specific antibodies were detected by assays using hMPV-infected monkey kidney cells. These methods require culture of primary monkey kidney cells and are subject to batch-to-batch variability.
The ELISA-based system has many distinct advantages over the methods used in prior studies. The amount of hMPV-specific antigen can be standardized for each assay, antibody to genotype-specific viral glycoproteins can be measured, and the results are based on defined criteria rather than subjective determinations of a positive result in an immunofluorescence study. Expression of hMPV F in recombinant VSV was validated by Western blot analysis by using a rabbit serum raised against a peptide corresponding to a hydrophilic domain of the C-terminal F1. The two bands detected by Western blotting are most likely cleaved and uncleaved forms of hMPV F. An understanding of the biochemical properties of hMPV F during infection is lacking, and therefore the relative amounts of the cleaved and uncleaved forms in hMPV-infected cells remain unknown. Our findings support the hypothesis that hMPV F is an antigenic virion glycoprotein.
The specificity and sensitivity of our ELISA were established. Antibodies to the F protein of the closely related paramyxovirus RSV were not detected with the recombinant hMPV F ELISA. The sensitivity of our assay was confirmed by using a whole-virus ELISA, based on hMPV-infected cell lysates, as a gold standard. Assays to detect virus-specific antibody are frequently based on immunodominant glycoprotein(s). In fact, a peptide-based enzyme immunoassay for detection of antibodies to the G protein of RSV is more sensitive than conventional serological tests (13). This indicates that the VSV-hMPV F-based ELISA is an ideal tool to screen for hMPV exposure and infection.
Several groups have determined that at least two major genotypes of hMPV circulate concurrently in the community (2, 7, 22, 26), and animal studies suggest that the two genotypes may represent distinct serotypes (23). The 100% concordance between the results of the hMPV F clade A ELISA and the hMPV clade B ELISA suggests that there is significant antigenic cross-reactivity between the F glycoprotein of the two clades. This is not surprising as the hMPV F proteins of clade A and clade B are >90% identical (unpublished data). This also suggests that the two putative serotypes cannot be distinguished based on the fusion glycoprotein.
The present study also demonstrates the usefulness of the VSV expression system for the development of diagnostic reagents. An additional advantage of the VSV expression system is that, in general, the prevalence of antibodies specific for the VSV vector in the human population is low. VSV is a virus that infects animals, mainly livestock and horses, and human infection with VSV is, for the most part, rare. However, there are regions in Central and South America where VSV infection is endemic in the human population. This could potentially limit the utility of the system for individuals from those areas (19). We controlled for the possibility that some individuals may have antibodies that bind to the viral vector. Each serum specimen tested was screened for antibodies for hMPV F and the VSV vector. VSV is considered a controlled organism by the United States Department of Agriculture and is considered a foreign animal disease by many countries throughout the world. Any use of the virus should include the necessary precautions.
In conclusion, our results indicate that infection with hMPV is common. Most individuals are likely infected during childhood. Specific antibodies to hMPV F are transmitted transplacentally and also develop after infection with hMPV. Further studies are required to determine the immunogenic domains of hMPV F and whether hMPV F-specific antibodies are protective.
Acknowledgments
We are indebted to George Miller for his continued support, intellectual and scientific input and exchange and critical review of the data and manuscript. We are indebted to Linda Buonocore for her technical assistance, the staff of the Clinical Chemistry Laboratory at Yale-New Haven Hospital for their assistance in identification of serum specimens, and to Eugene D. Shapiro for his thoughtful review of the manuscript. The collection of specimens and demographic data was approved by the Yale University Human Investigation Committee.
This work was supported by the Patrick and Catherine Weldon Donaghue Medical Research Foundation. This work was also supported in part by the Yale Children's Clinical Research Center grant M01-RR06022, General Clinical Research Centers Program, National Center for Research Resources, National Institutes of Health, and by National Institutes of Health grants T32 AI07210-20 and F32 AI055151-01 (both to F.E.).
REFERENCES
- 1.Biacchesi, S., M. H. Skiadopoulos, G. Boivin, C. T. Hanson, B. R. Murphy, P. L. Collins, and U. J. Buchholz. 2003. Genetic diversity between human metapneumovirus subgroups. Virology 315:1-9. [DOI] [PubMed] [Google Scholar]
- 2.Boivin, G., Y. Abed, G. Pelletier, L. Ruel, D. Moisan, S. Cote, T. C. Peret, D. D. Erdman, and L. J. Anderson. 2002. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J. Infect. Dis. 186:1330-1334. [DOI] [PubMed] [Google Scholar]
- 3.Boivin, G., G. De Serres, S. Cote, R. Gilca, Y. Abed, L. Rochette, M. G. Bergeron, and P. Dery. 2003. Human metapneumovirus infections in hospitalized children. Emerg. Infect. Dis. 9:634-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan, P. K., J. S. Tam, C. W. Lam, E. Chan, A. Wu, C. K. Li, T. A. Buckley, K. C. Ng, G. M. Joynt, F. W. Cheng, K. F. To, N. Lee, D. S. Hui, J. L. Cheung, I. Chu, E. Liu, S. S. Chung, and J. J. Sung. 2003. Human metapneumovirus detection in patients with severe acute respiratory syndrome. Emerg. Infect. Dis. 9:1058-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ebihara, T., R. Endo, H. Kikuta, N. Ishiguro, M. Yoshioka, X. Ma, and K. Kobayashi. 2003. Seroprevalence of human metapneumovirus in Japan. J. Med. Virol. 70:281-283. [DOI] [PubMed] [Google Scholar]
- 6.Esper, F., D. Boucher, C. Weibel, R. A. Martinello, and J. S. Kahn. 2003. Human metapneumovirus infection in the United States: clinical manifestations associated with a newly emerging respiratory infection in children. Pediatrics 111:1407-1410. [DOI] [PubMed] [Google Scholar]
- 7.Esper, F., R. A. Martinello, D. Boucher, C. Weibel, D. Ferguson, M. L. Landry, and J. S. Kahn. 2004. A 1-year experience with human metapneumovirus in children aged <5 years. J. Infect. Dis. 189:1388-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falsey, A. R., D. Erdman, L. J. Anderson, and E. E. Walsh. 2003. Human metapneumovirus infections in young and elderly adults. J. Infect. Dis. 187:785-790. [DOI] [PubMed] [Google Scholar]
- 9.Glezen, W. P., A. Paredes, J. E. Allison, L. H. Taber, and A. L. Frank. 1981. Risk of respiratory syncytial virus infection for infants from low-income families in relationship to age, sex, ethnic group, and maternal antibody level. J. Pediatr. 98:708-715. [DOI] [PubMed] [Google Scholar]
- 10.Kahn, J. S., A. Roberts, C. Weibel, L. Buonocore, and J. K. Rose. 2001. Replication-competent or attenuated, nonpropagating vesicular stomatitis viruses expressing respiratory syncytial virus (RSV) antigens protect mice against RSV challenge. J. Virol. 75:11079-11087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kahn, J. S., M. J. Schnell, L. Buonocore, and J. K. Rose. 1999. Recombinant vesicular stomatitis virus expressing respiratory syncytial virus (RSV) glycoproteins: RSV fusion protein can mediate infection and cell fusion. Virology 254:81-91. [DOI] [PubMed] [Google Scholar]
- 12.Kretzschmar, E., L. Buonocore, M. J. Schnell, and J. K. Rose. 1997. High-efficiency incorporation of functional influenza virus glycoproteins into recombinant vesicular stomatitis viruses. J. Virol. 71:5982-5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langedijk, J. P., A. H. Brandenburg, W. G. Middel, A. Osterhaus, R. H. Meloen, and J. T. van Oirschot. 1997. A subtype-specific peptide-based enzyme immunoassay for detection of antibodies to the G protein of human respiratory syncytial virus is more sensitive than routine serological tests. J. Clin. Microbiol. 35:1656-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maggi, F., M. Pifferi, M. Vatteroni, C. Fornai, E. Tempestini, S. Anzilotti, L. Lanini, E. Andreoli, V. Ragazzo, M. Pistello, S. Specter, and M. Bendinelli. 2003. Human metapneumovirus associated with respiratory tract infections in a 3-year study of nasal swabs from infants in Italy. J. Clin. Microbiol. 41:2987-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nissen, M. D., D. J. Siebert, I. M. Mackay, T. P. Sloots, and S. J. Withers. 2002. Evidence of human metapneumovirus in Australian children. Med. J. Aust. 176:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Schnell, M. J., L. Buonocore, M. A. Whitt, and J. K. Rose. 1996. The minimal conserved transcription stop-start signal promotes stable expression of a foreign gene in vesicular stomatitis virus. J. Virol. 70:2318-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stockton, J., I. Stephenson, D. Fleming, and M. Zambon. 2002. Human metapneumovirus as a cause of community-acquired respiratory illness. Emerg. Infect. Dis. 8:897-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tesh, R. B., J. Boshell, G. B. Modi, A. Morales, D. G. Young, A. Corredor, C. Ferro de Carrasquilla, C. de Rodriguez, L. L. Walters, and M. O. Gaitan. 1987. Natural infection of humans, animals, and phlebotomine sand flies with the Alagoas serotype of vesicular stomatitis virus in Colombia. Am. J. Trop. Med. Hyg. 36:653-661. [DOI] [PubMed] [Google Scholar]
- 20.Tesh, R. B., P. H. Peralta, and K. M. Johnson. 1969. Ecologic studies of vesicular stomatitis virus. I. Prevalence of infection among animals and humans living in an area of endemic VSV activity. Am. J. Epidemiol. 90:255-261. [DOI] [PubMed] [Google Scholar]
- 21.van den Hoogen, B. G., T. M. Bestebroer, A. D. Osterhaus, and R. A. Fouchier. 2002. Analysis of the genomic sequence of a human metapneumovirus. Virology 295:119-132. [DOI] [PubMed] [Google Scholar]
- 22.van den Hoogen, B. G., J. C. de Jong, J. Groen, T. Kuiken, R. de Groot, R. A. Fouchier, and A. D. Osterhaus. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van den Hoogen, B. G., S. Herfst, L. Sprong, P. A. Cane, E. Forleo-Neto, R. L. de Swart, A. D. Osterhaus, and R. A. Fouchier. 2004. Antigenic and genetic variability of human metapneumoviruses. Emerg. Infect. Dis. 10:658-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van den Hoogen, B. G., G. J. van Doornum, J. C. Fockens, J. J. Cornelissen, W. E. Beyer, R. de Groot, A. D. Osterhaus, and R. A. Fouchier. 2003. Prevalence and clinical symptoms of human metapneumovirus infection in hospitalized patients. J. Infect. Dis. 188:1571-1577. [DOI] [PubMed] [Google Scholar]
- 25.Vicenti, D., G. Cilla, M. Montes, and E. Perez-Trallero. 2003. Human metapneumovirus and community-acquired respiratory illness in children. Emerg. Infect. Dis. 9:602-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams, J. V., P. A. Harris, S. J. Tollefson, L. L. Halburnt-Rush, J. M. Pingsterhaus, K. M. Edwards, P. F. Wright, and J. E. Crowe, Jr. 2004. Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N. Engl. J. Med. 350:443-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolf, D. G., Z. Zakay-Rones, A. Fadeela, D. Greenberg, and R. Dagan. 2003. High seroprevalence of human metapneumovirus among young children in Israel. J. Infect. Dis. 188:1865-1867. [DOI] [PubMed] [Google Scholar]