Abstract
The PulseNet National Database, established by the Centers for Disease Control and Prevention in 1996, consists of pulsed-field gel electrophoresis (PFGE) patterns obtained from isolates of food-borne pathogens (currently Escherichia coli O157:H7, Salmonella, Shigella, and Listeria) and textual information about the isolates. Electronic images and accompanying text are submitted from over 60 U.S. public health and food regulatory agency laboratories. The PFGE patterns are generated according to highly standardized PFGE protocols. Normalization and accurate comparison of gel images require the use of a well-characterized size standard in at least three lanes of each gel. Originally, a well-characterized strain of each organism was chosen as the reference standard for that particular database. The increasing number of databases, difficulty in identifying an organism-specific standard for each database, the increased range of band sizes generated by the use of additional restriction endonucleases, and the maintenance of many different organism-specific strains encouraged us to search for a more versatile and universal DNA size marker. A Salmonella serotype Braenderup strain (H9812) was chosen as the universal size standard. This strain was subjected to rigorous testing in our laboratories to ensure that it met the desired criteria, including coverage of a wide range of DNA fragment sizes, even distribution of bands, and stability of the PFGE pattern. The strategy used to convert and compare data generated by the new and old reference standards is described.
The use of molecular subtyping methods for the characterization and grouping of organisms based on their genotypic characteristics has become commonplace in public health laboratories. Of the many molecular methods currently available, macrorestriction analysis by pulsed-field gel electrophoresis (PFGE) has been shown to be particularly useful for the clustering and differentiation of many bacterial pathogens (3, 5, 7, 8, 10-12, 15, 18, 20). Although the sensitivity and discriminatory power of PFGE depend on the organism being subtyped and the restriction enzyme used, its high epidemiologic relevance has made this technique the “gold standard” for subtyping foodborne bacterial pathogens (2, 4, 8, 13, 14, 19-21).
Since 1996, PulseNet (the National Molecular Subtyping Network for Foodborne Disease Surveillance) has used PFGE analysis to rapidly identify clusters of food-borne illness in the United States (21). PulseNet includes public health laboratories and federal food regulatory agencies (Food and Drug Administration and U.S. Department of Agriculture-Food Safety and Inspection Service), which submit patterns via the Internet to the PulseNet National Database maintained at the Centers for Disease Control and Prevention (CDC) in Atlanta, Ga. Comparison of patterns is possible because all the PulseNet participating laboratories perform PFGE analysis using the same highly standardized protocols, including the requirement to use the same reference or size standard, to generate the DNA fingerprints from pathogens such as E. coli O157:H7, Salmonella, Shigella, and Listeria monocytogenes. The software, analysis parameters, and the nomenclature used to name patterns also are standardized (21).
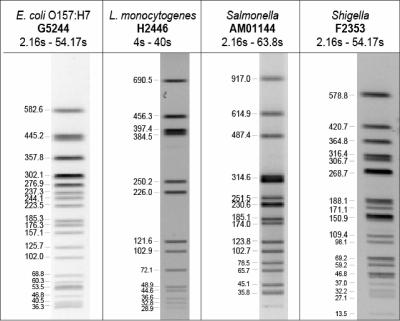
To ensure that PFGE patterns from different gels and even different laboratories can be compared accurately in the national database, a molecular size standard is included in at least three lanes of every PFGE gel that is run. The original standard strains were E. coli O157:H7 strain G5244, Salmonella serotype Newport strain AM01144, Shigella sonnei strain F2353, and L. monocytogenes strain H2446 for the E. coli, Salmonella, Shigella, and Listeria databases respectively; all were restricted with XbaI (Roche Applied Sciences, Indianapolis, Ind.), except H2446, which was restricted with AscI (New England BioLabs, Beverly, Mass.) (Fig. 1). These are the primary endonucleases used for each of the pathogens. The purpose of the size standard is to establish reference positions (visible restriction fragments in the pattern), which are then used to normalize the electronic images (tiff format) of the PFGE patterns of the standard strain to match the PFGE pattern of the reference standard established in the national database. The reference standard pattern is the PFGE pattern of the standard strain used to create the reference system in the analysis software (1). Fragment positions on this pattern were chosen as reference positions to be used for normalization; adjustments are made to the PFGE gel images by matching the standard in the gels to the electronic reference standard. The reference standard image and marked positions to be used for normalization are included in the PulseNet customization database setup files provided to all PulseNet participating laboratories. Once the positions are identified in the standard lanes, it is possible to correct minor variations that may exist between gels, such as differences in the length of the electrophoresis run. When gel images are properly normalized, the position of restriction fragments in a given “test” strain relative to the fragment positions in the standard strain should be the same regardless of which laboratory generated the data. All lanes from normalized tiff images run by the standardized protocol are comparable in the national database.
FIG. 1.
Original standards used for the normalization and analysis of E. coli O157:H7, L. monocytogenes, Salmonella, and Shigella. Approximate kilobase values for positions used for normalization are indicated. Lanes are from tiffs of gels run using the PulseNet standardized methods for that organism. Electrophoresis switch times are indicated for each organism.
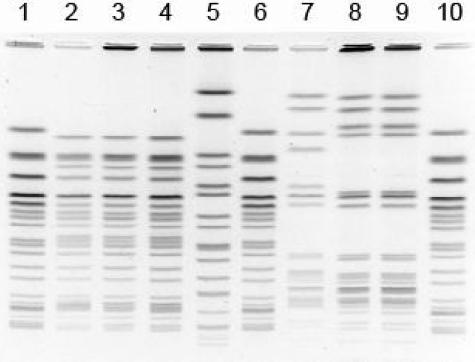
The addition of new organisms (17) to the PulseNet databases and the use of additional restriction enzymes in the subtyping scheme, which included AvrII (BlnI) and SpeI (Roche Applied Science) for E. coli O157:H7, Salmonella, and Shigella, and ApaI (Roche Applied Science) and SmaI for Listeria created new problems for image normalization and comparison. The PFGE patterns of new organisms or organisms analyzed with additional enzymes sometimes had bands that were larger in size than the largest fragment in the original organism-specific size standards used, which were approximately 580 kb in G5244 and F2353, 690 kb in H2446, and 917 kb in AM01144 (Fig. 1). This limited PulseNet participants' ability to normalize, analyze, and compare the PFGE patterns because the positions of the bands above the top band of the standard could not be accurately determined (Fig. 2, lanes 7 to 9). Thus, it was extremely important for us to find an adequate solution to this problem. Two different approaches were considered in our search for better size standards: (i) to identify new standards for each of the groups of organisms included in PulseNet (genus-specific approach) and (when necessary) different standards for each enzyme used or (ii) to identify a single strain that could be used as a “universal” size marker for all the organisms currently represented in the PulseNet National Database. We pursued the latter approach because it would eliminate the need to maintain multiple standard strain cultures and prepare multiple standard plugs. We began looking for a suitable candidate by examining patterns submitted to the national database. In 2000, the PFGE pattern of a strain of Salmonella serotype Braenderup observed on a gel submitted by one of the PulseNet participating laboratories was identified as a potential candidate.
FIG. 2.
PFGE image showing the E. coli O157:H7 (G5244) size standard (lanes 1, 6, and 10), three test strains restricted with XbaI (lanes 2 to 4) and BlnI (lanes 7 to 9), and the new universal standard (Salmonella serotype Braenderup H9812; lane 5).
Replacing the original size standards with the universal standard presented us with many challenges. Our foremost requirement was that PFGE patterns of pathogen isolates generated with the original organism-specific standards should be comparable to patterns of isolates run with the new universal standard. This meant that the pattern from the same isolate run and normalized with either standard should be indistinguishable after normalization. The following is a description of the process associated with the selection and evaluation of a strain of Salmonella serotype Braenderup (H9812) as the universal size standard for PFGE analysis of the foodborne bacterial pathogens tracked by PulseNet. The software issues associated with the conversion of the database also are addressed.
MATERIALS AND METHODS
PulseNet standard strains.
Selection of all the original PulseNet strain-specific standard strains, G5244 (E. coli O157:H7), AM01144 (Salmonella), F2353 (Shigella), and H2446 (L. monocytogenes) was based on the distribution of DNA fragments seen in PFGE patterns obtained with these strains when run with the appropriate PulseNet standardized PFGE protocol (6, 9, 17). The criteria used in the selection of these strains focused on fragment size range, distribution, and overall complexity of the pattern. Strains that were chosen covered the range of bands being seen from routine isolates at that time and also had an adequate distribution of bands for good normalization of tiff images. All of the original strain standards were obtained from the culture collection maintained by the Foodborne and Diarrheal Diseases Branch at the CDC in Atlanta. Salmonella serotype Braenderup strain H9812 was provided by the Health and Environmental Testing Laboratory of the Maine State Department of Human Services and has been deposited at the American Type Culture Collection under accession no. BAA-664.
Growth and storage of cultures.
Strains of E. coli O157:H7, Salmonella, and Shigella were grown on Trypticase soy agar with 5% sheep blood (Becton Dickinson and Company, Sparks, Md.); L. monocytogenes strains were grown on brain heart infusion agar plates. All strains were incubated at 37°C for 16 to 18 h as described previously (6, 9).
Preparation of plugs and PFGE methods.
Plug preparation, restriction digestion, electrophoresis conditions, and staining of gels for E. coli, Salmonella, and Shigella isolates were carried out as described by Ribot et al. (17) with the following modifications. The cell suspension buffer was 100 mM Tris and 100 mM EDTA at pH 8. The cell suspensions were adjusted to a turbidity reading of 0.48 to 0.52 on the digital output of a Dade Microscan turbidity meter (Baxter Diagnostics, Inc., McGraw Park, Ill.). In addition, the agarose used to make the plugs contained 1% sodium dodecyl sulfate. While the lysis buffer composition did not change, the lysis time for E. coli, Salmonella, and Shigella was increased to 1.5 to 2 h. A total of six washes (twice with sterile ultrapure water and four times with 0.01 M Tris-EDTA buffer, pH 8.0) were used to remove excess reagents and cell debris from the lysed plugs. The primary restriction enzyme used for E. coli, Salmonella, and Shigella is XbaI (40 to 50 U of enzyme at 37°C for 2 h). The electrophoresis conditions for E. coli, Salmonella, and Shigella had an initial switch time of 2.16 s, with final switch times of 54.17 s for E. coli and Shigella and 63.8 s for Salmonella. Plug preparation, restriction digestion, electrophoresis conditions, and staining of gels for L. monocytogenes isolates were performed with the PulseNet standardized protocol as described by Graves and Swaminathan (9). Gel images were captured on a Gel Doc 1000 or Gel Doc 2000 imaging system (Bio-Rad, Hercules, Calif.), converted to a tiff file, and analyzed with Molecular Analyst Fingerprinting Plus (Bio-Rad, Hercules, Calif.) or BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium).
Commercial size markers (nucleic acid standards).
The λ ladder (48.5 to 1,018.5 kb) was purchased from either New England Biolabs (Beverly, Mass.) or Bio-Rad (Hercules, Calif.). The 8.3- to 48.5-kb ladder, sometimes referred to as the high-molecular-weight standard, was purchased from Bio-Rad. Saccharomyces cerevisiae (yeast) chromosomal DNA (225 to 2,200 kb) was purchased from Bio-Rad. The 10-kb ladder was purchased from Stratagene (La Jolla, Calif.).
Susceptibility testing.
Since we preferred to have a standard strain that was susceptible to common therapeutic agents, the universal standard candidate strain (H9812) was tested for susceptibility to a panel of 17 antimicrobial agents (amikacin, amoxicillin-clavulanic acid, ampicillin, apramycin, ceftiofur, ceftriaxone, cephalothin, chloramphenicol, ciprofloxacin, gentamicin, kanamycin, nalidixic acid, streptomycin, sulfamethoxazole, tetracycline, ticarcillin, and trimethoprim-sulfamethoxazole) with the Sensititre system (TREK Diagnostic Systems, Westlake, Ohio).
Determination of kilobase values for band positions of original size standards for E. coli O157:H7, Salmonella, Shigella, and L. monocytogenes, and Salmonella serotype Braenderup H9812, the proposed universal standard.
The band sizes for the original standards for E. coli (G5244), Salmonella (AM01144), Shigella (F2353), and Listeria (H2446) were determined when these databases were originally established. Band position sizes for the proposed universal standard (H9812) strain were determined during the conversion process. DNA in agarose plug slices was digested with the appropriate primary enzyme and loaded into agarose gels along with the commercial molecular size markers (λ, 8.3- to 48.5-kb and 10-kb ladders, and S. cerevisiae chromosomal DNA) in alternating lane positions. DNA fragments were separated by PFGE on the Bio-Rad CHEF Mapper XA system (Bio-Rad, Hercules, Calif.) under the appropriate electrophoresis conditions for each of the test organisms and commercial standards. For example, switch times of 2.12 to 63.8 s and a run time of 19 h were used for gels with the λ ladder and standard strain AM01144, switch times of 0.22 to 12.9 s and a run time of 19 h were used for the 8.3- to 48.5-kb ladder and AM01144, and switch times of 25 to 93.7 s and a run time of 22 h were used when AM01144 was run with the yeast chromosomal DNA size marker. PFGE data from at least 24 lanes of each standard strain were used to calculate the size of the bands in the strains by using the Comparative Quantification/Polymorphism analysis feature of the Molecular Analyst Fingerprinting Plus software (Bio-Rad).
Standard curve correction for E. coli O157:H7, Salmonella, Shigella, and L. monocytogenes.
BioNumerics software has the capability of comparing patterns normalized by different standards using a feature called remapping, which is based on the fragment length calibration curves of both standards. These curves specify a fragment length (in kilobases) for each normalized run length position of a given standard. The process of remapping between different standards works by linking positions between both standards that share the same fragment length on their calibration curve. Hence, one can modify one of the calibration curves to maximize the compatibility between both standards. Since, for PulseNet data, these curves are obtained by applying cubic spline interpolation (16) of the calibration points (determined by a set of bands with fragment lengths for which an accurate estimate is known), this is equivalent to adjusting the calibration points used. We chose to leave the calibration points of the H9812 standard unchanged and optimize the remapping by modifying those of the original standard. A cross-comparison experiment was set up, consisting of a series of PFGE gels containing the original standard strains (G5244, AM01144, F2353, and H2446) and Salmonella serotype Braenderup H9812 (restricted with XbaI) in alternating lanes. The restriction enzyme and the electrophoresis conditions appropriate for each standard strain were used as described above and in the legend to Fig. 1. These gels were normalized twice: once using the original standard and once using the new standard (H9812). The optimized calibration points of the original standards were then obtained by taking their averaged positions determined by the normalization with H9812, resulting in a minimization of the distortions in a least-square sense.
The optimization was tested by analyzing gels that contained test isolates flanked by both standards run in alternate positions. Plugs were prepared and gels were run as described above. The resulting images were normalized twice, once with each reference standard, and the normalized patterns were compared.
RESULTS
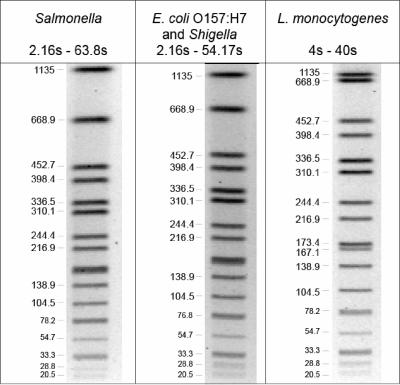
The size of each band in the XbaI restriction digestion pattern of H9812 was determined as described above. The estimated band size range for strain H9812 is 20.5 to 1,135 kb, which covers most of the largest and smallest bands seen in the strains tracked by PulseNet, regardless of the organism or the restriction enzyme used. Figure 1 shows the switch times and approximate band sizes for the original strain standards used for the E. coli (and Shigella), Salmonella, and L. monocytogenes databases. Figure 3 shows the switch times and estimated kilobase values for each of the DNA fragments in the PFGE pattern of strain H9812 used for normalization when they were run under the PulseNet standard electrophoresis conditions for E. coli (and Shigella), Salmonella, and L. monocytogenes.
FIG. 3.
Approximate band sizes in kilobases of the Salmonella serotype Braenderup reference standard (H9812) restricted with XbaI and run under the PulseNet standardized electrophoresis conditions specific for each organism.
Antimicrobial susceptibility data.
H9812 was susceptible to all 17 antimicrobials tested by the Sensititre system, including those commonly used to treat severe Salmonella infections.
Standard curve remapping for E. coli O157:H7, Salmonella, and L. monocytogenes.
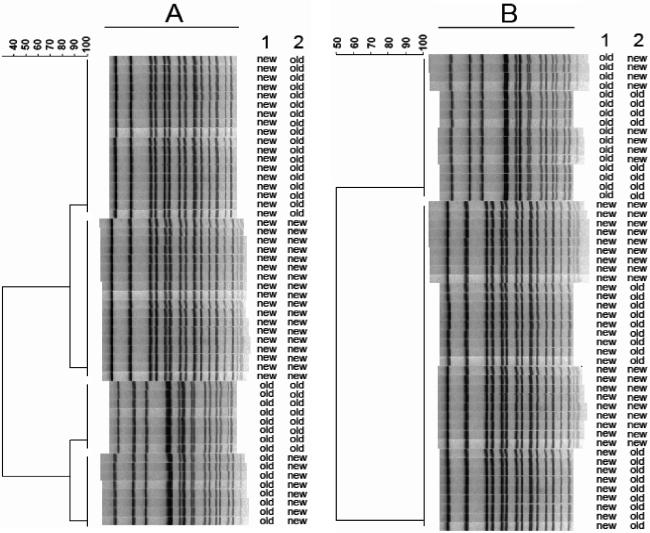
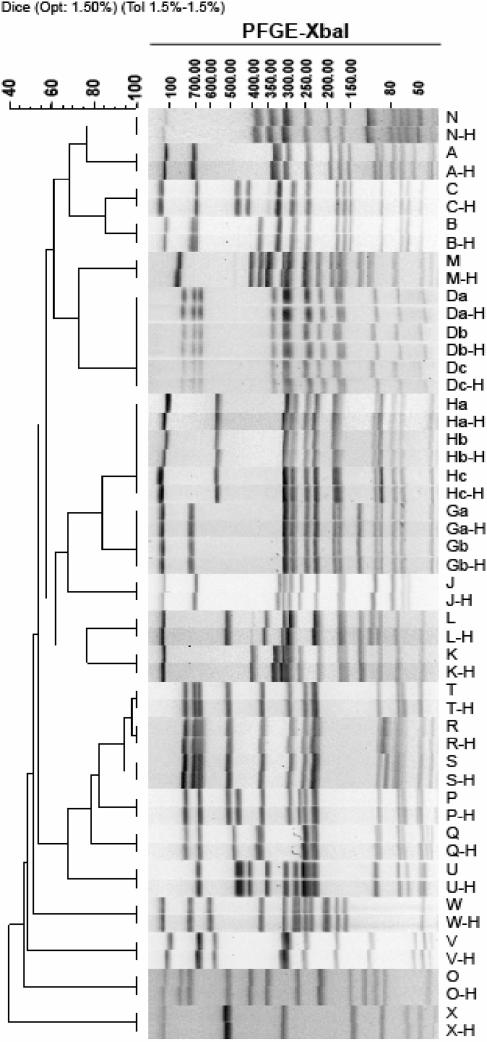
Figure 4 shows dendrograms of gel lanes containing AM01144 from tiff images of alternating lanes of standards analyzed with both the AM01144 and H9812 standards before (Fig. 4A) and after (Fig. 4B) the calibration curve optimizations. Similar gels were run and analysis was performed on the other standard strains: G5244, F2353, and H2446 (data not shown). Figure 5 shows a dendrogram of 28 Salmonella isolates run with both the old standard (AM01144) and H9812, the new universal standard, on the same gel, with each isolate normalized with both standards. The normalized patterns of each isolate matched at 100% similarity. Similar results were also obtained with E. coli, Shigella, and Listeria standards and test isolates (data not shown). These results indicate that gel images normalized with the old and new standards can be accurately compared.
FIG. 4.
Dendrograms of tiff images of gel lanes containing AM01144 (old Salmonella standard), alternating with H9812 (new standard), and normalized with both old and new standards. (The first column to the right of the dendrogram indicates the pattern for the standard, and the second column indicates which reference system was used for normalization.) Shown are results before optimization (A) and after the calibration curve optimizations (B) for alternating lanes of AM01144 and H9812.
FIG. 5.
Dendrogram showing 26 Salmonella isolates run with both the AM01144 and the H9812 standards on same gel; the tiff was renamed and normalized with each standard. Isolates normalized with the original Salmonella standard strain (AM01144) are designated by one or two letters; the same isolate normalized with the universal standard (H9812) is designated by the same combination of letters followed by -H. Isolates designated with the same uppercase letter but a different lowercase letter are epidemiologically related.
DISCUSSION
We originally considered using the λ ladder (48.5 to 1,018.5 kb) as the size standard for all PulseNet databases. However, we soon found the usefulness of the λ ladder as a DNA size standard to be limited by variability in both the concentration of DNA and the quality of commercial preparations from different vendors and even different lots from the same vendor. We felt that a better approach would be to use organism (genus or species)-specific strain standards as size standards. Selection of the original size standards used by PulseNet was based on the evaluation of PFGE patterns that had an adequate number of visible bands (∼10 to 20 fragments), even distribution of the restriction fragments without major gaps, and coverage of a wide range of fragment sizes (from ∼20 kilobases to >1 Mb). In addition to serving as size standards, organisms can also serve as internal controls for the process of plug preparation and restriction digestion. This approach worked well until the addition of new organisms and additional restriction enzymes necessitated a change to a universal standard.
Several steps had to be undertaken before the new size standards could be adopted. First, gels were run to show that the band sizes seen in the XbaI digest pattern from the proposed standard included the range of band sizes that were expected for primary and secondary enzyme restriction patterns for all of the organisms currently included in the PulseNet databases. Second, after band sizes of the proposed standard were determined, we needed to show that isolates on gels run with either the old or new standard could be accurately compared. It is essential for data currently in the database to be accurately compared with future data on gels generated with the new standard. Areas above the top band and below the lowest band in the original standards cannot be accurately compared because it was not possible to accurately normalize in these areas of the tiff, and these bands were not included in the comparison.
PulseNet participants use BioNumerics (Applied Maths, Sint-Martens-Latem, Belgium) software for database maintenance, tiff image normalization, and analysis and pattern comparisons. For each restriction enzyme experiment type, there can be one or multiple reference standards. For E. coli, Salmonella, Listeria, and Shigella, PulseNet strain-specific standards, with band sizes determined and specific positions marked to use as references, were originally used (Fig. 1). When the H9812 standard was introduced, the remapping feature in the BioNumerics software, which allows images normalized with different reference standards to be compared, was used to compare gels run with the old standard with gels run with the new standard. Some small differences, predominately at the ends of the gel, could be seen when comparing lanes normalized with each standard (Fig. 4A). The standard curve remapping was optimized to minimize these differences (Fig. 4B). After optimization, patterns normalized with either standard can be accurately compared (Fig. 5).
Since each participating laboratory has its own local database, program script files are sent to each laboratory to create a client (local) BioNumerics database customized for each of the organisms tracked by PulseNet. These client scripts create BioNumerics experiments with reference standards for each restriction enzyme used and include the original reference standard strains for E. coli O157:H7, Salmonella, L. monocytogenes, and Shigella, as well as the new Salmonella serotype Braenderup H9812 standard. The changes resulting from optimization were added to the PulseNet setup program scripts so that all participants would have exactly the same reference systems and lanes would remap accurately regardless of the standard used to normalize them.
Although there are many advantages to using Salmonella serotype Braenderup strain H9812 as the PulseNet “universal” standard, there are also some disadvantages. The PFGE plugs of H9812 have to be prepared and tested with the Salmonella PulseNet standardized protocol, which may be different from the protocol used for the organism of interest: e.g., L. monocytogenes (9) or Campylobacter (7, 17). Additionally, XbaI is used to restrict the H9812 standards, and it is not always the enzyme used to subtype the test organism. For example, the primary and secondary enzymes used to PFGE subtype L. monocytogenes are AscI and ApaI, respectively (9). This problem can be solved by including a pretested plug as a control to test the efficiency of the restriction digestion reactions. The appearance of the H9812 standard pattern will also be dependent on the electrophoresis conditions used, which will vary depending on the organism (Fig. 3). It is important to emphasize that the PFGE pattern of H9812 must be examined carefully when standard plugs of H9812 are prepared and tested with the Salmonella PulseNet standardized protocol to be certain that no observable genetic changes have occurred in the standard strain. The easiest and most accurate way to verify this is to analyze the restriction digests of the newly prepared PFGE plugs of H9812 on the same gel with PFGE plugs of H9812 that were tested previously and found to have the correct pattern and concentration.
In summary, a strain of Salmonella serotype Braenderup (H9812) restricted with XbaI was selected as a “universal” standard strain because of its even distribution of bands over the entire range of band sizes normally seen in the foodborne pathogens tracked by PulseNet. This strain was found to have a stable PFGE pattern upon subculture and was susceptible to antimicrobial agents commonly used to treat serious Salmonella infections. Data obtained with the new standard can be easily and accurately compared with data generated with the old standards. A universal standard means that PFGE plugs of only one standard strain need to be made and tested, instead of a different strain for each organism in the database. Strain H9812 is currently being used successfully by PulseNet for PFGE analysis of E. coli O157:H7, Salmonella, L. monocytogenes, Shigella, Campylobacter jejuni, and Campylobacter coli. We are therefore recommending that H9812 be used as a universal strain standard for PulseNet PFGE gels and have deposited the strain with the American Type Culture Collection (BAA-664) to make it available for other laboratories to use.
Acknowledgments
We thank John Allan for assistance with the analysis of PFGE patterns from the Salmonella serotype Braenderup strain H9812 and Kevin Joyce for performing the sensitivity testing. We thank Kelley Hise for assistance with figures. We also express our gratitude to Bala Swaminathan for guidance and comments during the course of this project.
Use of trade names is for identification only and does not imply endorsement by the Centers for Disease Control and Prevention or by the U.S. Department of Health and Human Services.
REFERENCES
- 1.Applied Maths BVBA. 2002. BioNumerics version 3.0 manual. Applied Maths BVBA, Sint-Martens-Latem, Belgium.
- 2.Arbeit, R. D., M. Arthur, R. Dunn, C. Kim, R. K. Selander, and R. Goldstein. 1990. Resolution of recent evolutionary divergence among Escherichia coli from related lineages: the application of pulsed field electrophoresis to molecular epidemiology. J. Infect. Dis. 161:230-235. [DOI] [PubMed] [Google Scholar]
- 3.Barrett, T. J., H. Lior, J. H. Green, R. Khakhria, and J. G. Wells. 1994. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J. Clin. Microbiol. 32:3013-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender, J. B., C. W. Hedberg, J. M. Besser, D. J. Boxrud, K. L. MacDonald, and M. T. Osterholm. 1997. Surveillance by molecular subtype for Escherichia coli O157:H7 infections in Minnesota by molecular subtyping. N. Engl. J. Med. 337:388-394. [DOI] [PubMed] [Google Scholar]
- 5.Branchini, M. L. 1993. Application of genomic DNA subtyping by pulsed field gel electrophoresis and restriction enzyme analysis of plasmid DNA to characterize methicillin-resistant Staphylococcus aureus from two nosocomial outbreaks. J. Hosp. Infect. 25:109-116. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2000. Standardized molecular subtyping of foodborne bacterial pathogen by pulsed-field gel electrophoresis: training manual. Centers for Disease Control and Prevention, Atlanta, Ga.
- 7.Fitzgerald, C., K. Stanley, S. Andrew, and K. Jones. 2001. Use of pulsed-field gel electrophoresis and flagellin gene typing in identifying clonal groups of Campylobacter jejuni and Campylobacter coli in farm and clinical environments. Appl. Environ. Microbiol. 67:1429-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goering, R. V. 2004. Pulsed-field gel electrophoresis, p. 185-196. In D. H. Persing, F. C. Tenover, J. Versalovic, Y.-W. Tang, E. R. Unger, D. A. Relman, and T. J. White (ed.), Molecular microbiology, diagnostic principles and practice. ASM Press, Washington, D.C.
- 9.Graves, L. M., and B. Swaminathan. 2001. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 65:55-62. [DOI] [PubMed] [Google Scholar]
- 10.Herrmann, J. L. 1992. Pulsed-field gel electrophoresis of NotI digests of leptospiral DNA: a new rapid method of serovar identification. Biochem. Biophys. Res. Commun. 185:1055-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahalingam, S., Y. M. Cheong, S. Kan, R. M. Yassin, J. Vadivelu, and T. Pang. 1994. Molecular epidemiologic analysis of Vibrio cholerae O1 isolates by pulsed-field gel electrophoresis. J. Clin. Microbiol. 32:2975-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nair, S., C. L. Poh, Y. S. Lim, L. Tay, and K. T. Goh. 1994. Genome fingerprinting of Salmonella typhi by pulsed-field gel electrophoresis for subtyping common phage types. Epidemiol. Infect. 113:391-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Najdenski, H. 1994. Efficient subtyping of pathogenic Yersinia enterocolitica strains by pulsed-field gel electrophoresis. Eur. J. Epidemiol. 10:325-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olive, M. D., and P. Bean. 1999. Principles and applications of methods for DNA-based typing of microbial organisms. J. Clin. Microbiol. 37:1661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Press, W. H., S. A. Teukolsky, W. T. Vetterling, and B. P. Flannery. 1992. Numerical recipes in C. Cambridge University Press, Cambridge, United Kingdom.
- 17.Ribot, E. M., C. Fitzgerald, K. Kubota, B. Swaminathan, and T. J. Barrett. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saito, Y., K. Seki, T. Ohara, C. Shimauchi, Y. Honma, M. Hayashi, S. Masuda, and M. Nakano. 1998. Epidemiologic typing of methicillin-resistant Staphylococcus aureus in neonate intensive care units using pulsed-field gel electrophoresis. Microbiol. Immunol. 42:723-729. [DOI] [PubMed] [Google Scholar]
- 19.Soldati, L. 1991. Molecular typing of Shigella strains using pulsed field gel electrophoresis and genome hybridization with insertion sequences. J. Hosp. Infect. 17:255-269. [DOI] [PubMed] [Google Scholar]
- 20.Streulens, M. J., R. DeRyck, and A. Deplano. 2001. Analysis of microbial genomic macrorestriction patterns by pulse-field gel electrophoresis (PFGE) typing, p. 159-176. In L. Dijkshoorn, K. J. Towner, and M. Strulens (ed.), New approaches for the generation and analysis of microbial typing data. Elsevier Science B.V., Amsterdam, The Netherlands.
- 21.Swaminathan, B., T. J. Barrett, S. B. Hunter, R. V. Tauxe et al. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]