Abstract
Streptococcus agalactiae causes severe invasive disease in humans and mastitis in cattle. Temporally matched bovine milk isolates and clinical human invasive isolates (52 each) collected in New York State over 18 months were characterized by molecular subtyping and phenotypic methods to probe the interspecies transmission potential of this species. EcoRI ribotyping differentiated 17 ribotypes, and DNA sequencing of the housekeeping gene sodA and the putative virulence gene hylB differentiated 7 and 17 allelic types, respectively. Human and bovine isolates were not randomly distributed between ribotypes or hylB and sodA clusters. The combined analysis of all subtyping data allowed the differentiation of 39 clonal groups; 26 groups contained only bovine isolates, and 2 groups contained both human and bovine isolates. The EcoRI ribotype diversity among bovine isolates (Simpson's numerical index of discrimination [mean ± standard deviation], 0.90 ± 0.05) being significantly higher than that among human isolates (0.42 ± 0.15) further supports that these isolates represent distinct populations. Eight human isolates, but no bovine isolates, showed an IS1548 transposon insertion in hylB, which encodes a hyaluronidase. Based on data for 43 representative isolates, human isolates, on average, showed lower hyaluronidase activities than bovine isolates. Isolates with the IS1548 insertion in hylB showed no hyaluronidase activity. Human and bovine isolates did not differ in their abilities to invade HeLa human epithelial cells. Our data show that (i) EcoRI ribotyping, combined with hylB and sodA sequencing, provides a discriminatory subtype analysis of S. agalactiae; (ii) most human invasive and bovine S. agalactiae isolates represent distinct subtypes, suggesting limited interspecies transmission; and (iii) hyaluronidase activity is not required for all human infections.
Streptococcus agalactiae (Lancefield group B) causes disease in both humans and cattle. For humans, S. agalactiae is responsible for severe invasive disease in adults and neonates, as well as asymptomatic infections in women (2). S. agalactiae has been recognized as the leading cause of neonatal septicemia and meningitis; it has been estimated to have caused 16,880 human cases, including 1,650 deaths, in the United States alone in 1998 (41). The colonization of humans can occur through a variety of mechanisms, including maternal vaginal carriage and breast-feeding (2). In cattle, S. agalactiae causes clinical and subclinical infections of the mammary gland (mastitis) (27). Initial studies have suggested that interspecies transmission between cattle and humans is possible. For example, experimental inoculation studies have shown that human S. agalactiae isolates can infect and cause clinical mastitis in cattle (23, 46). Tissue culture studies have also shown that both bovine and human S. agalactiae isolates can adhere to bovine epithelial cells (5). These data did not provide any information on epidemiological mechanisms or the likelihood of interspecies transmission of S. agalactiae, nor do they provide any conclusive evidence as to the ability of bovine-associated S. agalactiae to cause human invasive infections.
S. agalactiae isolates of human origin have been characterized by using a variety of methods, including biochemical characterization, serotyping, multilocus enzyme electrophoresis, randomly amplified polymorphic DNA (RAPD) analysis, ribotyping, and, more recently, multilocus sequence typing (MLST) (4, 8, 18, 20, 25, 39). Based on multilocus enzyme electrophoresis data, Helmig et al. (19) were able to divide 118 human S. agalactiae isolates from Denmark into two major clusters. One cluster contained isolates that were almost all from patients with clinical symptoms, while the other cluster contained isolates mostly from asymptomatic carriers. Comparative subtyping studies using both classical phenotypic (e.g., serotyping) and DNA sequence-based methods to examine human and bovine S. agalactiae isolates have provided only limited insight into the likelihood of cattle serving as a reservoir or direct source of S. agalactiae strains in human invasive infections (4, 24, 32). For example, serotyping and ribotyping provided some evidence that cattle and asymptomatic human carriers that are present in the same locality can carry indistinguishable S. agalactiae subtypes (24). RAPD typing showed that 223 S. agalactiae isolates from asymptomatic human carriers and from bovines generally represented different subtypes; however, this study also found one human isolate and one bovine isolate that shared an identical RAPD type, even though these isolates had different serotypes (32). A recent MLST study also showed that animal and human invasive and carriage-associated isolates represent largely separate populations, even though a specific human hyperinvasive clone appears to have emerged from a bovine ancestor (4). Similarly, a subtyping study using phenotypic methods showed that human and bovine S. agalactiae strains isolated in Kenya appear to represent largely separate populations (34). Most of these previous studies have focused on (24, 32, 34) or included (4) human carriage isolates, thus providing limited insight into the likelihood of cattle serving as a source of human invasive disease-associated S. agalactiae. Thus, the goal of this study was to characterize temporally and geographically matched S. agalactiae isolates from human invasive and bovine udder infections by using standardized and reproducible molecular subtyping methods. While the use of S. agalactiae isolate sets that include human invasive and animal isolates collected in different regions, countries, or continents and/or during different times can provide important insights into the evolution of this pathogen (4), the characterization of temporally and geographically matched isolates is critical to assess the likelihood and potential for interspecies transmission of S. agalactiae.
MATERIALS AND METHODS
S. agalactiae isolates.
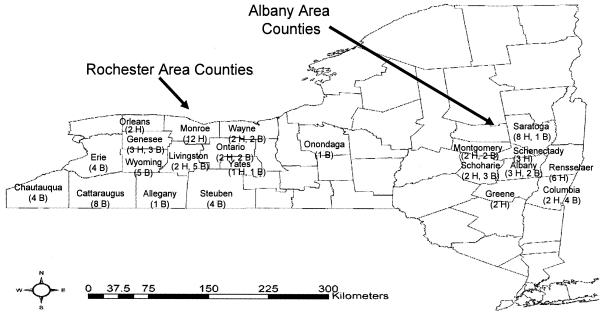
A total of 194 human clinical isolates of S. agalactiae were obtained through the U.S. Centers for Disease Control and Prevention Emerging Infections Program (EIP) conducted by the New York State Department of Health. As part of this active surveillance program, group B Streptococcus isolates from invasive human infections were collected from different counties in two distinct geographic regions of New York State (Rochester and Albany). A total of 109 isolates were obtained from counties participating in the Rochester area surveillance program, including Genesee, Livingston, Monroe, Ontario, Orleans, Wayne, and Yates counties. The other 85 isolates were obtained from counties participating in the Albany area surveillance program, including Albany, Columbia, Greene, Montgomery, Rensselaer, Saratoga, Schenectady, and Schoharie counties (Fig. 1). A total of 236 bovine S. agalactiae isolates, representing 52 farms, were obtained from the Cornell University Quality Milk Production Services mastitis control program. As part of this program, S. agalactiae strains were isolated from cows as previously described by Wilson et al. (48). The S. agalactiae strains isolated for this study represented the predominant organism found in a given quarter milk sample and are thus the likely causative agent of clinical or subclinical mastitis in these animals. Geographically, these isolates were gathered from the aforementioned EIP surveillance counties, as well as from geographically close non-EIP counties (i.e., Allegany, Cattaraugus, Chautauqua, Erie, Onondaga, Steuben, and Wyoming counties) (Fig. 1). A total of 52 bovine isolates, 1 per farm, were chosen from the larger pool of 236 isolates for this study. An equal number of human isolates were temporally matched with these 52 bovine isolates and selected as representative of the geographic diversity for the entire pool of human isolates. Thus, a total of 104 isolates were used for this study (Fig. 1). All isolates were stored at −80°C in brain heart infusion (BHI) broth (Becton Dickinson, Sparks, Md.) with 15% glycerol.
FIG. 1.
Geographic distribution, across New York State, of the 104 S. agalactiae isolates included in this study. The numbers of human (H) and bovine isolates (B) are indicated in parentheses below the county names.
Automated ribotyping.
Automated EcoRI ribotyping was performed using the RiboPrinter (Qualicon Inc., Wilmington, Del.), as previously described (6), with bacterial isolates grown overnight on BHI agar. Template preparation, restriction enzyme digestion, gel electrophoresis, and Southern hybridization probing with an Escherichia coli rrnB rRNA operon probe were carried out with the RiboPrinter system. Images were acquired with a charge-coupled-device camera and processed using the RiboPrinter's custom software. This software normalizes fragment pattern data for the band intensity and relative band position (6). Similarity coefficients for ribotype patterns from different isolates were calculated using the RiboPrinter's proprietary algorithm. Similarity coefficients and visual evaluation were used to define distinct ribotypes.
PCR and DNA sequencing.
While traditional MLST schemes focus on the sequencing of housekeeping genes, which diversify slowly and are not heavily influenced by evolutionary forces other than point mutations (i.e., positive selection and recombination) (31), we chose to include a secreted and putative virulence gene (hylB) as a target for DNA sequencing-based subtyping, since the sequencing of virulence genes often provides higher discriminatory power and can provide insight into the evolution of virulence-related characteristics (7). Cell lysates for PCR were prepared using lysozyme and proteinase K as described previously (13). Primers Sa-hylF (5′-CAT ACC TTA ACA AAG ATA TAT AAC CCA AA-3′) and Sa-hylR (5′-AGA TTT TTT AGA GAA TGA GAA GTT TTT T-3′) were designed from known sequences (GenBank accession numbers Y15903 and U15050) to amplify a 950-bp fragment of hylB. Primers sodA-F (′5-GCC CTA ATT AAA GAA ACA AAA GAG T-3′) and sodA-R (5′-CTG TTA TCA GCT TGC TTA CCT TAA A-3′) were designed to amplify a 911-bp fragment that included the complete sodA coding region (609 bp). Primers sodA-F2 (5′-GAT TTA GCC TTA TTA AAA ATT GCT CAT-3′) and sodA-R2 (5′-ATT CGC TTA ATT GAA GAT AAT AGG ACT GT-3′) were used to amplify an 849-bp fragment for three bovine isolates (FSL S3-038, FSL S3-066, and FSL S3-081), which showed no amplification product with sodA-F and sodA-R. hylB and sodA PCR conditions consisted of an initial 5-min denaturation step at 95°C, 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, and a final 10-min extension cycle at 72°C. PCR products were purified using the QIAquick PCR purification kit (QIAGEN, Inc., Chatsworth, Calif.). Following gel electrophoresis, DNA was quantified by using the LabImage gel analysis software (version 2.62; Kapelan, Saale, Germany) to compare the band intensities of PCR products to marker band intensities of known concentrations (pGEM marker; Promega, Madison, Wis.).
The sequencing of PCR products was performed at Cornell University's BioResource Center by using the respective PCR primers, Big Dye Terminator chemistry, and AmpliTaq-FS DNA polymerase; the sequencing reactions were run on the ABI 3700 automated sequencer (Perkin-Elmer Biosystems, Foster City, Calif.). DNA sequence data obtained with both forward and reverse primers were proofread and assembled with the Seqman software program (DNAStar, Madison, Wis.) to yield the final hylB and sodA sequences used for analyses.
DNA sequence analysis.
hylB and sodA sequences were aligned using the Clustal W algorithm in MegAlign (DNAStar). The sodA alignments used for the analyses described below included the full sodA coding region (609 bp); hylB alignments encompassed an 886-bp hylB fragment. Alignments were used to assign hylB and sodA alleles; individual gene sequences that differed by a single nucleotide were assigned an allelic type (e.g., hyl-1.0, hyl-1.1, or hyl-3.0). Allelic types were designated so that groups of closely related allelic types that differed from each other by single nucleotides were assigned decimals of the same number (e.g., hyl-1.0 and hyl-1.1).
The TOPAL software program (33) was used to probe for indications of intragenic recombination events. Neighbor-joining phylogenetic trees were generated with MEGA software version 2.1 (28). Phylogenetic trees based on maximum parsimony with bootstrapped sequence data were constructed using the SEQBOOT, DNAPARS, CONSENSE, and DRAWTREE programs of the PHYLIP version 3.5c software package (12). Phylogenetic trees were also used to categorize closely related sodA and hylB allelic types into clusters.
The dN (the number of nonsynonymous substitutions per nonsynonymous site)/dS (the number of synonymous substitutions per synonymous site) ratios for hylB and sodA were also calculated using MEGA version 2.1 (28). The LIAN 3.0 software program (17) was used to perform a significance test for linkage disequilibrium among subtypes generated by ribotyping, hylB allelic typing, and sodA allelic typing.
Simpson's index of discrimination.
Simpson's numerical index of discrimination (SID) provides an indication of the discriminatory power of a given subtyping method (21), as well as an estimate of the subtype diversity within a given population (16). By use of a method described by Hunter and Gaston (21), the SID was calculated in order to determine the discriminatory power of the subtyping methods used. The 95% confidence intervals surrounding the corresponding indices were calculated using the method described by Grundmann et al. (16).
Microplate assay for hyaluronidase activity.
Hyaluronidase activities were determined for a set of 43 S. agalactiae isolates, representative of the genetic diversity of our original set of 104 isolates. Isolates were specifically selected to encompass all EcoRI ribotypes and hylB clusters represented in our isolate set. Hyaluronic acid (from human umbilical cord), cetylpyridinium chloride, and sodium azide were all purchased from Sigma Chemical Co. (St. Louis, Mo.). To measure hyaluronidase activity, S. agalactiae isolates were grown overnight in BHI broth at 37°C without shaking. Hyaluronidase activity was determined according to a method previously described by Tung et al. (44), with some modifications. Hyaluronic acid (5 mg/ml) was dissolved in water overnight at 4°C and then stored at −20°C. A 1% (wt/vol) sodium azide solution was prepared in water and stored at room temperature. Agarose was dissolved in 0.3 M sodium phosphate buffer (pH 7.0) in a microwave oven and maintained at 55°C before use. Hyaluronic acid and sodium azide solutions were preheated to 55°C and mixed with the agarose to give final concentrations of 0.8 mg of hyaluronic acid/ml, 0.1% (wt/vol) sodium azide, and 0.8% (wt/vol) agarose. The warm mixture was dispensed in 100-μl aliquots into each well of a 96-well microtiter plate. Once the gel solidified, each well was filled with 100 μl of a 1:20 dilution of S. agalactiae broth culture or 100 μl of sodium phosphate as a blank. After incubation at 37°C for 18 h, the supernatant was removed from each well. Wells were washed three times with 150 μl of sodium phosphate buffer and then filled with 100 μl of 10% (wt/vol) aqueous cetylpyridinium chloride to precipitate the hyaluronic acid. After incubation at room temperature for 2 h, the absorbance at 600 nm was measured using a Fusion automated microplate reader (Perkin-Elmer). Absorbance values were standardized using the formula 1 − (Asample/Ablank), where Asample is the absorbance of the culture sample and Ablank is the absorbance of the blank. The enzyme activity for each bacterial isolate was determined in duplicate by measuring the absorbance for two wells loaded with aliquots of the same S. agalactiae broth culture. The assay was replicated twice for each isolate.
Tissue culture assays.
Three human isolates and three bovine isolates representative of the most common S. agalactiae hylB allelic types (hyl-3.0, hyl-4.0, and hyl-7.0 for human isolates and hyl-1.0, hyl-5.0, and hyl-6.0 for bovine isolates) (Table 1) were tested for their invasion phenotypes in HeLa cells. Invasion assays were performed as previously described (40) with minor modifications. Briefly, S. agalactiae isolates were passaged twice to a final optical density at 600 nm of 0.4 in BHI broth. Bacterial cells were inoculated (at approximately 106 CFU per well) onto a monolayer of HeLa cells grown on coverslips in six-well plates with Dulbecco's modified Eagle medium (Invitrogen Co., Carlsbad, Calif.) and 10% fetal bovine serum. Six-well plates were then centrifuged at 800 × g for 10 min at 25°C to bring S. agalactiae cells into contact with the HeLa cell monolayer. Plates were allowed to incubate for 2 h at 37°C in 5% CO2. Cells were then washed three times with phosphate-buffered saline, followed by addition of Dulbecco's modified Eagle medium containing penicillin (5 μg/ml) and gentamicin (100 μg/ml). Infected cells were incubated for another 2 h at 37°C in 5% CO2 to kill extracellular bacterial cells. Coverslips were then removed, lysed in 0.1% Triton X-100, and vortexed for 30 s. The number of CFU invaded per coverslip for each S. agalactiae isolate was determined by plating serial dilutions on BHI agar. Four independent repetitions of this invasion assay were performed.
TABLE 1.
Frequency distribution of S. agalactiae isolates among EcoRI ribotypes, hylB allelic types, and sodA allelic types
| Subtype | Incidence (prevalence [%]) among indicated type of isolates
|
P valuea | |
|---|---|---|---|
| Human (n = 52) | Bovine (n = 52) | ||
| EcoRI ribotype | |||
| 106-1501-S-8 | 39 (75.0) | 1 (1.9) | <0.01 |
| 116-549-S-1 | 7 (13.5) | 3 (5.8) | 0.50 |
| 106-1501-S-5 | 4 (7.7) | 12 (23.1) | 0.01 |
| 116-611-S-7 | 2 (3.8) | 3 (5.8) | 0.65 |
| 106-1502-S-6 | 0 | 9 (17.3) | <0.01 |
| 106-1502-S-2 | 0 | 6 (11.5) | 0.01 |
| 106-1502-S-3 | 0 | 4 (7.7) | 0.03 |
| 106-1504-S-1 | 0 | 3 (5.8) | 0.08 |
| 116-522-S-1 | 0 | 2 (3.8) | NA |
| 106-1501-S-4 | 0 | 2 (3.8) | NA |
| 116-549-S-7 | 0 | 1 (1.9) | NA |
| 116-574-S-2 | 0 | 1 (1.9) | NA |
| 116-607-S-6 | 0 | 1 (1.9) | NA |
| 116-622-S-8 | 0 | 1 (1.9) | NA |
| 116-638-S-1 | 0 | 1 (1.9) | NA |
| 116-646-S-2 | 0 | 1 (1.9) | NA |
| 106-1502-S-1 | 0 | 1 (1.9) | NA |
| hylB clusters and allelic typesb | |||
| hyl-1 cluster | 0 | 31 (59.6) | <0.01 |
| hyl-1.0 | 0 | 24 (46.2) | |
| hyl-1.1 | 0 | 5 (9.6) | |
| hyl-1.2 | 0 | 1 (1.9) | |
| hyl-1.3 | 0 | 1 (1.9) | |
| hyl-2 cluster (hyl-2.0) | 2 (3.8) | 0 | 0.50 |
| hyl-3 cluster (hyl-3.0) | 7 (13.5) | 0 | 0.01 |
| hyl-4 cluster (hyl-4.0) | 26 (50.0) | 1 (1.9) | <0.01 |
| hyl-5 cluster | 0 | 17 (32.6) | <0.01 |
| hyl-5.0 | 0 | 13 (25.0) | |
| hyl-5.1 | 0 | 1 (1.9) | |
| hyl-5.2 | 0 | 1 (1.9) | |
| hyl-5.3 | 0 | 1 (1.9) | |
| hyl-5.4 | 0 | 1 (1.9) | |
| hyl-6 cluster | 9 (17.3) | 3 (5.7) | 0.12 |
| hyl-6.0 | 7 (13.5) | 2 (3.8) | |
| hyl-6.1 | 2 (3.8) | 0 | |
| hyl-6.2 | 0 | 1 (1.9) | |
| hyl-7 cluster | 8 (15.4) | 0 | 0.01 |
| hyl-7.0 | 7 (13.5) | 0 | |
| hyl-7.1 | 1 (1.9) | 0 | |
| sodA clusters and allelic typesb | |||
| sod-1 cluster | 41 (78.8) | 5 (9.6) | <0.01 |
| sod-1.0 | 40 (76.9) | 4 (7.7) | |
| sod-1.1 | 1 (1.9) | 0 | |
| sod-1.2 | 0 | 1 (1.9) | |
| sod-2 cluster (sod-2.0) | 3 (5.8) | 0 | 0.24 |
| sod-3 cluster (sod-3.0) | 6 (11.5) | 0 | 0.03 |
| sod-4 cluster | 2 (3.8) | 47 (90.4) | <0.01 |
| sod-4.0 | 2 (3.8) | 46 (88.5) | |
| sod-4.1 | 0 | 1 (1.9) | |
All P values (between human and bovine isolates) were computed using a two-tailed Fisher's exact test. NA, Fisher's exact test was not applicable.
P values were computed for hylB and sodA clusters only (e.g., hyl-1, hyl-2, sod-1, or sod-2).
Statistical methods.
For all statistical analyses, isolates with closely related hylB and sodA allelic types were grouped into hylB and sodA clusters, which included closely related alleles as determined by phylogenetic analyses (see below). The frequency distributions of each genetic characteristic (i.e., ribotype, hylB cluster, or sodA cluster) for human and bovine isolates were compared by using a χ2 test of independence. For comparisons in which one or more of the expected values were <5, Fisher's exact test was performed using the SAS program (version 8.01; SAS Institute, Cary, N.C.). P values of <0.05 were considered statistically significant.
To determine associations between either the host species or the hylB cluster and hyaluronidase activity, a nonparametric analysis of variance (Kruskal-Wallis) was conducted with the Statistix software program (Analytical Software, Tallahassee, Fla.). Nonparametric analyses were deemed appropriate, as the activity data deviated significantly from normality. Associations were analyzed based on averaged assay results for each isolate. Data for isolates in clusters hyl-7 and hyl-2 were excluded from the analysis of association between hylB clusters and activity levels because these two clusters were represented by two isolates and one isolate only, respectively, in the hyaluronidase activity assays.
To determine whether human and bovine strains differed significantly in their abilities to invade HeLa cells, a one-way analysis of variance was conducted using the Minitab software program (version 14; Minitab, Inc., State College, Pa.).
World Wide Web-based data access.
Detailed isolate source information and raw data from this study (hylB and sodA sequences and EcoRI ribotype patterns) are accessible through the PathogenTracker version 2.0 website (http://www.pathogentracker.net).
RESULTS
Ribotype pattern analysis.
A total of 17 distinct EcoRI ribotypes were identified among the 104 human and bovine S. agalactiae isolates characterized (Table 1 and Fig. 2). Human isolates clustered into four ribotypes; ribotype 106-1501-S-8 represented the majority of human isolates (75%). All four EcoRI ribotypes that contained human isolates also contained bovine isolates; however, 13 ribotypes contained bovine isolates exclusively. Eight ribotypes contained only one bovine isolate each. A two-tailed Fisher's exact test demonstrated significant associations between five ribotypes and isolation from either human or bovine hosts (Table 1).
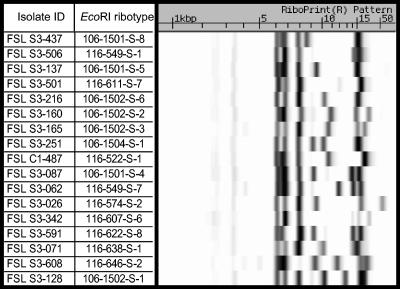
FIG. 2.
EcoRI ribotype patterns obtained for human and bovine S. agalactiae isolates. The left-hand side of the figure includes the identification codes (ID; e.g., FSL S3-437) of the isolates representative of the 17 EcoRI ribotypes encountered, along with the corresponding ribotype designations (e.g., 106-1501-S-8). Ribotype patterns are shown on the right-hand side and correspond to a gel run from right to left (high molecular weight bands are thus shown at the far right). Ribotype patterns for all 104 isolates can be viewed at the PathogenTracker website (http://www.pathogentracker.net).
hylB allelic and phylogenetic analysis.
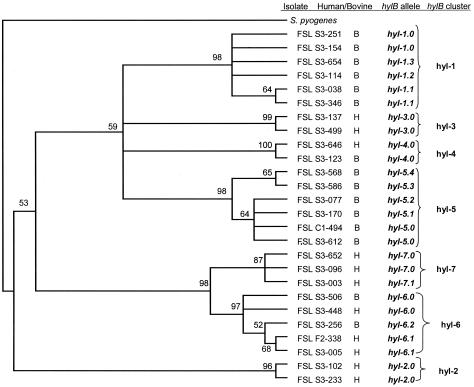
Initial analyses using TOPAL (33) revealed no indications of recombination events in the 886-bp hylB fragment analyzed. A total of 17 allelic types were found among the 104 S. agalactiae isolates characterized. A neighbor-joining tree constructed by using two representatives of each hylB allelic type (where available) revealed the phylogenetic relationships among allelic types and was used to group hylB allelic types into seven hylB clusters, which included closely related hylB alleles (Fig. 3). A parsimony tree created by using DNAPARS in the PHYLIP program showed topology and clustering of human and bovine isolates similar to those of the neighbor-joining phylogenetic tree (not shown). A two-tailed Fisher's exact test demonstrated significant associations between five hylB clusters and isolation from either human or bovine hosts (Table 1). Statistical analyses were not conducted for each hylB allelic type, due to the small numbers of isolates for some allelic types (Table 1).
FIG. 3.
Consensus neighbor-joining tree based on partial hylB sequences. Two representative sequences of each hylB allelic type (where available) were used for the construction of this phylogenetic tree. An S. pyogenes hylA sequence (encoding hyaluronate lyase) was used as an outgroup (GenBank accession number NC002737). The tree was built by using the Jukes-Cantor model of nucleotide substitution. A total of 1,000 bootstrap replicates were generated. Bootstrap values of >50% are shown at the nodes. Closely related hylB allelic types were assigned an hylB cluster (e.g., cluster hyl-2), which is also indicated in this figure.
PCR amplification of hylB revealed a PCR product approximately 1,300 bp larger than expected for 8 of the 52 human isolates and none of the 52 bovine isolates. DNA sequencing of these PCR products revealed a putative insertion sequence within hylB, which showed >98% homology with IS1548 according to the basic local alignment search tool (1). The IS1548 insertion was mapped to the same hylB location in all eight isolates. All eight human isolates with the IS1548 insertion in hylB share the same ribotype (106-1501-S-8). After the removal of the IS1548 sequence, the hylB sequences for these eight isolates were included in the overall hylB alignment for allele assignment and phylogenetic analysis. All eight isolates were shown to share the same hylB allele (hyl-7.0), with the exception of one isolate (FSL S3-003), which is the only isolate with allele hyl-7.1.
To further probe the evolution of the hylB fragments sequenced among human and bovine isolates, the dN/dS ratios for the sequenced fragments were calculated. The dN/dS ratio obtained for human isolates (0.425) was slightly higher than the ratio obtained for the bovine isolates (0.357). The overall dN/dS ratio for hylB was 0.382. A total of 39 polymorphic nucleotide sites were found among the 52 human isolates, including 27 sites with nonsynonymous substitutions. Among the bovine isolates, there were 40 polymorphic sites, including 23 sites with nonsynonymous substitutions.
sodA allelic analysis.
A total of seven different sodA allelic types were identified (Table 1). Based on phylogenetic analysis, these types could be grouped into four sodA clusters, which contain allelic types that differ by single-nucleotide differences. The majority of human isolates (76.9%) clustered into allelic type sod-1.0 and cluster sod-1, while the majority of bovine isolates (88.5%) clustered into allelic type sod-4.0 and cluster sod-4. As a result, these sod clusters (sod-1 and sod-4) were significantly host associated (P < 0.01 for both alleles) (Table 1).
Direct nucleotide sequence analysis of the complete sodA coding sequence revealed limited diversity among the 104 S. agalactiae isolates characterized. Within the 609-nucleotide coding sequence, a total of 12 polymorphic nucleotide sites were identified. The dN/dS ratio for sodA was 0.250 (0.333 and 0.200 for human and bovine isolates, respectively).
Distribution of S. agalactiae clonal groups among human and bovine isolates.
The standardized index of association (IA), a measure of linkage disequilibrium (17), was calculated based on subtypes generated by the three subtyping methods for all 104 S. agalactiae isolates. The standardized IA was calculated as 0.41, which represents a highly significant (P < 0.0001) indication of linkage disequilibrium (and, thus, of a clonal population structure) among the isolates characterized.
Clonal groups were defined as unique combinations of subtypes (EcoRI ribotypes and hylB and sodA allelic types) as previously proposed by van Belkum et al. (45). A total of 39 clonal groups were found among the 104 human and bovine isolates (Table 2). Eleven clonal groups were unique to human isolates, and 26 clonal groups were unique to bovine isolates. Only two clonal groups shared both human and bovine isolates; clonal group A included 25 human isolates and 1 bovine isolate, and clonal group G included 1 human isolate and 1 bovine isolate.
TABLE 2.
Genetic characteristics and distributions of S. agalactiae clonal groups
| Clonal group | EcoRI ribotype | Allele of indicated type
|
Incidence in indicated type(s) of isolates
|
|||
|---|---|---|---|---|---|---|
| hylB | sodA | Human (n = 52) | Bovine (n = 52) | Human and Bovine (n = 104) | ||
| A | 106-1501-S-8 | hyl-4.0 | sod-1.0 | 24 | 1 | 25 |
| B | 106-1501-S-8 | hyl-7.0 | sod-1.0 | 7 | 0 | 7 |
| C | 116-549-S-1 | hyl-6.0 | sod-3.0 | 6 | 0 | 6 |
| D | 106-1501-S-8 | hyl-3.0 | sod-1.0 | 4 | 0 | 4 |
| E | 106-1501-S-8 | hyl-2.0 | sod-2.0 | 2 | 0 | 2 |
| F | 116-611-S-7 | hyl-3.0 | sod-1.0 | 2 | 0 | 2 |
| G | 116-549-S-1 | hyl-6.0 | sod-1.0 | 1 | 1 | 2 |
| H | 106-1501-S-5 | hyl-3.0 | sod-1.0 | 1 | 0 | 1 |
| I | 106-1501-S-5 | hyl-4.0 | sod-4.0 | 1 | 0 | 1 |
| J | 106-1501-S-5 | hyl-6.1 | sod-2.0 | 1 | 0 | 1 |
| K | 106-1501-S-5 | hyl-6.1 | sod-4.0 | 1 | 0 | 1 |
| L | 106-1501-S-8 | hyl-4.0 | sod-1.1 | 1 | 0 | 1 |
| M | 106-1501-S-8 | hyl-7.1 | sod-1.0 | 1 | 0 | 1 |
| N | 106-1502-S-6 | hyl-1.0 | sod-4.0 | 0 | 7 | 7 |
| O | 106-1501-S-5 | hyl-1.0 | sod-4.0 | 0 | 5 | 5 |
| P | 106-1501-S-5 | hyl-1.1 | sod-4.0 | 0 | 5 | 5 |
| Q | 106-1502-S-2 | hyl-5.0 | sod-4.0 | 0 | 5 | 5 |
| R | 106-1502-S-3 | hyl-5.0 | sod-4.0 | 0 | 3 | 3 |
| S | 106-1504-S-1 | hyl-1.0 | sod-4.0 | 0 | 3 | 3 |
| T | 106-1501-S-4 | hyl-1.0 | sod-4.0 | 0 | 2 | 2 |
| U | 116-611-S-7 | hyl-1.0 | sod-4.0 | 0 | 2 | 2 |
| V | 106-1501-S-5 | hyl-1.2 | sod-4.0 | 0 | 1 | 1 |
| W | 106-1501-S-5 | hyl-5.0 | sod-4.0 | 0 | 1 | 1 |
| X | 106-1502-S-1 | hyl-1.0 | sod-4.0 | 0 | 1 | 1 |
| Y | 106-1502-S-2 | hyl-5.1 | sod-4.0 | 0 | 1 | 1 |
| Z | 106-1502-S-3 | hyl-5.4 | sod-4.0 | 0 | 1 | 1 |
| AA | 106-1502-S-6 | hyl-5.0 | sod-1.2 | 0 | 1 | 1 |
| AB | 106-1502-S-6 | hyl-5.0 | sod-4.0 | 0 | 1 | 1 |
| AC | 116-522-S-1 | hyl-5.0 | sod-4.0 | 0 | 1 | 1 |
| AD | 116-522-S-1 | hyl-5.2 | sod-4.0 | 0 | 1 | 1 |
| AE | 116-549-S-1 | hyl-1.3 | sod-4.1 | 0 | 1 | 1 |
| AF | 116-549-S-1 | hyl-6.2 | sod-1.0 | 0 | 1 | 1 |
| AG | 116-549-S-7 | hyl-1.0 | sod-4.0 | 0 | 1 | 1 |
| AH | 116-574-S-2 | hyl-5.0 | sod-4.0 | 0 | 1 | 1 |
| AI | 116-607-S-6 | hyl-6.0 | sod-1.0 | 0 | 1 | 1 |
| AJ | 116-611-S-7 | hyl-5.3 | sod-4.0 | 0 | 1 | 1 |
| AK | 116-622-S-8 | hyl-1.0 | sod-4.0 | 0 | 1 | 1 |
| AL | 116-638-S-1 | hyl-1.0 | sod-4.0 | 0 | 1 | 1 |
| AM | 116-646-S-2 | hyl-1.0 | sod-4.0 | 0 | 1 | 1 |
Discriminatory power of subtyping methods and genetic diversity.
The SIDs, along with the corresponding confidence intervals, were determined for all subtyping methods (EcoRI ribotyping and hylB and sodA allelic types) across three populations of isolates (human only, bovine only, and human and bovine) (Table 3). The combination of all three methods allowed for an index of discrimination (SID [mean ± standard deviation], 0.93 ± 0.03) that was almost identical to the SID obtained by EcoRI ribotyping and hylB sequence typing combined (0.92 ± 0.03). The SID obtained for EcoRI ribotyping alone, as well as that for EcoRI ribotyping combined with hylB and/or sodA sequencing, showed a significantly higher genetic diversity among bovine isolates than among human isolates (Table 3).
TABLE 3.
SIDs for different S. agalactiae subtyping methods
| Method(s) | SID (95% confidence interval) for indicated type(s) of isolates
|
||
|---|---|---|---|
| Human (n = 52) | Bovine (n = 52) | Human and bovine (n = 104) | |
| EcoRI ribotyping | 0.42 (0.27, 0.57) | 0.90 (0.85, 0.94) | 0.81 (0.75, 0.87) |
| hylB allelic typing | 0.71 (0.60, 0.82) | 0.73 (0.64, 0.81) | 0.85 (0.82, 0.89) |
| sodA allelic typing | 0.40 (0.23, 0.56) | 0.22 (0.07, 0.36) | 0.61 (0.56, 0.66) |
| EcoRI ribotyping and sodA allelic typing | 0.51 (0.35, 0.67) | 0.91 (0.86, 0.95) | 0.84 (0.79, 0.90) |
| EcoRI ribotyping and hylB allelic typing | 0.74 (0.63, 0.84) | 0.96 (0.94, 0.98) | 0.92 (0.88, 0.95) |
| hylB allelic typing and sodA allelic typing | 0.75 (0.65, 0.85) | 0.73 (0.63, 0.83) | 0.87 (0.83, 0.90) |
| EcoRI ribotyping and hylB allelic typing and sodA allelic typing | 0.76 (0.65, 0.87) | 0.96 (0.94, 0.97) | 0.93 (0.89, 0.96) |
Hyaluronidase activity among selected S. agalactiae isolates.
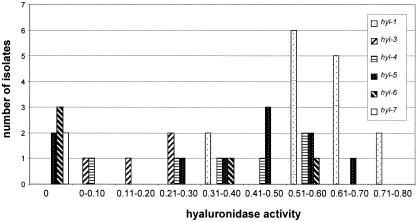
Hyaluronidase activities were determined for 43 representative S. agalactiae isolates. The average hyaluronidase activity of human isolates (n = 15) was significantly lower than the average hyaluronidase activity for bovine isolates (n = 28) (P < 0.01). Frequency distributions of average hyaluronidase activities for hylB clusters (Fig. 4) revealed that most isolates in cluster hyl-1 had activities towards the upper range of the data, whereas isolates in clusters hyl-3 and hyl-4 were distributed across the lower and middle ranges of the data, respectively. Clusters hyl-5 and hyl-6 showed evidence of a bimodal distribution of activities; some isolates had no activity, while others had medium to high activity. There was no apparent correlation between hyl alleles (e.g., hyl-5.1 and hyl-5.2) and hyaluronidase activities, but no formal statistical analyses for associations could be conducted, due to the small number of isolates assayed for each suballele. All isolates in cluster hyl-7, which contained insertions of the transposable element IS1548 in hylB, showed no hyaluronidase activity. In addition, two bovine isolates and one human isolate in cluster hyl-6 and two animal isolates in cluster hyl-5 also showed no detectable hyaluronidase activity in this in vitro assay. Based on the Kruskal-Wallis analysis of variance, there is a statistically significant difference in hyaluronidase activity between hylB clusters. Specifically, isolates in cluster hyl-1, which is associated with bovine hosts, showed a significantly higher average level of hyaluronidase activity than isolates in cluster hyl-3, which is associated with human hosts, and isolates in cluster hyl-6, which is numerically but not statistically associated with human hosts. Activities for isolates in clusters hyl-4 and hyl-5 overlap with those for isolates in cluster hyl-1 (the high end of the activity range for both cluster hyl-4 and cluster hyl-5) as well as with activities for isolates in clusters hyl-3 and hyl-6 (the low end of the activity range for both cluster hyl-4 and cluster hyl-5).
FIG. 4.
Frequency distribution of hyaluronidase activity levels for isolates grouping into different hylB clusters. Labels on the x axis indicate activities higher than the lower bound of the range and up to and including the upper bound of the range. Hyaluronidase activity levels are expressed as absorbance values normalized to the negative control absorbance values; higher values indicate higher levels of hyaluronidase activity.
Tissue culture invasion assay.
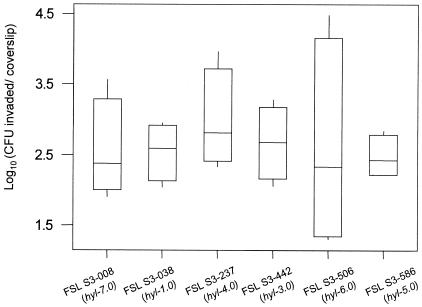
Representative human (n = 3) and bovine (n = 3) S. agalactiae isolates were evaluated for their invasion phenotypes in the HeLa human epithelial cell line. There was no significant difference in relative invasions of human and bovine isolates (P = 0.5; one-way analysis of variance) (Fig. 5). One of the isolates tested (FSL S3-008) contained allele hyl-7.0, which is characterized by an IS1548 insertion in hylB. This isolate did not differ from the other isolates, which did not carry an IS1548 insertion in the sequenced hylB fragment, in HeLa invasion properties.
FIG. 5.
Box plot of HeLa cell invasion efficiency of three human S. agalactiae isolates and three bovine S. agalactiae isolates. Invasion phenotypes as shown on the y axis were measured as log10 (CFU invaded per coverslip). Values represent the medians and interquartile ranges from four independent experiments. Isolates tested are indicated on the x axis; isolates FSL-S3-008, FSL-S3-237, and FSL-S3-442 represent isolates from humans with invasive infections, and isolates FSL-S3-038, FSL-S3-506, and FSL-S3-586 were obtained from bovine hosts.
DISCUSSION
While it has been well established that bacteria classified in the species S. agalactiae can cause severe human invasive disease as well as clinical and subclinical mastitis in cattle, the likelihood and potential of S. agalactiae transmission between these two host species has not been clearly defined. Based on our data, we conclude that (i) EcoRI ribotyping in combination with hylB and sodA sequencing provides for discriminatory subtype analysis of S. agalactiae; (ii) human invasive and bovine S. agalactiae strains represent distinct populations with limited interspecies transmission, as determined by both molecular subtype data and phenotypic data (i.e., hyaluronidase activities); and (iii) hyaluronidase activity is not required for all human infections.
EcoRI ribotyping in combination with hylB and sodA sequencing provides a discriminatory subtyping approach for S. agalactiae.
While banding pattern-based DNA subtyping methods (e.g., RAPD analysis) have been shown to successfully differentiate among S. agalactiae strains (32), difficulties in standardization and reproducibility of results can limit the usefulness of some of these subtyping methods. DNA sequencing-based subtyping methods, on the other hand, provide highly standardized and portable subtype data, which are also suitable for phylogenetic analyses (31). Thus, we used automated EcoRI ribotyping, one of the most standardized and reproducible DNA banding-based subtyping methods currently available (6, 47), and DNA sequencing of the putative virulence gene hylB (encoding hyaluronidase) and the housekeeping gene sodA (encoding superoxide dismutase) as subtyping methods for S. agalactiae in this study. Ribotyping has been applied to various species and has been reported to be useful for population genetics and transmission studies because this method scores evolutionary changes that appear to accumulate slowly enough to allow large-scale surveillance yet occur frequently enough to provide sufficient discrimination to track epidemic clones (43). DNA sequencing-based subtyping methods, on the other hand, produce not only highly reproducible and unambiguous data but also, unlike banding-based DNA subtyping methods, provide data suitable for evolutionary analysis (47).
While our data showed that EcoRI ribotyping or allelic analysis of sodA or hylB alone provided only limited subtype discrimination, the combination of the different subtyping methods provided sufficient discrimination for our studies. Interestingly, sodA, which encodes a superoxide dismutase (14), appeared highly conserved and of limited subtype diversity, while hylB, a putative virulence gene (30), showed significantly higher diversity. These data further support our choice of including the sequencing of a putative virulence gene (hylB) in a DNA sequencing-based typing scheme and are consistent with previous observations that the sequencing of virulence genes provides higher subtype discrimination than the sequencing of housekeeping genes (7). Sequencing data from housekeeping genes are necessary for long-term epidemiological analyses, though, since these genes are unlikely to be under positive selection (31, 42). In a previous study, ribotyping using enzymes PstI, HindIII, and CfoI was able to differentiate between unrelated S. agalactiae isolates from neonates, with an SID of >0.95 when data for different enzymes were combined; single-enzyme ribotyping provided discrimination similar to that reported here for EcoRI ribotyping (8). EcoRI ribotyping provided easily interpretable and distinct patterns (Fig. 2), and we thus conclude that the combination of EcoRI ribotyping and allelic analysis of sodA or hylB provides a suitable subtyping approach for studying the transmission and epidemiology of human and bovine S. agalactiae isolates.
Human and bovine S. agalactiae isolates represent distinct subtypes and populations.
Studies on transmission and epidemiology require sensitive and reproducible subtyping approaches, as well as truly representative and unbiased isolate collections appropriate for the hypothesis tested. Our hypothesis was that bovine S. agalactiae isolates in milk represent distinct subtypes from those S. agalactiae isolates that cause human invasive disease. To test this hypothesis, we conducted a prospective study of temporally and geographically matched S. agalactiae isolates collected through existing surveillance systems in New York. While many previous subtyping studies on human S. agalactiae strains have used and/or included isolates from asymptomatic carriers (4, 18, 32, 38), the human isolates used here were exclusively from human invasive infections. Molecular subtyping data for the human clinical and bovine isolates characterized here were used in two separate and independent statistical analyses (i.e., a frequency distribution of subtypes and a comparison of the genetic diversities of human and bovine populations) to probe whether human clinical isolates and bovine milk isolates represent separate populations.
Each of the three subtyping approaches used clearly provided strain discrimination into subtypes that largely showed host-specific distribution patterns. Although associations were not always exclusive, our data revealed several statistically significant and exclusive associations between subtypes and host species. Utilizing an approach proposed by van Belkum et al. (45), we also used a combination of all the subtyping methods to define S. agalactiae clonal groups. Human and bovine isolates largely grouped into separate clonal groups; only 2 of the 39 clonal groups contained both human and bovine isolates. These data are consistent with an RAPD subtyping study on 223 S. agalactiae isolates from bovines and human asymptomatic carriers (32), which found that human and bovine S. agalactiae isolates largely represented separate subtypes, even though one human isolate and one bovine isolate had identical RAPD types. Our data are also consistent with a recent MLST study on bovine and human invasive and carrier isolates, which showed that human and bovine isolates represent largely separate populations and in which only 3 of 50 sequence types were found in both humans and cattle (4). While this study found that a single hyperinvasive clonal group linked to neonatal infections grouped with bovine isolates and has likely arisen from a bovine ancestor, this clonal group was unique to humans and was not isolated from any bovine cases (4).
A comparison of the genetic diversities (as determined by EcoRI ribotyping) of human and bovine S. agalactiae populations performed by using Simpson's index of diversity as described by Grundmann et al. (16) showed that these S. agalactiae isolates represent distinct populations. Interestingly, bovine isolates showed a significantly higher EcoRI ribotype diversity than human isolates, while hylB and sodA allelic diversities did not differ significantly between human and bovine isolates. The relatively low degree of genetic diversity among human isolates is consistent with previous studies, which showed that S. agalactiae isolates from humans with invasive infections had less diversity than isolates from asymptomatic human carriers, which had a high degree of genetic diversity (19, 32). While a recent MLST study showed that human isolates were more diverse than bovine isolates, human isolates in this study were obtained from multiple countries and continents and represented both patients with invasive cases and carriers, whereas bovine isolates were from a single country (4). While our data thus support the idea that bovine milk isolates and human invasive isolates of S. agalactiae represent distinct populations with very little overlap, further studies using temporally and geographically matched isolate populations and standardized and reproducible subtyping methods (including, for example, MLST) (25) will be necessary to determine the relationships between S. agalactiae isolates found in human asymptomatic carriers and bovines, particularly since preliminary data provide some evidence for an overlap between these populations (24). MLST studies on temporally and geographically matched isolate populations will also be necessary to further probe the transmission of the hyperinvasive human neonatal S. agalactiae clone (10), which appears to have arisen from a bovine ancestor (4).
Human and bovine isolates differ in hyaluronidase activity but not in ability to invade human epithelial cells.
Based on our preliminary hylB DNA sequence data and prior studies with S. agalactiae isolates from human patients with neonatal meningitis, which identified a single clone that produces unusually high amounts of hyaluronidase (29, 36), we focused further investigations on the role of hyaluronidase activity in S. agalactiae host specificity. S. agalactiae hylB encodes hyaluronate lyase (hyaluronidase), a putative virulence factor that is involved in cleaving hyaluronic acid, a component of the extracellular matrix in many tissues (30). Hyaluronidases have also been found to be important virulence factors in other gram-positive organisms, such as Streptococcus pyogenes (22). Overall, human isolates showed a significantly lower average hyaluronidase activity than bovine isolates, indicating that S. agalactiae isolates from cattle and human invasive cases also represent phenotypically distinct populations. Our data also show significant differences in hyaluronidase activities for hylB clusters, which are consistent with the host-specific distribution of hylB clusters that we observed. Interestingly, we found that 8 of the 52 human isolates and none of the 52 bovine isolates characterized in our study carried an IS1548 insertion in hylB, which was associated with a hyaluronidase-negative phenotype. Our findings are consistent with previous studies that showed the presence of the IS1548 insertion in hylB in S. agalactiae isolates from human endocarditis and septicemia patients and from human asymptomatic carriers (15) as well as in human and feline isolates collected in Europe (49). These data further support the idea that hyaluronidase may not be required for the ability of S. agalactiae to cause certain infections (e.g., endocarditis) in human hosts. Interestingly, we also identified an additional five isolates (including four from bovines) with a hyaluronidase-negative phenotype without an apparent IS1548 in the hylB fragment sequenced. This fact may indicate that hyaluronidase activity is also not always required for animal infection or colonization, or it may suggest that these strains regulate hyaluronidase activity more tightly so that it cannot be detected in vitro.
While early studies have shown that both human and bovine S. agalactiae isolates can adhere to bovine mammary gland epithelial cells (5), we were not able to find any prior data on the ability of bovine isolates to adhere to and invade human epithelial cells. Using an established tissue culture model of human S. agalactiae invasion (40), we did not find any significant difference between the invasion phenotypes of three selected bovine S. agalactiae isolates and three human S. agalactiae isolates. One isolate, which contained the IS1548 insertion in the sequenced hylB, did not differ from the other isolates in its HeLa invasion properties. This fact indicates that hylB is not essential for the invasion of human epithelial cells.
To further probe for differences in the role of hyaluronidase in the pathogenesis of bovine mastitis and human invasive infections, we performed a preliminary analysis of the ratio of nonsynonymous nucleotide changes to synonymous nucleotide changes within the hylB fragment sequenced. The higher ratio for hylB sequences from human invasive isolates and the presence of IS1548 in only human isolates add evidence to the hypothesis of reduced functional constraints on hylB in human isolates compared to that in bovine isolates. While hyaluronidase appears to play an important role in the pathogenesis of bovine mammary infections, this enzyme may not be required for at least some forms of human infections and may thus have accumulated deleterious mutations (including IS1548 insertions). Similar observations have been reported for other bacterial species. For example, in Salmonella enterica, a loss of gene function has been observed for some serotypes that have adapted to an avian host range (3). Reductive evolution has also been documented in obligate intracellular parasites, such as Rickettsia and Chlamydia spp. (9).
Conclusions.
Our data indicate the existence of distinct S. agalactiae subtypes and clonal groups that appear to differ in their host specificities as well as their pathogenic potentials. These findings provide evidence that the potential for transmission of S. agalactiae from a bovine host to a human host or vice versa is low. In spite of the low transmission potential between human and bovine species predicted by our results, it appears that human-associated isolates have the necessary virulence factors to occasionally cause bovine infections, as indicated by the rare occurrence of some human-associated subtypes among bovine isolates found in both this study and a recent MLST study (4). Further studies using additional human and bovine tissue culture cell lines and appropriate animal models will be necessary, though, to understand whether specific functional barriers restrict the interspecies transmission of S. agalactiae. Ultimately, research of this nature should lead to the development of specifically targeted disease prevention strategies for human- and bovine-associated S. agalactiae subtypes. Our findings also add to the increasing recognition of host-specific bacterial clones among pathogens that otherwise might be considered candidates for interspecies transmission in the absence of appropriate subtyping data (26). For example, evidence for host and/or tissue specificity among clonal groups has been described for Pasteurella haemolytica, which, as a species, is associated with infections of sheep and cattle (11); for Bordetella bronchiseptica, which is associated with infections of pigs and dogs (35); and for Staphylococcus aureus, which is associated with infections in human and different animal species (37).
Acknowledgments
This publication was developed under the auspices of the Cornell University Center for Biotechnology, a New York State Center for Advanced Technology supported by New York State, and West Agro Inc., located in Hamilton, N.Y.
We thank the staff at the Quality Milk Production Services at Cornell University for providing bovine isolates and the staff at the New York State Department of Health for providing human isolates. We also thank Courtney E. Bolger, Severino W. Cuison, Ivy Tan, Magenta Sim, and Rachel M. Willems for their dedicated service to this project. Special thanks to Céline A. Nadon and Barbara M. Bowen for their help with revisions and Mario Roma for his help with the tissue culture study. Finally, we thank Bernhard Haubold for his time and assistance with LIAN 3.0.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Balter, S., C. G. Whitney, and A. Schuchat. 2000. Epidemiology of group B streptococcal infections, p. 154-162. In V. A. Fischetti, R. P. Novick, J. J. Ferreti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 3.Baumler, A. J., R. M. Tsolis, T. A. Ficht, and L. G. Adams. 1998. Evolution of host adaptation in Salmonella enterica. Infect. Immun. 66:4579-4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisharat, N., D. W. Crook, J. Leigh, R. M. Harding, P. N. Ward, T. J. Coffey, M. C. Maiden, T. Peto, and N. Jones. 2004. Hyperinvasive neonatal group B streptococcus has arisen from a bovine ancestor. J. Clin. Microbiol. 42:2161-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bramley, A. J., and E. M. Hogben. 1982. The adhesion of human and bovine isolates of Streptococcus agalactiae (group B) to bovine mammary gland epithelial cells. J. Comp. Pathol. 92:131-136. [DOI] [PubMed] [Google Scholar]
- 6.Bruce, J. L. 1996. Automated system rapidly identifies and characterizes micro-organisms in food. Food Technol. 50:77-81. [Google Scholar]
- 7.Cai, S., D. Y. Kabuki, A. Y. Kuaye, T. G. Cargioli, M. S. Chung, R. Nielsen, and M. Wiedmann. 2002. Rational design of DNA sequence-based strategies for subtyping Listeria monocytogenes. J. Clin. Microbiol. 40:3319-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatellier, S., H. Huet, S. Kenzi, A. Rosenau, P. Geslin, and R. Quentin. 1996. Genetic diversity of rRNA operons of unrelated Streptococcus agalactiae strains isolated from cerebrospinal fluid of neonates suffering from meningitis. J. Clin. Microbiol. 34:2741-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole, S. T., K. Eiglmeier, J. Parkhill, K. D. James, N. R. Thomson, P. R. Wheeler, N. Honore, T. Garnier, C. Churcher, D. Harris, K. Mungall, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. M. Davies, K. Devlin, S. Duthoy, T. Feltwell, A. Fraser, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, C. Lacroix, J. Maclean, S. Moule, L. Murphy, K. Oliver, M. A. Quail, M. A. Rajandream, K. M. Rutherford, S. Rutter, K. Seeger, S. Simon, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, K. Taylor, S. Whitehead, J. R. Woodward, and B. G. Barrell. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007-1011. [DOI] [PubMed] [Google Scholar]
- 10.Davies, H. D., N. Jones, T. S. Whittam, S. Elsayed, N. Bisharat, and C. J. Baker. 2004. Multilocus sequence typing of serotype III group B streptococcus and correlation with pathogenic potential. J. Infect. Dis. 189:1097-1102. [DOI] [PubMed] [Google Scholar]
- 11.Davies, R. L., S. Arkinsaw, and R. K. Selander. 1997. Evolutionary genetics of Pasteurella haemolytica isolates recovered from cattle and sheep. Infect. Immun. 65:3585-3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics. 5:164-166. [Google Scholar]
- 13.Furrer, B., U. Candrian, C. Hoefelein, and J. Luethy. 1991. Detection and identification of Listeria monocytogenes in cooked sausage products and in milk by in vitro amplification of haemolysin fragments. J. Appl. Bact. 70:372-379. [DOI] [PubMed] [Google Scholar]
- 14.Galliot, O., C. Poyart, P. Berche, and P. Trieu-Cuot. 1997. Molecular characterization and expression analysis of the superoxide dismutase gene from Streptococcus agalactiae. Gene. 204:213-218. [DOI] [PubMed] [Google Scholar]
- 15.Granlund, M., L. Oberg, M. Sellin, and M. Norgren. 1998. Identification of a novel insertion element, IS1548, in group B streptococci, predominantly in strains causing endocarditis. J. Infect. Dis. 177:967-976. [DOI] [PubMed] [Google Scholar]
- 16.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haubold, B., and R. R. Hudson. 2000. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Bioinformatics 16:847-848. [DOI] [PubMed] [Google Scholar]
- 18.Hauge, M., C. Jespersgaard, K. Poulsen, and M. Kilian. 1996. Population structure of Streptococcus agalactiae reveals an association between specific evolutionary lineages and putative virulence factors but not disease. Infect. Immun. 64:919-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helmig, R., N. Uldbjerg, J. Boris, and M. Kilian. 1993. Clonal analysis of Streptococcus agalactiae isolated from infants with neonatal sepsis or meningitis and their mothers and from healthy pregnant women. J. Infect. Dis. 168:904-909. [DOI] [PubMed] [Google Scholar]
- 20.Huet, H., C. Martin, P. Geslin, F. Grimont, and R. Quentin. 1993. Ribotyping of Streptococcus agalactiae strains isolated from vaginas of asymptomatic women. Res. Microbiol. 144:457-465. [DOI] [PubMed] [Google Scholar]
- 21.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hynes, W. L., A. R. Dixon, S. L. Walton, and L. J. Aridgides. 2000. The extracellular hyaluronidase gene (hylA) of Streptococcus pyogenes. FEMS Microbiol. Lett. 184:109-112. [DOI] [PubMed] [Google Scholar]
- 23.Jensen, N. E. 1982. Experimental bovine group-B streptococcal mastitis induced by strains of human and bovine origin. Nord. Vet. Med. 34:441-450. [PubMed] [Google Scholar]
- 24.Jensen, N. E., and F. M. Aarestrup. 1996. Epidemiological aspects of group B streptococci of bovine and human origin. Epidemiol. Infect. 117:417-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, N., J. F. Bohnsack, S. Takahashi, K. A. Oliver, M. S. Chan, F. Kunst, P. Glaser, C. Rusniok, D. W. Crook, R. M. Harding, N. Bisharat, and B. G. Spratt. 2003. Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 41:2530-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapur, V., W. M. Sischo, R. S. Greer, T. S. Whittam, and J. M. Musser. 1995. Molecular population genetic analysis of Staphylococcus aureus recovered from cows. J. Clin. Microbiol. 33:376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keefe, G. P. 1997. Streptococcus agalactiae mastitis: a review. Can. Vet. J. 38:429-437. [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 29.Li, S., and M. J. Jedrzejas. 2001. Hyaluronan binding and degradation by Streptococcus agalactiae hyaluronate lyase. J. Biol. Chem. 276:41407-41416. [DOI] [PubMed] [Google Scholar]
- 30.Lin, B., S. K. Hollingshead, J. E. Coligan, M. L. Egan, J. R. Baker, and D. G. Pritchard. 1994. Cloning and expression of the gene for group B streptococcal hyaluronate lyase. J. Biol. Chem. 269:30113-30116. [PubMed] [Google Scholar]
- 31.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez, G., J. Harel, R. Higgins, S. Lacouture, D. Daignault, and M. Gottschalk. 2000. Characterization of Streptococcus agalactiae isolates of bovine and human origin by randomly amplified polymorphic DNA analysis. J. Clin. Microbiol. 38:71-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGuire, G., and F. Wright. 1998. TOPAL: recombination detection in DNA and protein sequences. Bioinformatics 14:219-220. [DOI] [PubMed] [Google Scholar]
- 34.Mosabi, J. M., S. M. Arimi, and E. K. Kang'ethe. 1997. Isolation and characterization of group B streptococci from human and bovine sources within and around Nairobi. Epidemiol. Infect. 118:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Musser, J. M., D. A. Bemis, H. Ishikawa, and R. K. Selander. 1987. Clonal diversity and host distribution in Bordetella bronchiseptica. J. Bacteriol. 169:2793-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Musser, J. M., S. J. Mattingly, R. Quentin, A. Goudeau, and R. K. Selander. 1989. Identification of a high-virulence clone of type III Streptococcus agalactiae (group B streptococcus) causing invasive neonatal disease. Proc. Natl. Acad. Sci. USA 86:4731-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Musser, J. M., P. M. Schlievert, A. W. Chow, P. Ewan, B. N. Kreiswirth, V. T. Rosdahl, A. S. Naidu, W. Witte, and R. K. Selander. 1990. A single clone of Staphylococcus aureus causes the majority of cases of toxic shock syndrome. Proc. Natl. Acad. Sci. USA 87:225-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quentin, R., H. Huet, F. S. Wang, P. Geslin, A. Goudeau, and R. K. Selander. 1995. Characterization of Streptococcus agalactiae strains by multilocus enzyme genotype and serotype: identification of multiple virulent clone families that cause invasive neonatal disease. J. Clin. Microbiol. 33:2576-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rivas, A. L., R. N. Gonzalez, M. Wiedmann, J. L. Bruce, E. M. Cole, G. J. Bennett, H. F. Schulte III, D. J. Wilson, H. O. Mohammed, and C. A. Batt. 1997. Diversity of Streptococcus agalactiae and Staphylococcus aureus ribotypes recovered from New York dairy herds. Am. J. Vet. Res. 58:482-487. [PubMed] [Google Scholar]
- 40.Rubens, C. E., S. Smith, M. Hulse, E. Y. Chi, and G. van Belle. 1992. Respiratory epithelial cell invasion by group B streptococci. Infect. Immun. 60:5157-5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schrag, S. J., S. Zywicki, M. M. Farley, A. L. Reingold, L. H. Harrison, L. B. Lefkowitz, J. L. Hadler, R. Danila, P. R. Cieslak, and A. Schuchat. 2000. Group B streptococcal disease in the era of intrapartum antibiotic prophylaxis. N. Engl. J. Med. 342:15-20. [DOI] [PubMed] [Google Scholar]
- 42.Selander, R. K., D. A. Caugant, H. Ochman, J. M. Musser, M. N. Gilmour, and T. S. Whittam. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51:873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Struelens, M. J., Y. De Gheldre, and A. Deplano. 1998. Comparative and library epidemiological typing systems: outbreak investigations versus surveillance systems. Infect. Control Hosp. Epidemiol. 19:565-569. [DOI] [PubMed] [Google Scholar]
- 44.Tung, J. S., G. E. Mark, and G. F. Hollis. 1994. A microplate assay for hyaluronidase and hyaluronidase inhibitors. Anal. Biochem. 223:149-152. [DOI] [PubMed] [Google Scholar]
- 45.van Belkum, A., M. Struelens, A. de Visser, H. Verbrugh, and M. Tibayrenc. 2001. Role of genomic typing in taxonomy, evolutionary genetics, and microbial epidemiology. Clin. Microbiol. Rev. 14:547-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van den Heever, L. W., and W. H. Giesecke. 1980. Experimental induction of bovine mastitis with human strains of group B streptococci (Streptococcus agalactiae). J. S. Afr. Vet. Assoc. 51:107-109. [PubMed] [Google Scholar]
- 47.Wiedmann, M. 2002. Subtyping of bacterial foodborne pathogens. Nutr. Rev. 60:201-208. [DOI] [PubMed] [Google Scholar]
- 48.Wilson, D., R. Gonzalez, and H. Das. 1997. Bovine mastitis pathogens in New York and Pennsylvania: prevalence and effects on somatic cell count and milk production. J. Dairy Sci. 80:2592-2598. [DOI] [PubMed] [Google Scholar]
- 49.Yildirim, A. O., C. Lammler, R. Weiss, and P. Kopp. 2002. Pheno- and genotypic properties of streptococci of serological group B of canine and feline origin. FEMS Microbiol. Lett. 212:187-192. [DOI] [PubMed] [Google Scholar]