Abstract
With six separate wards accommodating more than 1,600 patients, V Nursing Center (VNC) is a long-stay psychiatric nursing center in eastern Taiwan. During 2001 to 2003, 39 shigellosis cases occurred in VNC. Different from the notion that most cases of shigellosis are caused by Shigella sonnei, all except one of these cases were caused by S. flexneri, with the remaining one caused by an S. sonnei isolate. O-antigen serotyping showed that the 38 S. flexneri strains were of either type 1a (n = 20) or 4a (n = 18), two less prevalent serotypes in Taiwan. NotI-based pulsed-field gel electrophoresis analyses performed with 8 type 1a non-VNC strains and 9 type 4a non-VNC strains isolated from 1996 to 2003 for comparison divided the 28 type 1a strains and the 27 type 4a strains into 7 and 10 subtypes, designated subtypes P1A to P1G and subtypes P4A to P4J, respectively. Subtypes P1A and P4A, which appeared in three consecutive years in VNC as well as outside of VNC, are the most prevalent subtypes. Analyses of the relatedness of the VNC strains on the basis of the banding patterns grouped the type 1a and 4a strains into four and five clusters, respectively. All except one of the type 1a strains had 95% similarity, indicating that they had a common parent, whereas the type 4a strains had similarities that ranged from 77 to 93%, suggesting that they were of diverse origins. In two of the outbreaks, less related subtypes of the type 4a strains were found in the same VNC wards in consecutive years, suggesting the possible existence of different subtypes in VNC all the time. Antibiotic susceptibility testing showed that all except one of the S. flexneri strains were sensitive to at least seven antibiotics; the remaining isolate was sensitive to three antibiotics. The data from the latter tests should be helpful for selection of proper treatments for S. flexneri infections in Taiwan.
Shigellosis, which is caused by Shigella species, including Shigella dysenteriae, S. flexneri, S. boydii, and S. sonnei, is recognized as a global problem and continues to be one of the important causes of diarrheal diseases. It causes an estimated 165 millions cases and results in 1.1 millions deaths per year worldwide (14). Epidemic and endemic cases are most frequently caused by S. dysenteriae and S. flexneri in developing countries and S. sonnei in industrialized countries (1, 5, 10, 14, 26). Shigella transmission occurs via the fecal-oral route, usually from person to person, or the intake of contaminated food or drinking water. Shigellosis outbreaks involving S. sonnei are mostly associated with crowding and poor sanitation in institutions such as schools, day care centers, and nursing centers and also occur among members of tour groups (4, 7, 12, 13, 15, 16, 19, 25, 29). In the United States, 70% of the infections are caused by S. sonnei and 24% are caused by S. flexneri, with the majority (49%) of infections affecting children younger than 9 years of age (5, 22). In Taiwan, 200 to 500 cases of shigellosis were identified annually from 1998 to 2002; in 2001, however, there were with 1,327 cases, for an average annual incidence rate of 2.42 per 100,000 population. The majority (51%) of infections occurred in children younger than 9 years of age and were caused by S. sonnei or S. flexneri, which accounted for 73.3 and 26.5% of the total Shigella strains isolated in Taiwan, respectively (9).
Shigella organisms are highly contagious, with infectious doses of as low as 10 to 100 viable bacterial cells and with infections having an incubation period of 1 to 5 days (14, 28). Patients who recover from acute shigellosis might shed Shigella organisms for as long as 12 days if they remain untreated, but excretion might last for more than 1 year in individuals with chronic cases (28). The most prevalent serotypes of S. flexneri are 1b, 2a, 3a, 4a, and 6 in developing countries and 2a in developed countries (14). In Taiwan, the most prevalent serotype of S. flexneri is 2a, followed by 4a, 2b, and then 1a, with 1a being only found in eastern Taiwan in recent years (9).
During 2001 to 2003, four outbreaks and 12 sporadic shigellosis cases occurred in a long-stay psychiatric nursing center (V Nursing Center [VNC]) in Hualien County in eastern Taiwan (Fig. 1). These cases resulted in the isolation of 38 strains of S. flexneri and 1 strain of S. sonnei. In this study, to illustrate the correlation among these VNC strains and to trace the possible sources of infection, the 38 S. flexneri isolates were subjected to serotyping, antibiogram, plasmid profile, and pulsed-field gel electrophoresis (PFGE) analyses, with 17 strains isolated elsewhere in eastern Taiwan during 1996 to 2003 (non-VNC strains) used for comparison.
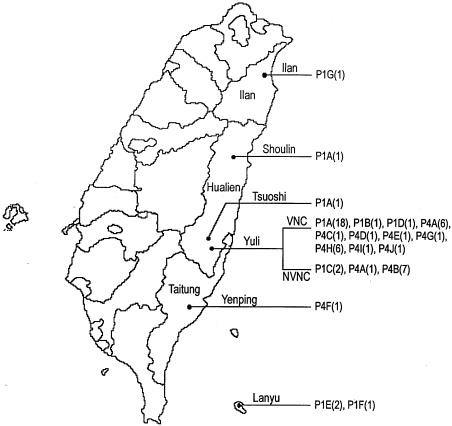
FIG. 1.
Map of Taiwan. The three counties in the east are labeled. The distributions of the strains are indicated by P1A to P1G and P4A to P4J, which represent the subtypes of S. flexneri types 1a and 4a, respectively, obtained by NotI-based PFGE. The numbers in parentheses indicate the numbers of isolate obtained.
MATERIALS AND METHODS
Bacterial strains.
The 55 Shigella strains used in this study were isolated from 1996 to 2003 from stool samples of patients with shigellosis and were kept at −70°C until use (Table 1). They included 38 S. flexneri strains and 1 S. sonnei strain isolated from patients involved in outbreaks or with sporadic cases that occurred in VNC (VNC strains) and 17 strains isolated from patients elsewhere in eastern Taiwan (non-VNC strains). The identities of all isolates were verified by Gram staining, conventional biochemical tests (3), and slide agglutination tests with commercial polyclonal antiserum (Denka Seiken, Tokyo, Japan). Escherichia coli ATCC 25922 was used as the reference strain for antimicrobial susceptibility tests. Salmonella enterica serovar Braenderup H9812 was used as the reference strain in the PFGE analyses.
TABLE 1.
Characteristics of the 38 VNC isolates and 17 non-VNC isolates of S. flexneri
| Isolate | Onset date (mo/day/yr) | Source | PFGE pattern | Plasmid profile | Antibiogram type | Outbreak or sporadic case no.a |
|---|---|---|---|---|---|---|
| S. flexneri type 1a | ||||||
| VNC | ||||||
| 90e4802 | 09/23/01 | W4 | P1A | 1aI | A | OB1 |
| 90e4808 | 09/23/01 | W4 | P1A | 1aI | A | OB1 |
| 90e4947 | 09/23/01 | W4 | P1A | 1aI | A | OB1 |
| 90e4807 | 09/24/01 | W4 | P1B | 1aI | A | OB1 |
| 90e4939 | 10/04/01 | W4 | P1A | 1aI | A | OB1 |
| 90e4940 | 10/04/01 | W4 | P1A | 1aI | A | OB1 |
| 90e4982 | 10/05/01 | W4 | P1A | 1aI | A | OB1 |
| 90e5047 | 10/05/01 | W4 | P1A | 1aI | A | OB1 |
| 90e5053 | 10/05/01 | W4 | P1A | 1aI | A | OB1 |
| 90e5119 | 10/05/01 | W4 | P1A | 1aI | A | OB1 |
| 90e5041 | 10/05/01 | W4 | P1A | 1aI | A | OB1 |
| 91e0463 | 03/06/02 | W4 | P1A | 1aI | A | SR1 |
| 91e0833 | 03/26/02 | W4 | P1A | 1aI | A | SR2 |
| 92e0465 | 04/19/03 | W2 | P1D | 1aI | A | SR3 |
| 92e1065 | 09/20/03 | W4 | P1A | 1aI | A | SR4 |
| 92e1572 | 12/09/03 | W4 | P1A | 1aI | B | OB4 |
| 92e1587 | 12/17/03 | W4 | P1A | 1aI | B | OB4 |
| 92e1582 | 12/18/03 | W4 | P1A | 1aI | B | OB4 |
| 92e1577 | 12/19/03 | W4 | P1A | 1aI | B | OB4 |
| 92e1588 | 12/19/03 | W4 | P1A | 1aI | B | OB4 |
| Non-VNC | ||||||
| 85e47153 | 10/11/96 | Lanyu | P1E | 1aIII | A | SR5 |
| 86e47955 | 05/10/97 | Tsuoshi | P1A | 1aI | A | SR6 |
| 86e48398 | 08/16/97 | Lanyu | P1F | 1aIII | A | SR7 |
| 87e49704 | 01/13/98 | Lanyu | P1E | 1aIII | A | SR8 |
| 89e0903 | 09/07/00 | Shoulin | P1A | 1aI | A | SR9 |
| 91e0110 | 01/13/02 | Yuli | P1C | 1aII | A | SR10 |
| 91e0164 | 01/19/02 | Yuli | P1C | 1aI | A | SR11 |
| 91e0115 | 12/17/02 | Ilan | P1G | 1aIV | C | SR12 |
| S. flexneri type 4a | ||||||
| VNC | ||||||
| 90e5710 | 10/08/01 | W1 | P4J | 4aI | E | SR13 |
| 90e5256 | 10/08/01 | W2 | P4H | 4aI | D | OB2 |
| 90e5969 | 10/12/01 | W2 | P4E | 4aI | D | OB2 |
| 90e5967 | 10/15/01 | W2 | P4H | 4aI | D | OB2 |
| 90e5968 | 10/17/01 | W2 | P4H | 4aI | D | OB2 |
| 90e5999 | 10/25/01 | W2 | P4H | 4aI | D | OB2 |
| 90e6312 | 11/05/01 | W16 | P4A | 4aII | I | SR14 |
| 91e0488 | 03/11/02 | W2 | P4H | 4aI | D | OB3 |
| 91e0498 | 03/11/02 | W2 | P4H | 4aI | D | OB3 |
| 91e0593 | 03/15/02 | W16 | P4A | 4aI | D | OB3 |
| 91e0604 | 03/15/02 | W16 | P4A | 4aI | D | OB3 |
| 91e0612 | 03/15/02 | W16 | P4A | 4aI | D | OB3 |
| 91e0594 | 03/18/02 | W16 | P4I | 4aI | D | OB3 |
| 91e0955 | 05/06/02 | WC | P4A | 4aI | D | SR15 |
| 91e1351 | 08/23/02 | W3 | P4G | 4aIII | E | SR16 |
| 91e1875 | 12/16/02 | W3 | P4D | 4aIII | E | SR17 |
| 92e1179 | 11/04/03 | W16 | P4A | 4aI | D | SR18 |
| 92e1190 | 11/07/03 | W16 | P4C | 4aI | H | SR19 |
| Non-VNC | ||||||
| 90e2459 | 06/27/01 | Yenping | P4F | 4aIV | G | SR20 |
| 92e0537 | 05/19/03 | Yuli | P4B | 4aI | D | OB5 |
| 92e0538 | 05/19/03 | Yuli | P4B | 4aI | D | OB5 |
| 92e0539 | 05/19/03 | Yuli | P4B | 4aI | D | OB5 |
| 92e0548 | 05/19/03 | Yuli | P4B | 4aI | D | OB5 |
| 92e0555 | 05/19/03 | Yuli | P4B | 4aI | D | OB5 |
| 92e0569 | 05/19/03 | Yuli | P4B | 4aI | F | OB5 |
| 92e0601 | 05/19/03 | Yuli | P4B | 4aI | D | OB5 |
| 92e1100 | 10/09/03 | Yuli | P4A | 4aI | D | SR21 |
OB, outbreak; SR, sporadic case.
PFGE and banding pattern analyses.
The standard protocol described by Gautom (11) was followed for preparation of intact chromosomal DNA, in-gel digestion, and PFGE of the NotI digests in a 1% agarose gel with 0.5× TBE (Tris-borate-EDTA) buffer with a CHEF Mapper apparatus (Bio-Rad Laboratories, Hercules, Calif.) at 6 V/cm and an angle of 120° with pulses of 2.16 to 54.17 s for 19 h. The PFGE banding patterns were digitized with AlphaEase software (Applied Innotech Co., San Leandro, Calif.) and were then compared for similarity by using Bionumerics software (version 3.0; Applied Math, Austin, Tex.). On the basis of the Pearson correlation coefficient, a dendrogram was constructed by the unweighted pair group method with arithmetic averages clustering. A tolerance for the banding migration distance of 1.0% was applied during the comparison of the banding patterns. Isolates that differed by one or more bands were assigned to different subtypes.
Plasmid profile analysis.
The plasmids of the S. flexneri strains were prepared by using a Qiaprep Spin Miniprep kit (Qiagen, Hilden, Germany) and were separated by electrophoresis in a horizontal agarose gel (0.8% in TAE [Tris-acetate-EDTA] buffer). A mixture containing 11 supercoiled plasmids (Sigma, St. Louis, Mo.) was used as a molecular size marker for plasmids of 20 kb or smaller.
Antibiotic sensitivity test.
Disk diffusion tests were performed as recommended by the National Committee for Clinical Laboratory Standards (21) with disks, purchased from Oxoid (Basingstoke, United Kingdom), containing antibiotics of known concentrations. The antibiotics tested were amikacin, ampicillin, cefazolin, cefotaxime, cephalothin, chloramphenicol, gentamicin, nalidixic acid, ofloxacin, streptomycin, tetracycline, and trimethoprim-sulfamethoxazole.
Epidemiological data.
The information on the patients involved in the outbreaks and patients with sporadic cases were obtained from infectious diseases reporting forms filled out by the bureaus of public health of the relevant counties. The internal epidemiological investigation reports were recorded by the Sixth Branch of the Center for Disease Control, Taipei, Taiwan.
RESULTS AND DISCUSSION
Environmental descriptions.
Taiwan is an island geographically separated into eastern and western parts by the Central Mountains, which cross the country longitudinally (Fig. 1). The eastern part, which is less developed and much less populated than the western part, consists of three mountainous counties, Ilan, Hualien, and Taitung, from north to south. VNC is in Yuli, a township in Hualien County (Fig. 1). VNC has six separate three-story buildings, designated W1, W2, W3, W4, W16, and WC. The numbers of beds for chronic patients in the wards were 189 in W1, 248 in W2, 216 in W3, 360 in W4, 500 in W16, and 196 in WC, with an occupation rate of about 95% during 2001 to 2003 and an average period of patient hospitalization of about 9.8 years. The patients were all physically capable adults (age range, 40 to 68 years) but were unable to take care of themselves. Each ward, including the facilities and the nursing staffs, was managed separately, and each ward took care of about 28 patients. Even though the communities were isolated, patients in the same building still had direct or indirect contact with one another in the public areas, such as the exercise yards, bathrooms, and restaurants. Visitors and patients entering and leaving VNC were registered, and the records were retained. The periodic surveillance of the tap water indicated that the residual chlorine content was maintained within an effective range (0.2 to 0.8 ppm).
Occurrence of shigellosis in VNC.
Shigellosis is a notifiable disease in Taiwan, and all cases must be reported and the isolated strains must be sent to the Center for Disease Control of Taiwan. During 2001 to 2003, 39 culture-positive shigellosis cases occurred in VNC (Table 1). These included four outbreaks with 11, 5, 6, and 5 cases that occurred in 2001, 2001, 2002, and 2003, respectively; and a total of 12 sporadic cases occurred during 2001 to 2003. The incidences in W1, W2, W3, W4, W16, and WC of VNC were 1, 8, 2, 19, 7, and 1, respectively (Table 1). We collected 39 isolates from the patients involved in the outbreaks and the patients with sporadic cases described above. Strain identification indicated that, except for one S. sonnei isolate (which occurred in WC in 2001), all strains were S. flexneri (Table 1).
A few small-scale shigellosis outbreaks affecting psychogeriatric patients and elderly individuals in long-stay nursing centers have been reported. These outbreaks were all caused by S. sonnei, which is the same as the information in the records that the predominant pathogen responsible for the shigellosis outbreaks was S. sonnei (12, 13, 19). In contrast, the cases that occurred in VNC were different, in that the responsible agent was S. flexneri.
The 38 S. flexneri strains were each subjected to O-antigen serotyping by a slide agglutination test with commercial polyclonal antisera. Surprisingly, they belonged to either serotype 1a (20 strains) or serotype 4a (18 strains) (Table 1). The records of the Center for Disease Control of Taiwan indicate that type 1a strains had not been found in western Taiwan in recent years (6, 7), indicating that VNC might have been a reservoir for S. flexneri type 1a. Two points concerning the distribution of the serotypes were noticed (Table 1). First, while both serotypes appeared in W2, only one of the serotypes appeared in the other five wards. Second, all except one of the type 1a strains appeared in W4; one type 1a occurred in W2 (and was isolated in 2003), whereas the type 4a strains primarily appeared in W2 (n = 7) and W16 (n = 8). These observations suggest that the serotypes had been confined to the wards where they were isolated and rarely spread because the wards were managed separately.
During 1996 to 2003, while other serotypes also appeared, eight type 1a strains and nine type 4a strains (non-VNC strains) were isolated elsewhere in eastern Taiwan (Table 1). These strains were used for comparison in further studies.
PFGE typing and dendrograms of the S. flexneri isolates.
Serotyping is a simple and quick method for the identification of Shigella strains and is very useful for epidemiological surveillance. However, new DNA molecular subtyping methods may further differentiate a serotype into molecular subtypes and facilitate analysis and tracing of the source of infections (6, 7, 23, 24). NotI-based PFGE was previously shown (6, 7) to be a useful method for the molecular differentiation of S. flexneri strains isolated in Taiwan. Therefore, to elucidate the correlation among the VNC strains and to trace the possible source of the infections, NotI-based PFGE analyses were performed by using the eight type 1a non-VNC strains and the nine type 4a non-VNC strains for comparison.
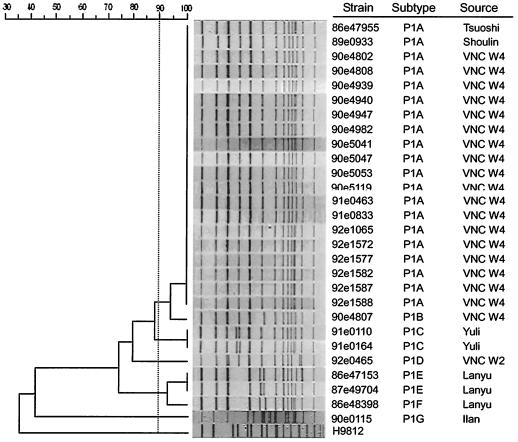
Separation of NotI digests of the S. flexneri 1a genomic DNA by PFGE displayed 13 to 14 DNA bands with sizes ranging from 20 to 600 kb. According to the banding patterns, the 28 type 1a strains were divided into the following seven subtypes: P1A (n = 20), P1B (n = 1), P1C (n = 2), P1D (n = 1), P1E (n = 2), P1F (n = 1), and P1G (n = 1) (Table 1; Fig. 2). The band patterns were subjected to relatedness analyses. In these analyses, isolates that showed similarities that were ≥90% were grouped into one cluster. Thus, the 28 S. flexneri type 1a strains were divided into five clusters. P1A and P1B formed one cluster and P1E and P1F grouped in one cluster, whereas P1C, P1D, and P1G each diversified into a separate cluster (Fig. 2). On the basis of the dendrogram, more than 75% similarity was shared among the type 1a isolates.
FIG. 2.
Dendrogram of the S. flexneri type 1a strains determined on the basis of NotI-based PFGE patterns. Twenty VNC isolates and 8 non-VNC strains are included.
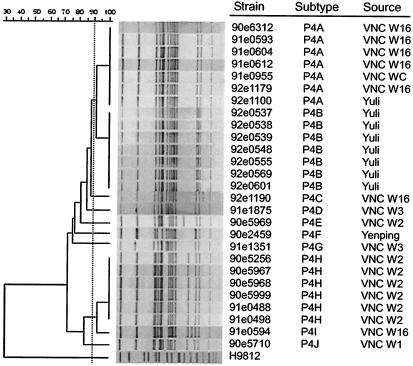
Digestion of the genomic DNA from the 27 S. flexneri type 4a strains gave 13 to 16 fragments that ranged in size from 20 to 600 kb. These type 4a strains were grouped into the following 10 subtypes: P4A (n = 7), P4B (n = 7), P4C (n = 1), P4D (n = 1), P4E (n = 1), P4F (n = 1), P4G (n = 1), P4H (n = 6), P4I (n = 1), and P4J (n = 1) (Table 1; Fig. 3). Relatedness analyses divided these subtypes into eight clusters. Cluster 1 included P4A and P4B; clusters 2, 3, 4, 5, and 6 contained P4C, P4D, P4E, P4F, and P4G, respectively; cluster 7 consisted of P4H and P4I; and cluster 8 included P4J (Table 1; Fig. 3). On the basis of the data from the dendrogram, more than 77% similarity was shared among all type 4a isolates except for the P1G strain isolated in Ilan.
FIG. 3.
Dendrogram of the S. flexneri type 4a strains determined on the basis of the NotI-based PFGE patterns. Eighteen VNC isolates and 9 non-VNC strains are included.
Epidemiology revealed by relatedness analysis.
Subtype P1A was the most frequently occurring subtype among the type 1a strains (20 of 28 isolates) and was the only subtype that appeared in both VNC (exclusively in W4) and elsewhere in Hualien. More importantly, in addition to causing shigellosis in three consecutive years (2001 to 2003), strains of this subtype had in fact previously caused two sporadic shigellosis cases outside VNC, one in 1997 (strain 86e47955, isolated in Tsuoshi) and the other in 2000 (strain 89e0903, isolated in Shoulin) (Fig. 1).
Several points were noticed from the outbreak in W4 (outbreak OB4) caused by subtype P1A, which occurred in December 2003 (Table 1). During this period, stool samples were taken from individuals who lived or worked in W4 and were subjected to organism isolation and identification. The samples were taken from patients with diarrhea, 26 individuals in close contact with the patients, 25 members of the nursing staff, and 73 kitchen workers. Additionally, swab samples taken from restaurants, toilets, and kitchens were also tested. The results showed that S. flexneri was detected only in the patients, whereas all other samples were Shigella negative. These data suggest that the bacteria were spread among the patients in W4 but not the other individuals in VNC. Furthermore, no visitors or patients were recorded to have entered or left VNC within the 1 month before the outbreak. These data suggest that a subtype P1A strain existed in an unnoticed carrier or a patient(s) with chronic shigellosis who shed organisms from time to time in W4 during 2001 to 2003.
Ten subtype P1A isolates and one subtype P1B isolate were involved in the outbreak that occurred in 2001 (outbreak OB1 in W4) (Table 1). Three interesting points were noticed here. First, the outbreak had two onset dates: the first on 23 and 24 September 2001 and the second on 4 and 5 October 2001 (Table 1). Most individuals who are infected with Shigella develop diarrhea, fever, and stomach cramps starting a day or two after they are exposed to the bacterium (28). An interval of 10 days is unusual. This suggests that there might have been an asymptomatic carrier(s) or an unnoticed infected patient(s) between the two onset dates. Second, because subtypes P1A and P1B were 95% similar (Fig. 2), they were most closely related. Third, the NotI-based PFGE patterns showed that subtype P1B differed from subtype P1A in a reduction of the size of fragment 6, suggesting that a new NotI restriction site had been generated within this fragment (Fig. 2).
Two subtype P1C strains were isolated in 2002 from patients in Yuli (Table 1), the same township where VNC is located. These two strains shared 89% similarity with subtypes P1A and P1B, suggesting that they were derived from a common parent. P1D, a VNC isolate with two band differences from subtypes P1A and P1B, is still closely related to the other VNC strains. Subtypes P1E and P1F, which formed a cluster, were found only in Lanyu, a small island with a few villages. Three strains of these two subtypes appeared in Lanyu in three consecutive years from 1996 to 1998. As they have greater than 93% similarity to each other, they are closely related. This is another example showing that shigellosis is transmitted in small communities. Subtype P1G, which was isolated in Ilan and which showed less than 40% similarity to the VNC strains, was apparently from a relatively distant source.
The 27 S. flexneri type 4a strains caused three outbreaks and nine sporadic cases from 2001 to 2003 (Table 1). They consisted of 26 VNC or non-VNC strains that were isolated in Yuli and 1 subtype P4F strain that was isolated in Yenping (Fig. 1). Notably, among the type 4a strains, subtype P4A appeared in three consecutive years and was the only subtype that appeared in more than one of the VNC wards (W16 and WC) as well as in Yuli but outside VNC (Fig. 1; Table 1). In other words, subtype P4A was the most widely spread of the type 4a strains. The subtype P4B strains, isolated exclusively outside VNC, shared about 93% similarity with the subtype P4A strains. The high degree of similarity suggests a recent divergence of the two subtypes, and the occurrence of the two strains in the same township indicates possible transmission between VNC and the communities in the vicinity.
In addition to subtype P4A, two other subtypes were also isolated in W16, subtypes P4C and P4I. Subtype P4C had 89% similarity with subtype P4A, and the two subtypes appeared to be from a common parent. However, subtype P4I formed a cluster different from that containing subtype P4A, indicating that different subtypes had existed in W16. Subtype P4D (isolated in W3) had 87% similarity to members of cluster 1, suggesting that they were more closely related to each other than to members of clusters F to I. In W2, two relatively distant subtypes were found, P4E and P4H, also indicating the existence of different subtypes. Subtype P4J (isolated in W1) was classified into cluster 8 and was more closely related to the members of subtypes P4H and P4I, thus suggesting a closer relationship to these strains.
Finally, it is worth noting that outbreak 2 occurred in W2 and outbreak 3 occurred in W2 and W16 and that the two outbreaks involved two (P4E and P4H) and three (P4B, P4H, and P4I) subtypes of type 4a strains, respectively (Table 1). It was not realized that these outbreaks were caused by different subtypes until the data from the NotI-based PFGE analyses were obtained. These findings demonstrate the usefulness of PFGE subtyping on the one hand (6, 7, 15) and suggest the potential existence of different subtypes in VNC all the time on the other.
Antimicrobial activity.
In developing countries, the rates of resistance of S. flexneri to antimicrobial drugs has been increasing in recent years (2, 10, 14, 18, 20, 26, 30). Therefore, we were curious about the resistance of the type 1a and 4a S. flexneri strains isolated in eastern Taiwan. The 38 VNC strains and the 17 non-VNC strains were separately tested for their susceptibilities to the 13 antibiotics listed in Materials and Methods. The testing differentiated the strains into nine antibiogram types (types A to I). As shown in Table 2, the situation can be summarized as follows. First, 22, 5, and 1 type 1a strains belonging to subtypes A, B, and C, respectively, were sensitive to 8, 9, and 11 antibiotics, respectively. Second, 20, 3, 1, 1, 1, and 1 of the type 4a strains belonging to subtypes D, E, F, G, H, and I, respectively, were sensitive to 11, 10, 9, 7, 7, and 3 antibiotics, respectively. Third, all of them were sensitive to amikacin and ofloxacin (data not shown), and except for the subtype H strain (strain 92e1190, a subtype P4C strain isolated in W16; Tables 1 and 2), they were all sensitive to cefotaxime, indicating that amikacin, ofloxacin, and cefotaxime were the most potent antibiotics. Taken together, the data presented above suggest that, except for the type I strain (strain 90e6312, a subtype P4B strain isolated in W16; Tables 1 and 2), the resistance situation is not as serious as that in developing countries. Finally, strain 90e6312, which was the true multiple-drug-resistant strain, was sensitive only to amikacin, ofloxacin, and cefotaxime. Although it appeared only once in 2001 and did not appear in the subsequent years, caution against a possible recurrence of this strain in the future must be taken.
TABLE 2.
Antimicrobial susceptibility phenotypes of the 28 type 1a and the 27 type 4a strains of S. flexneri
| Serotype | Antibiogram type | No. of strains | Antibiotic resistancea
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | KFZ | CEF | CHL | GEM | NAF | SXT | TET | STR | CTX | AMK | OFX | |||
| 1a | A | 22 | R | S | S | R | S | S | S | R | R | S | S | S |
| 1a | B | 5 | R | S | S | R | S | S | S | S | R | S | S | S |
| 1a | C | 1 | S | S | S | S | S | S | S | S | I | S | S | S |
| 4a | D | 20 | S | S | S | S | S | S | S | S | R | S | S | S |
| 4a | E | 3 | R | S | S | S | S | S | S | S | R | S | S | S |
| 4a | F | 1 | R | S | S | S | S | S | R | S | R | S | S | S |
| 4a | G | 1 | R | S | S | R | S | S | R | R | R | S | S | S |
| 4a | H | 1 | R | R | R | S | S | S | S | S | R | R | S | S |
| 4a | I | 1 | R | R | R | R | R | R | R | R | R | S | S | S |
AMK, amikacin; AMP, ampicillin; CEF, cephalothin; CHL, chloramphenicol; CTX, cefotaxime; GEM, gentamycin; KFZ, cefazolin; NAF, nalidixic acid; OFX, ofloxacin; STR, streptomycin; SXT, trimethoprim-sulphamethoxazole; TET, tetracycline; R, resistance; S, susceptible; I, intermediate.
Our data obtained in the present study may provide a strategy for the treatment of infections caused by these multiple-drug-resistant strains. The results of genetic clustering analysis indicated that infection with a specific strain was confined to a specific ward; infection between wards should not occur infrequently. A ward should be used as a control unit; and long-term control measures should be taken, with emphasis on environmental hygiene in the wards, since personal hygiene is not easily accomplished by the psychiatric patients and asymptomatic carriers can be a main source of infection. In addition, regular monitoring for diarrhea should be performed for the early detection of Shigella infection.
In recent years, the rates of integron-associated multidrug resistance have increased rapidly, such as resistance type ACSSuT (ampicillin, chloramphenicol, streptomycin, trimethoprim-sulfamethoxazole, and tetracycline resistance) in the family Enterobacteriaceae, primarily in S. enterica serovar Typhimurium DT104, but also in Shigella spp. (8, 17, 27). As shown in Table 2, among the S. flexneri isolates, 2 type 4a strains belonging to antibiogram types G and H had the ACSSuT resistance type, 22 type 1a strains (antibiogram type A) had the ACST resistance type (resistance to ampicillin, chloramphenicol, streptomycin, and tetracycline), and 5 type 1a strains (antibiogram type B) had the ACS resistance type (resistance to ampicillin, chloramphenicol, and streptomycin). Whether the antibiotic resistance of these S. flexneri isolates is integron associated remains to be investigated. Should it be true, the rapid and wide spread of resistance is anticipated in the future, because the mobile gene cassettes may contribute to the acquisition and dissemination of antibiotic resistance.
Plasmid profiles.
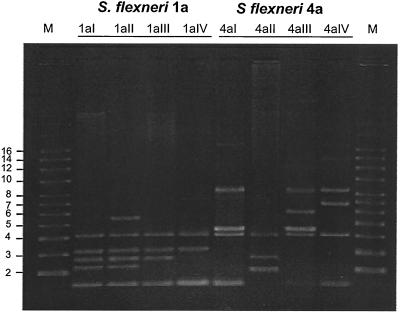
To examine whether correlations exist between antibiotic resistance and the presence of specific plasmids, all the S. flexneri strains described above were subjected to plasmid profile analysis. Since large plasmids are unstable and tend to be lost during storage, cultivation, and plasmid preparation, only those of less than 20 kb were taken into account for assignment to a plasmid profile. The profiles are shown in Fig. 4. Each of the strains contained plasmids that ranged in size from about 1 to 12 kb. One plasmid of about 3 kb was common in all strains. The type 1a and 4a strains were each differentiated into four plasmid subtypes. Among the type 1a strains, all isolates of subtypes P1A, P1B, and P1D and one of the two subtype P1C isolates belonged to plasmid subtype 1aI; the other subtype P1C isolate was differentiated into subtype 1aII. The subtype P1E and P1F isolates were grouped into subtype 1aIII, and the subtype P1G isolates belonged to subtype 1aIV (Table 1). Subtype 4aI included all strains of subtypes P4A, P4B (except one), P4C, and P4H; subtype 4aII contained only one of the subtype P4B strains; subtype 4aIII contained the subtype P4D and P4G strains; and subtype 4aIV contained the subtype P4F strains (Table 1). Multiple-drug-resistant strain 90e6312 (subtype 4aII) harbored three plasmids which were also found in strains of other the plasmid subtypes. In other words, no possible correlations were found between the presence of small plasmids and multiple-antibiotic resistance.
FIG. 4.
Profiles of plasmids extracted from representative type 1a and 4a S. flexneri strains. Plasmids were prepared as described in Materials and Methods and subjected to agarose gel (0.8%) electrophoresis. Lanes: M, markers containing 11 supercoiled plasmids (numbers to the left of the gel are in kilobases); 1aI, S. flexneri 90e4802; 1aII, S. flexneri 91e110; 1aIII, S. flexneri 86e48398; 1aIV, S. flexneri N02.115; 4aI, S. flexneri 92e0537; 4aII, S. flexneri 90e6312; 4aIII, S. flexneri 91e1351; 4aIV, S. flexneri 90e2459.
Conclusions.
This study describes the epidemiology of shigellosis cases that occurred in a long-stay psychiatric nursing center (VNC). Several points can be made from the findings from this study. First, since few shigellosis outbreaks in long-stay nursing centers have been documented, this appears to be the first study to evaluate large-scale cases of shigellosis occurring in a long-stay psychiatric nursing center in consecutive years. Second, different from the notion that the majority of shigellosis cases are caused by S. sonnei, the VNC cases involved 38 S. flexneri isolates and only 1 S. sonnei isolate. Third, all the patients in VNC were adults, different from the majority of cases, in which the patients are mostly younger than 9 years of age. Fourth, types 1a and 4a of S. flexneri responsible for the cases in VNC are less prevalent serotypes in Taiwan, with type 1a being only found in eastern Taiwan and not elsewhere in Taiwan in recent years. Fifth, the type 1a strains isolated in VNC are closely related, indicating that they were possibly derived from a common parent strain. Sixth, subtypes P4A and P4B are highly similar (93%), indicating a common parent, whereas the other type 4a strains shared lower degrees of similarity, suggesting that they might have been derived from divergent sources. Seventh, the antibiogram analysis indicated that all except one of the S. flexneri strains studied here are sensitive to at least seven antibiotics. The resistance situation does not seem to be as serious in Taiwan as it is in other areas in the world. The data that we obtained here might provide a strategy for the treatment of infections caused by type 1a and 4a S. flexneri strains in Taiwan.
Acknowledgments
This study was supported by a research grant (grant DOH93-DC-2002) from the Center for Disease Control, Department of Health, Taipei, Taiwan.
We thank Chung-Sheng Chao and Wan-Ching Chen from the Sixth Branch Office of the Center for Disease Control and the staffs of the Bureau of Public Health of Hualien County for performing the epidemiological investigation. We are grateful to Chien-Shun Chiou for helpful discussions and technical assistance. We also thank Ching-Mo Chueh, Nien-Tsung Lin, and Pei-Jane Tsai for technical assistance.
REFERENCES
- 1.Ahmed, K., F. R. Shakoori, and A. R. Shakoori. 2003. Aetiology of shigellosis in northern Pakistan. J. Health Popul. Nutr. 21:32-39. [PubMed] [Google Scholar]
- 2.Ashkenazi, S., I. Levy, V. Kazaronovski, and Z. Samra. 2003. Growing antimicrobial resistance of Shigella isolates. J. Antimicrob. Chemother. 51:427-429. [DOI] [PubMed] [Google Scholar]
- 3.Bopp, C. A., F. W. Brenner, P. A. Fields, J. G. Wells, and N. A. Strockbine. 2003. Escherichia, Shigella, and Salmonella, p. 654-671. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. ASM Press, Washington, D.C.
- 4.Centers for Disease Control and Prevention. 1996. Shigella sonnei outbreak associated with contaminated drinking water—Island Park, Idaho, August 1995. JAMA 275:1071. [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2003. Shigella surveillance: annual summary, 2002. Centers for Disease Control and Prevention, U.S. Department of Health and Human Services, Atlanta, Ga.
- 6.Chen, J. H., C. S. Chiou, P. C. Chen, T. L. Liao, J. M. Li, and W. B. Hsu. 2003. Molecular epidemiology of Shigella in a Taiwan township during 1996 to 2000. J. Clin. Microbiol. 41:3078-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiou, C. S., W. B. Hsu, H. L. Wei, and J. H. Chen. 2001. Molecular epidemiology of a Shigella flexneri outbreak in a mountainous township in Taiwan, Republic of China. J. Clin. Microbiol. 39:1048-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLappe, N., F. O'Halloran, S. Fanning, G. Corbett-Feency, T. Cheasty, and M. Cormican. 2003. Antimicrobial resistance and genetic diversity of Shigella sonnei isolates from western Ireland, an area of low incidence of infection. J. Clin. Microbiol. 41:1919-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Department of Health, Taiwan. 2003. Statistics of communicable diseases and surveillance report in Taiwan area. Department of Health, The Executive Yuan, Taipei, Taiwan.
- 10.Egah, D. Z., E. B. Banwat, E. S. Audu, J. A. Allanana, M. L. Danung, J. G. Damen, and B. P. Badung. 2003. Multiple drug resistant strains of Shigella isolated in Jos, central Nigeria. Niger. Postgrad. Med. J. 10:154-156. [PubMed] [Google Scholar]
- 11.Gautom, R. K. 1997. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J. Clin. Microbiol. 35:2977-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horan, M. A., R. S. Gulati, R. A. Fox, E. Glew, L. Ganguli, and M. Kaeney. 1984. Outbreak of Shigella sonnei dysentery on a geriatric assessment ward. J. Hosp. Infect. 5:210-212. [DOI] [PubMed] [Google Scholar]
- 13.Hunter, P. R., and P. G. Hutchings. 1987. Outbreak of Shigella sonnei dysentery on a long stay psychogeriatric ward. J. Hosp. Infect. 10:73-76. [DOI] [PubMed] [Google Scholar]
- 14.Kotloff, K. L., J. P. Winickoff, B. Ivanoff, J. D. Clemens, D. L. Swerdlow, P. J. Sansonetti, G. K. Adak, and M. M. Levine. 1999. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull. W. H. O. 77:651-666. [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, T. M., C. Y. Chang, L. L. Chang, W. M. Chen, T. K. Wang, and S. F. Chang. 2003. One predominant type of genetically closely related Shigella sonnei prevalent in four sequential outbreaks in school children. Diagn. Microbiol. Infect. Dis. 45:173-181. [DOI] [PubMed] [Google Scholar]
- 16.Maguire, H. C., C. Seng, S. Chambers, T. Cheasty, G. Double, N. Soltanpoor, and D. Morse. 1998. Shigella outbreak in a school associated with eating canteen food and person to person spread. Commun. Dis. Public Health 1:279-280. [PubMed] [Google Scholar]
- 17.Majtan, V., L. Majtanova, and L. Kovac. 2004. Analysis of integrons in human isolates of Salmonella enterica serovar Typhimurium isolated in the Slovak Republic. FEMS Microbiol. Lett. 239:23-31. [DOI] [PubMed] [Google Scholar]
- 18.Maraki, S., A. Georgiladakis, Y. Tselentis, and G. Samonis. 2003. A 5-year study of the bacterial pathogens associated with acute diarrhoea on the island of Crete, Greece, and their resistance to antibiotics. Eur. J. Epidemiol. 18:85-90. [DOI] [PubMed] [Google Scholar]
- 19.McCall, B., R. Stafford, S. Cherian, K. Heel, H. Smith, N. Corones, and S. Gilmore. 2000. An outbreak of multi-resistant Shigella sonnei in a long-stay geriatric nursing centre. Commun. Dis. Intell. 24:272-275. [PubMed] [Google Scholar]
- 20.MoezArdalan, K., M. R. Zali, M. M. Dallal, M. R. Hemami, and S. Salmanzadeh-Ahrabi. 2003. Prevalence and pattern of antimicrobial resistance of Shigella species among patients with acute diarrhoea in Karaj, Tehran, Iran. J. Health Popul. Nutr. 21:96-102. [PubMed] [Google Scholar]
- 21.National Committee for Clinical Laboratory Standards. 2000. Performance standards for antimicrobial disk susceptibility tests, 7th ed. Approved standard M2-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 22.Shiferaw, B., S. Shallow, R. Marcus, S. Segler, D. Soderlund, F. P. Hardnett, and T. Van Gilder. 2004. Trends in population-based active surveillance for shigellosis and demographic variability in FoodNet sites, 1996-1999. Clin. Infect. Dis. 38(Suppl. 3):S175-S180. [DOI] [PubMed] [Google Scholar]
- 23.Surdeanu, M., L. Ciudin, E. Pencu, and M. Straut. 2003. Comparative study of three different DNA fingerprint techniques for molecular typing of Shigella flexneri strains isolated in Romania. Eur. J. Epidemiol. 18:703-710. [DOI] [PubMed] [Google Scholar]
- 24.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Totaro, J., C. Tan, V. Reddy, K. Dail, M. Davies, P. Jenkins, J. M. Maillard, D. M. Toney, A. Beall, E. Mintz, M. Drees, and A. Shane. 2004. Day care-related outbreaks of rhamnose-negative Shigella sonnei—six states, June 2001-March 2003. Morb. Mortal. Wkly. Rep. 53:60-63. [PubMed] [Google Scholar]
- 26.van Pelt, W., M. A. de Wit, W. J. Wannet, E. J. Ligtvoet, M. A. Widdowson, and Y. T. van Duynhoven. 2003. Laboratory surveillance of bacterial gastroenteric pathogens in The Netherlands, 1991-2001. Epidemiol. Infect. 130:431-441. [PMC free article] [PubMed] [Google Scholar]
- 27.White, P. A., C. J. Mclver, and W. D. Rawlinson. 2001. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob. Agents Chemother. 45:2658-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.World Health Organization, Programme for Control of Diarrhoeal Diseases. 1988. Guidelines for the control of epidemics due to Shigella dysenteriae 1, p. 1-15. Report W.H.O./CDD/SER/88.12. World Health Organization, Geneva, Switzerland.
- 29.Yamada, S., K. Ogata, R. Kato, K. Morimoto, Y. Hayashi, T. Ito, S. Matushita, N. Konishi, A. Kai, and M. Endoh. 1999. Outbreak case caused by different colicin type of Shigella sonnei in a day nursery in Tokyo (1998). Kansenshogaku Zasshi 73:1130-1139. [DOI] [PubMed] [Google Scholar]
- 30.Yurdakok, K., N. Sahin, E. Ozmert, and E. Berkman. 1997. Shigella gastroenteritis: clinical and epidemiological aspects, and antibiotic susceptibility. Acta Paediatr. Jpn. 39:681-684. [DOI] [PubMed] [Google Scholar]