Abstract
The goal of the present study was to assess the contribution of real-time molecular typing, used alone or with clinical surveillance, to the prompt identification of clusters of Campylobacter enteritis. Potential poultry sources were sought by comparing the pulsed-field gel electrophoresis genotypes of human and fresh whole retail chicken isolates collected during the same study period. Among 183 human isolates, 82 (45%) had unique genotypes, 72 (39%) represented 26 clusters of 2 to 7 isolates each, and 29 (16%) represented three clusters of 8 to 11 isolates each. Molecular typing was useful for the confirmation of outbreaks suspected on the basis of epidemiological surveillance, but for most small clusters, no epidemiological link could be established. Thus, the added value of real-time molecular typing is questionable, since the numerous small clusters identified were of unclear public health significance. Among 177 chickens, 41 (23%) yielded campylobacter isolates; of these, 19 (46%) had genotypes similar to those of 41 (22%) human isolates. However, a temporal association was demonstrated in only a minority of cases, and most genotypes were present only in a single species, suggesting that sources other than chickens are important in human campylobacteriosis. Further investigation with samples from water and other possible environmental sources is needed to define the most efficient strategy for the application of molecular typing and identification of the source(s) of sporadic cases of campylobacteriosis.
Campylobacter enteritis is the third most frequent notifiable infectious disease in Québec Province and the fourth most frequent notifiable infectious disease in Canada. In Québec, nearly 3,000 cases of diarrheal illness are attributed annually to Campylobacter enteritis, more than the combined total caused by Salmonella and Shigella species, Escherichia coli O157:H7, and Yersinia enterocolitica. The epidemiology of campylobacteriosis is poorly understood. Campylobacters are endemic among domestic as well as wild animals and are ubiquitous in the environment (1). Raw milk, untreated water, and poultry have all been well documented as sources of Campylobacter outbreaks (5). Nevertheless, most clinical cases appear as isolated, sporadic infections for which the source is rarely identified.
Molecular strain typing methods have helped clarify the epidemiology of other bacterial infections. Pulsed-field gel electrophoresis (PFGE) is a highly reproducible and discriminatory technique for the molecular typing of C. jejuni (4). The combination of new protocols which provide results in 24 h (10, 14) and computerized systems for the analysis of numerous PFGE patterns across multiple gels (12) allow real-time molecular surveillance of Campylobacter enteritis. Canadian and American reference laboratories participating in the Centers for Disease Control and Prevention's PulseNet program already perform such surveillance for enteritis due to other bacterial pathogens (i.e., E. coli O157:H7, Salmonella spp., and Shigella spp.) (15), but its application to Campylobacter remains controversial, considering the large number of isolates that must be analyzed compared to the small number of outbreaks reported (7).
A recent preliminary molecular typing study of Campylobacter (12) combined with data regarding the dates and locations of isolation of the isolates suggested that 49% of the isolates from the Eastern Townships, Québec, and 39% of the isolates from Montreal belonged to clusters of potentially related isolates and that Campylobacter jejuni outbreaks may be more common than was previously suspected on the basis of traditional epidemiological data alone. That study (12) also indicated that clinical descriptive data were insensitive and unreliable for identification of the sources of sporadic cases of campylobacteriosis. Therefore, we hypothesized that continuous surveillance performed by combining clinical and molecular epidemiology analyses would identify related cases more rapidly and more accurately and could determine the sources of Campylobacter enteritis in the community.
This paper describes the application of PFGE to the real-time genotyping of isolates gathered over a 15-month period during a prospective case-control study of Campylobacter enteritis in the townships of the Eastern Townships, Québec, Canada. The genotypes of Campylobacter isolates from fresh whole retail chickens purchased in the Eastern Townships during the same study period were included for comparison.
MATERIALS AND METHODS
Isolates were collected through a case-control study and a survey of the prevalence of Campylobacter in fresh whole retail chickens, as described previously (11). In brief, all cases of Campylobacter enteritis reported by hospital microbiology laboratories to the regional public health department between 1 July 2000 and 30 September 2001 were eligible for inclusion in the case-control study. Cases were excluded if the infection was acquired outside Québec (i.e., travel outside the province during the entire 10-day period before the onset of symptoms) or if the interval between the onset of symptoms and reporting was longer than 6 weeks. Public health nurses administered a standardized epidemiological questionnaire by telephone to each case patient within 2 weeks of reporting of the case to capture demographic and clinical data, travel history, food consumption, water consumption, recreational water activity, animal contacts, and other illnesses during the 10 days before the onset of symptoms. For subjects reported on multiple occasions during the study period, only the first episode of infection was considered. The median interval from the onset of symptoms to the interview of the case patients was 13 days (range, 5 to 56 days; 90th percentile, 23 days). Microbiological laboratories were asked to send us all Campylobacter isolates identified between 1 July 2000 and 30 September 2001.
From November 2000 to November 2001, four fresh eviscerated whole chickens were bought weekly in different counties (one chicken per store); for each county, the number of chickens sampled monthly was proportional to the population. Retail chickens sold in the Eastern Townships are produced by multiple companies based elsewhere in Québec Province. Campylobacters were isolated from the whole retail chickens as described previously (11).
Molecular epidemiology study.
The molecular epidemiology study was performed in two phases to compare the identities of the isolates involved in putative outbreaks on the basis of clinical surveillance without and with molecular typing. Initially (phase I; 1 July 2000 to 30 April 2001), isolates were typed retrospectively and clinical and molecular epidemiological data were analyzed separately. During the second phase of the study (phase II; 1 May to 30 September 2001), all isolates were typed prospectively each week and clinical and molecular data were analyzed jointly, in collaboration with public health nurses.
PFGE.
C. jejuni isolates were grown on 5% sheep blood agar for 48 h at 37°C in a microaerobic atmosphere. Bacterial colonies were harvested and resuspended in 1,000 μl of cold suspension buffer (100 mM Tris, 100 mM EDTA [pH 8.0]). The optical densities of the bacterial suspensions were then adjusted to 1.9 to 2.0 μm at 405 nm, and 340-μl aliquots were gently mixed with 12.5 μl of proteinase K (20 mg/ml) and 170 μl of Seakem Gold agarose 1.5% (FMC BioProducts, Rockland, Maine) prepared in TE (10 mM Tris, 1 mM EDTA [pH 8.0]). The resulting mixture was poured into plug molds and allowed to solidify at 4°C for 20 min. The plugs were then incubated with 5 ml of cell lysis buffer (50 mM Tris, 50 mM EDTA [pH 8.0], 1% N-lauroyl sarcosine) supplemented with 25 μl of proteinase K (20 mg/ml) in a 50°C water bath with constant agitation (150 rpm) for 1 h, transferred to 40-ml polypropylene flat-bottom tubes, and washed six times for 10 min for each wash in a 50°C water bath with constant agitation (150 rpm): twice with 15 ml of preheated (50°C) water and four times with 10 ml of preheated (50°C) TE (10 mM Tris, 0.1 mM EDTA [pH 8.0]). Individual plugs were then washed twice for 10 min each time at room temperature with agitation in 300 μl of 1× NE 1 buffer (New England Biolabs, Inc., Beverly, Mass.) and transferred to 300 μl of fresh buffer-bovine serum albumin (0.1 mg/ml), and the DNA was digested with 20 U of KpnI for 2 h in a 37°C water bath. The digests were electrophoresed at 200 V in a 1% SeaKem Gold agarose gel (FMC BioProducts) in 0.5× TBE (Tris-borate-EDTA) buffer at 14°C (CHEF Mapper; Bio-Rad Laboratories). The pulsing was set to ramp from 4 to 13.6 s over 14 h. The gels were stained for 20 min in 1 liter of sterile water containing ethidium bromide (1 mg/ml), destained by two washes of 30 min each in 1 liter of sterile water, and photographed under UV light by using a digital camera.
Each gel comprised 15 lanes and included SmaI digests of Staphylococcus aureus NCTC 8325 in lanes 2, 8, and 14 as a reference standard and a KpnI digest of C. jejuni strain 153B-80 in lane 13 as a reproducibility control. Lanes 1 and 15 were left blank; the remaining lanes were used for the study isolates.
BioNumerics software analysis.
The PFGE fingerprinting patterns were analyzed with BioNumerics software (version 2.0 for Windows; Applied Maths, Kortrijk, Belgium). Restriction fragments were identified visually, and the PFGE patterns were normalized by interpolation to the nearest reference lane. The molecular sizes of the fragments detected for the study isolates were calculated on the basis of the sizes of the fragments of S. aureus NCTC 8325. Only fragments in the size range from 80 to 674 kb were analyzed; smaller fragments were not consistently resolved. Optimization of 1.0% and a position tolerance of 1.25% were applied. Dice similarity coefficients (SCs) were calculated on the basis of pairwise comparisons of the PFGE profiles of the study isolates. The matrix of coefficients was used to generate dendrograms based on the unweighted pair group method with arithmetic averages.
Criteria used to define clusters.
Three different sets of criteria were used to define clusters of related study isolates. (i) Isolates were considered to have closely related genotypes on the basis of the molecular typing results if their PFGE profiles were related at a level equal to or greater than 0.90, as determined by the BioNumerics software analysis. (ii) Genotypically related isolates were considered clustered in space if they were cultured from patients whose infection was acquired in Québec Province; infections acquired in a foreign country were excluded. (iii) Isolates that were genotypically and geographically related were also considered clustered in time if there was less than 2 months between the times of infection with sequential isolates. Hypotheses regarding putative sources of infections were generated by analyzing the epidemiological questionnaires for related cases.
RESULTS
Clinical epidemiology data.
Between July 2000 and October 2001, 201 cases of campylobacteriosis were reported, of which 43 were excluded: 18 case patients acquired the infection outside Québec, 18 resided outside the Eastern Townships, 6 could not be interviewed within 6 weeks after the onset of symptoms, and 1 declined to participate. During the study period, the mean crude incidence of campylobacteriosis was 63.1 per 100,000 population in the Eastern Townships. The median age of the case patients was 31 years (range, 11 days to 91 years), with 30 children and 128 adults. The incidence of campylobacteriosis varied considerably by age, with the highest rates occurring among children 0 to 4 years of age (169.2 per 100,000 population) and young adults 15 to 34 years of age (mean, 79.4 per 100,000 population). Additional demographic and epidemiological data are detailed elsewhere (11).
In the questionnaire, the case patients were specifically asked, as an open-ended question, what they considered to be the probable source of their infection. The responses were chicken (10%), contaminated water (9%), an animal contact (9%), raw milk (7%), beef (4%), other food (6%), traveling abroad (3%), an infectious contact (2%), and other sources (5%). However, 45% of the case patients could not identify any putative source of infection.
Molecular typing of Campylobacter isolates from patients with enteritis.
A total of 184 of the 201 Campylobacter isolates of human origin were sent to our laboratory and typed by PFGE; 172 (93.4%) of the isolates represented C. jejuni; the remainder included C. coli (n = 7), C. lari (n = 2), C. fetus (n = 1), and C. upsaliensis (n = 2). Overall, 144 isolates belonged to patients included in the case-control study (102 isolates during phase I of the study and 42 isolates during phase II) and 40 isolates belonged to excluded cases (29 isolates during phase I and 11 isolates during phase II).
Among the 43 gels analyzed, there was a 100% SC between the 43 reproducibility control isolates and a 91% SC between the 124 S. aureus NCTC 8325 isolates (dendrogram not shown). Only one Campylobacter isolate was untypeable by PFGE. We identified 101 different PFGE patterns among the 183 isolates analyzed, with an overall SC of 11.7%.
Molecular typing of Campylobacter isolates from fresh whole retail chickens.
A total of 177 chickens from 58 different food stores were cultured (median number per month, 16; range, 8 to 20) (11). Campylobacter spp. were cultured from 41 of the chickens (23%; C. jejuni, n = 37; C. coli, n = 4). There was no correlation between the monthly prevalence of campylobacters in chickens and the incidence of disease in humans. The prevalence of campylobacters in chickens peaked 1 month after the peak incidence of disease in humans and was not followed by an increased number of infections in humans.
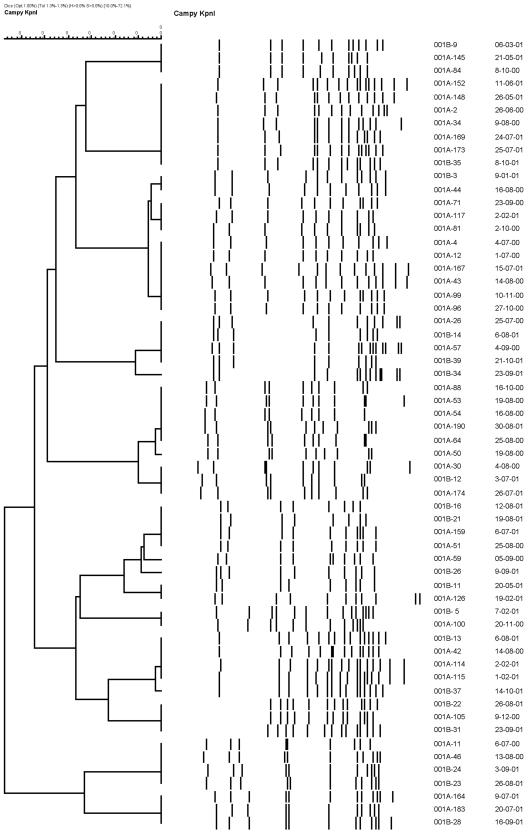
Molecular typing identified 34 different PFGE patterns among the 41 chicken isolates analyzed, with an SC of 20.4%. Overall, 19 (46%) chicken isolates had a PFGE pattern similar to one or more of the PFGE patterns of 41 (22%) human isolates (Fig. 1). However, only six of these chicken isolates were isolated within 2 months before or during the same week as the five human isolates. Some form of chicken exposure was documented in each of these five patients, but chicken was the suspected source of infection in only two of them.
FIG. 1.
Dendrograms representing relatedness among PFGE profiles of KpnI digests of 41 human (001A prefix) and 19 chicken (001B prefix) isolates of C. jejuni.
Analysis of clusters defined by molecular typing and time criteria.
On the basis of molecular typing only, we identified 29 clusters of 2 to 11 isolates each representing 101 (55%) cases (Table 1). One cluster of two isolates was excluded because it included one isolate of C. upsaliensis and one isolate of C. jejuni. No clustered isolate was excluded for geographical reasons (i.e., an infection acquired outside Québec Province).
TABLE 1.
Number and size of clusters of Campylobacter isolates identified by PFGE by using different criteria
| No. of isolates per cluster | No. of clusters:
|
||
|---|---|---|---|
| By use of molecular typing criteriaa | By use of molecular typing and time criteriab | Suspected by public health officials | |
| 2 | 15 | 21 | 2 |
| 3 | 7 | 3 | 2 |
| 4 | 2 | 3 | 0 |
| 5 | 0 | 1 | 0 |
| 6 | 1 | 1 | 0 |
| 7 | 1 | 0 | 0 |
| 8 | 1 | 1 | 0 |
| 9 | 0 | 0 | 0 |
| 10 | 1 | 0 | 0 |
| 11 | 1 | 0 | 0 |
| Total no. of clusters | 29 | 30 | 4 |
| Total no. of cases | 101 | 82 | 10 |
One cluster of two isolates was excluded because it included different species of Campylobacter.
The combination of molecular typing, time, and space criteria yielded the same results.
When molecular typing was combined with the time criteria, the number of clustered isolates was reduced to 82 (45%) isolates distributed among 30 clusters of 2 to 8 isolates each. Twenty-three clusters were identified during phase I of the study (1 July 2000 to 30 April 2001), and seven clusters were identified during phase II (1 May to 30 September 2001) (Table 2). Overall, 57% of the cases and 60% of the clusters occurred in July, August, and September. All age groups were represented among the case patients whose isolates belonged to clusters, with the highest proportion detected among young adults 15 to 34 years of age (44%) and children 0 to 4 years of age (13%).
TABLE 2.
Description of clusters identified by use of molecular typing (PFGE) and time criteria
| Study phase and cluster no. | Isolate no. | Patient age (yr) | Date (day/mo/yr) | City of residence | Source suspected by patient | Source suspected by public health officials |
|---|---|---|---|---|---|---|
| Phase I | ||||||
| 1 | 001A-1 | 24 | 2/07/00 | Sherbrooke | Unknown | Unknown |
| 001A-6 | 21 | 4/07/00 | East Angus | Chicken | Chicken, cross-contamination | |
| 001A-7 | 19 | 6/07/00 | Fleurimont | Untreated ground water | Untreated groundwater, chicken | |
| 001A-25 | 41 | 28/07/00 | Montreal | Excluded | ||
| 2a | 001A-10 | 36 | 10/07/00 | Fleurimont | Unknown | Deep-well water contaminationb |
| 001A-13 | 12 | 17/07/00 | Fleurimont | Chicken or water | Deep-well water contaminationb | |
| 001A-23 | 19 | 26/07/00 | Sherbrooke | Contaminated water | Deep-well water contaminationb | |
| 3c | 001A-19 | 26 | 24/07/00 | Ste-Anne de Bellevue | Excluded | |
| 001A-20 | 25 | 24/07/00 | Ste-Anne de Bellevue | Excluded | ||
| 4 | 001A-12 | 28 | 1/07/00 | Blainville | Excluded | |
| 001A-4 | 34 | 4/07/00 | Rock Forest | Water, travel | Chicken, cross-contamination | |
| 001A-43 | 82 | 14/08/00 | Sherbrooke | Water or Chinese buffet | Untreated water at a camping site | |
| 001A-44 | 27 | 16/08/00 | Sherbrooke | Water contamination at work | Water contamination at work, chicken | |
| 001A-71 | 25 | 23/09/00 | Sherbrooke | Unknown | Cross-contamination | |
| 001A-81 | 31 | 2/10/00 | Bonsecours | Infectious contact | Infectious contact, cross-contamination | |
| 001A-96 | 18 | 27/10/00 | Sherbrooke | Chicken | Raw milk cheese, poorly cooked chicken | |
| 001A-99 | 54 | 10/11/00 | Windsor | Chicken | Poorly cooked chicken | |
| 5 | 001A-3 | 26 | 26/06/00 | Bromptonville | Club sandwich | Club sandwich |
| 001A-45 | 61 | 13/08/00 | Magog | Unknown | Chicken in a restaurant | |
| 6 | 001A-11 | 72 | 6/07/00 | Windsor | Unknown | Farm animals, deep well |
| 001A-46 | 48 | 13/08/00 | Magog | PÂté de campagne | Chicken in a restaurant | |
| 7 | 001A-21 | 78 | 24/07/00 | Ville St-Laurent | Excluded | |
| 001A-60 | 45 | 5/09/00 | Lennoxville | Brook water | Brook water | |
| 8 | 001A-26 | 15 | 25/07/00 | Audet | Unknown | Farm, deep well, raw milk |
| 001A-57 | 35 | 4/09/00 | Sherbrooke | Unknown | Farm, deep well | |
| 9 | 001A-37 | 14 | 5/08/00 | Lennoxville | Unknown | Horses, deep well, chicken |
| 001A-32 | 21 | 11/08/00 | Sherbrooke | Iguana | Cats, bird, iguana | |
| 10 | 001A-38 | 33 | 7/08/00 | St-François Xavier | Unknown | Raw milk |
| 001A-65 | 16 | 25/08/00 | Danville | Raw milk | Raw milk | |
| 11 | 001A-30 | 34 | 4/08/00 | Sherbrooke | Unknown | Chicken |
| 001A-54 | 75 | 16/08/00 | Windsor | Soup | Raw ground beef | |
| 001A-50 | 25 | 19/08/00 | Asbestos | Unknown | Chicken and mussels in a restaurant | |
| 001A-53 | 55 | 19/08/00 | Asbestos | Lasagna | Pork in a restaurant | |
| 001A-64 | 27 | 25/08/00 | Trois-Lacs | Unknown | Deep well, untreated water | |
| 001A-88 | 2 | 16/10/00 | Stoke | Excluded | ||
| 12 | 001A-35 | 16 | 9/08/00 | Sherbrooke | Unknown | Brook water, chicken in a restaurant |
| 001A-85 | 32 | 11/10/00 | Sherbrooke | Water | Poorly cooked chicken/raw milk | |
| 13 | 001A-2 | 33 | 26/06/00 | St-Camille | Unknown | Cook, deep well, chicken |
| 001A-34 | 15 | 9/08/00 | Richmond | Raw milk | Raw milk, chicken | |
| 14 | 001A-29 | 1 | 2/08/00 | Stoke | Unknown | Infectious contact, farm animals |
| 001A-52 | 30 | 25/08/00 | Ascot | Unknown | Work in kindergarden, cross-contamination | |
| 15 | 001A-51 | 16 | 25/08/00 | Rock Forest | Unknown | Abattoir, farm, chicken in a restaurant |
| 001A-59 | 42 | 5/09/00 | Rock Forest | Unknown | Poorly cooked chicken, chicken in a restaurant | |
| 16 | 001A-73 | 54 | 22/09/00 | Valcourt | Excluded (relapse) | |
| 001A-93 | 17 | 18/10/00 | Fleurimont | Poorly cooked chicken | Poorly cooked chicken | |
| 17 | 001A-76 | 75 | 1/10/00 | Cookshire | Unknown | Chicken in a restaurant |
| 001A-83 | 68 | 10/10/00 | Cookshire | Unknown | Untreated water | |
| 18 | 001A-77 | 22 | 1/10/00 | Richmond | Unknown | Raw milk |
| 001A-79 | 1 | 2/10/00 | Sherbrooke | Contact with farm animals | Contact with farm animals | |
| 19 | 001A-101 | 19 | 25/11/00 | Sherbrooke | Hamburger | Infectious contact, chicken in a restaurant |
| 001A-112 | 4 | 20/01/01 | Sherbrooke | Unknown | Chicken in a restaurant | |
| 20 | 001A-102 | 4 | 1/12/00 | Sherbrooke | Unknown | Cross-contamination, cats |
| 001A-103 | 33 | 4/12/00 | Fleurimont | Unknown | Infectious contact, work in a kindergarden, raw milk | |
| 001A-106 | 4 | 19/12/00 | Richmond | Unknown | Unknown | |
| 001A-108 | 55 | 27/12/00 | Richmond | Unknown | Unknown | |
| 001A-118 | 46 | 1/02/01 | Tingwick | Excluded | ||
| 21 | 001A-115 | 37 | 1/02/01 | Sherbrooke | Unknown | Assistant cook |
| 001A-114 | 9 | 2/02/01 | Valcourt | Turtles or water | Poorly cooked chicken | |
| 22 | 001A-116 | 3 | 4/02/01 | Ascot | Unknown | Unknown |
| 001A-122 | 32 | 4/03/01 | Valcourt | Travel | Raw milk cheese | |
| 001A-141 | 59 | 30/04/01 | Coaticook | Unknown | Chicken in a restaurant, animal auction, cross-contamination | |
| 001A-147 | 34 | 2/06/01 | Val-Joli | Raw milk cheese | Raw milk cheese | |
| 23 | 001A-128 | 43 | 21/03/01 | East Angus | Chicken | Sheep breeding, deep well, chicken in a restaurant |
| 001A-139 | 80 | 22/04/01 | Eastman | Water from a surface well | Well, groundwater, raw milk cheese, chicken in a restaurant | |
| Phase II | ||||||
| 24 | 001A-138 | 87 | 22/04/01 | Rock Forest | Unknown | Deep well, chicken in a restaurant |
| 001A-142 | 60 | 2/05/01 | Victoriaville | Excluded | ||
| 001A-143 | 3 | 7/05/01 | Windsor | Raw milk | Deep well, chicken breeding, raw milk | |
| 25 | 001A-148 | 18 | 26/05/01 | Sherbrooke | Chicken croquettes | Chicken croquettes |
| 001A-152 | 67 | 11/06/01 | Outside Eastern Townships | Excluded | ||
| 001A-169 | 25 | 24/07/01 | Weedon | Unknown | Chicken | |
| 001A-173 | 54 | 25/07/01 | Gartby Station | Excluded | ||
| 26a | 001A-163 | 7 | 23/06/01 | Wotton | Raw milk | Raw milk and secondary infectious contactd |
| 001A-162 | 33 | 27/06/01 | Wotton | Raw milk | Raw milk and secondary infectious contactd | |
| 001A-166 | 4 | 7/07/01 | Wotton | Infectious contact (same family) | Raw milk and secondary infectious contactd | |
| 27 | 001A-164 | 2 | 9/07/01 | Bromptonville | Animal contact | Zoo visit, chicken |
| 001A-183 | 51 | 20/07/01 | Sherbrooke | Unknown | Chicken, pink pork | |
| 28 | 001A-174 | 49 | 26/07/01 | Sherbrooke | Unknown | Chicken |
| 001A-190 | 46 | 30/08/01 | Ascot | Infectious contact | Infectious contact, chicken | |
| 29 | 001A-176 | 25/07/01 | Sherbrooke | Excluded | ||
| 001A-178 | 66 | 1/08/01 | Sherbrooke | Unknown | Poorly cooked chicken | |
| 30c | 001A-187 | 19/08/01 | Outside Eastern Townships | Excluded | ||
| 001A-188 | 19/08/01 | Outside Eastern Townships | Excluded |
Clusters suspected by public health officials.
Three cases occurred among members of the same family. Water analysis was requested.
Clusters not investigated by public health officials because cases resided outside the Eastern Townships.
Three cases in the same family.
Only two clusters had been suspected by public health nurses to be related to a common source of infection, and both clusters were confirmed by molecular typing to be a result of infection from common sources. One cluster involved three members from the same family who acquired infection after consuming raw milk, and the second cluster represented three patients infected after consuming contaminated water from a deep well. Two additional clusters of two isolates each included patients who lived outside the Eastern Townships and were not investigated by public health nurses. During phase I, the identification of cluster 10 suggested the consumption of raw milk as a common source of infection in two patients, an association not initially suspected by public health nurses.
For essentially all the remaining clusters that were defined by molecular typing and that met the time criteria, an epidemiological factor linking the cases could not be verified. The identification of common sources was difficult because most clusters included only two to three cases each and because no information for cases living outside the Eastern Townships was available.
Even during phase II, when molecular typing was performed in real time and related isolates were identified promptly, the clustered cases could not be linked to common sources by using the epidemiological data routinely collected. Additional investigations would have been necessary to confirm more speculative possibilities. Chicken consumption was often hypothesized as the source of infection, but this risk factor was poorly discriminatory since it was present in 89% of the cases and 93% of the controls in the case-control study.
Of note, some sets of isolates that were highly related by PFGE patterns (Table 1) were assigned to several different clusters on the basis of the time criteria (Table 2). For example, clusters 10, 20, and 24 occurred over 10 months and represented a single strain, as defined by PFGE. Similarly, clusters 13 and 25, which occurred approximately 1 year apart, represented a single strain (Fig. 1), which was also isolated from a chicken (strain 001B-35) during the period that the second cluster was identified. These observations suggest that there may be particular genotypes that could have a higher potential to cause outbreaks, either because of greater dissemination among sources such as water or domestic animals or because of increased virulence. Thus, the cases in cluster 13 may represent illnesses due to a single strain acquired from different sources (e.g., raw milk and chicken).
DISCUSSION
In this study, as previously (5), a classic epidemiological investigation indicated that most Campylobacter infections represented sporadic cases, with relatively few being part of well-defined outbreaks. Molecular strain typing effectively confirmed the outbreaks suspected by public health personnel, but it also identified many small clusters (two to seven cases) for which an epidemiological link could not be established. The analysis of clinical descriptive data was insensitive and unreliable for identification of the sources of sporadic cases of campylobacteriosis, partly due to the time delay between the onset of symptoms and the epidemiological investigation. Real-time molecular typing added little, if any, value, possibly due to the small size of the clusters identified or limitations of the questionnaire.
Hedberg et al. (7) evaluated the usefulness of molecular typing of all Campylobacter isolates submitted to the Minnesota Department of Health in 1994. A total of 673 isolates were grouped into 248 distinct PFGE patterns, 74% of which were represented by only 1 or 2 isolates each. Routine epidemiological methods identified two outbreaks and nine other case clusters involving 4% of all isolates. Use of PFGE revealed eight more temporal clusters involving 9% of all isolates. Cases that could not be linked with other cases by PFGE pattern, time, or geographic location accounted for 87% of the reported isolates. These results are consistent with our conclusion that molecular typing identifies relatively few additional cases representing potential common-source clusters. Perhaps more importantly, the observation that most clinical Campylobacter isolates represent unique genotypes suggests that there is little yield to using scarce public health resources to investigate sporadic cases.
Our study also indicated, as others have (2, 3, 6, 8, 9, 13), that humans and chickens can be infected by related genotypes of Campylobacter. Overall, the PFGE patterns of 19 (46%) of the chicken isolates matched those of 1 or more of the human isolates. However, a temporal association was demonstrated in only a minority of cases and the majority of genotypes causing clinical illness were never found among chicken isolates, suggesting that sources other than chickens make an important contribution to human campylobacteriosis.
Our results may have been influenced by several technical factors. The selective enrichment methods may have failed to recover strains with poor fitness in vitro. Since only a single isolate was analyzed per chicken, the presence of multiple different strains in individual animals may have been missed. Since our epidemiological surveillance was done on a regional basis, we had limited information for many case patients living outside the Eastern Townships. A provincial or national surveillance system might have identified additional clusters; however, typing and investigation of such a large number of isolates might be unwieldy.
The most efficient strategy for the surveillance and investigation of sporadic cases of campylobacteriosis remains undefined. Should molecular studies be reserved for case clusters putatively identified by epidemiological surveillance? Conversely, should all cases be analyzed first by molecular strain typing and then should the epidemiological investigation be directed only at genotypically related cases? Should small clusters of two to four cases be investigated at all, since the yield appears to be very low? Resolution of these questions will require a more comprehensive analysis of Campylobacter strains in all sources, including water (e.g., by less selective isolation methods and analysis of multiple isolates per sample), and by more precise and portable genotyping systems (e.g., multilocus sequence typing).
Acknowledgments
We thank Diane Dion and Danielle Proulx for data collection, Linda Billard and Mélanie Proulx for molecular typing, and Bruno Maynard for appreciable help in purchasing the chickens. We also thank all the Eastern Townships' hospital microbiology laboratories for collecting the Campylobacter isolates.
Financial support for this study was received from the Ministère de la Santé et des Services Sociaux du Québec, the Régie Régionale de la Santé et des Services Sociaux de l'Estrie, and the Centre de Recherche Clinique du Centre Hospitalier Universitaire de Sherbrooke.
REFERENCES
- 1.Altekruse, S. F., N. J. Stern, P. I. Fields, and D. L. Swerdlow. 1999. Campylobacter jejuni—an emerging foodborne pathogen. Emerg. Infect. Dis. 5:28-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chu, Y. W., M. Y. Chu, K. Y. Luey, Y.W. Ngan, K. L. Tsang, and K. M. Kam. 2004. Genetic relatedness and quinolone resistance of Campylobacter jejuni strains isolated in 2002 in Hong Kong. J. Clin. Microbiol. 42:3321-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dickins, M. A., S. Franklin, R. Stefanova, G. E. Schutze, K. D. Eisenach, I. Wesley, and M. D. Cave. 2002. Diversity of Campylobacter isolates from retail poultry carcasses and from humans as demonstrated by pulsed-field gel electrophoresis. J. Food Prot. 65:957-962. [DOI] [PubMed] [Google Scholar]
- 4.Fitzgerald, C., L. O. Helsel, M. A. Nicholson, S. J. Olsen, D. L. Swerlow, R. Flahart, J. Sexton, and P. I. Fields. 2001. Evaluation of methods for subtyping Campylobacter jejuni during an outbreak involving a food handler. J. Clin. Microbiol. 39:2386-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedman, C., J. Neimann, C. Wegener, and R. Tauxe. 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations, p. 121-138. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, D.C.
- 6.Hänninen, M. L., P. Perko-Mäkelä, A. Pitkälä, and H. Rautelin. 2000. A three-year study of Campylobacter jejuni genotypes in humans with domestically acquired infections and in chicken samples from the Helsinki area. J. Clin. Microbiol. 38:1998-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hedberg, C. W., K. E. Smith, J. M. Besser, D. J. Boxrud, T. W. Hennessy, J. B. Bender, F. A. Anderson, and M. T. Osterholm. 2001. Limitations of pulsed-field gel electrophoresis for the routine surveillance of Campylobacter infections. J. Infect. Dis. 184:242-244. [DOI] [PubMed] [Google Scholar]
- 8.Hook, H., M. B. Ekegren, H. Ericsson, I. Vagsholm, and M. L. Danielsson-Tham. 2004. Genetic and epidemiological relationships among Campylobacter isolates from humans. Scand. J. Infect. Dis. 36:435-442. [DOI] [PubMed] [Google Scholar]
- 9.Karenlampi, R., H. Rautelin, M. Hakkinen, and M. L. Hanninen. 2003. Temporal and geographical distribution and overlap of Penner heat-stable serotypes and pulsed-field gel electrophoresis genotypes of Campylobacter jejuni isolates collected from humans and chickens in Finland during a seasonal peak. J. Clin. Microbiol. 41:4870-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michaud, S., R. D. Arbeit, and C. Gaudreau. 2001. Molecular strain typing of Campylobacter jejuni by pulsed-field gel electrophoresis in a single day. Can. J. Microbiol. 47:667-669. [PubMed] [Google Scholar]
- 11.Michaud, S., S. Ménard, and R. D. Arbeit. 2004. Campylobacteriosis, Eastern Townships, Quebec. Emerg. Infect. Dis. 10:1844-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michaud, S., S. Ménard, C. Gaudreau, and R. D. Arbeit. 2001. Comparison of SmaI-defined genotypes of Campylobacter jejuni examined by KpnI: a population-based study. J. Med. Microbiol. 50:1075-1081. [DOI] [PubMed] [Google Scholar]
- 13.Nadeau, E., S. Messier, and S. Quessy. 2002. Prevalence and comparison of genetic profiles of Campylobacter strains isolated from poultry and sporadic cases of campylobacteriosis in humans. J. Food Prot. 65:73-78. [DOI] [PubMed] [Google Scholar]
- 14.Ribot, E. M., C. Fitzgerald, K. Kubota, B. Swaminathan, and T. J. Barrett. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swaminathan B., T. J. Barrett, and the CDC PulseNet Task Force. 2000. A national molecular subtyping network for food-borne bacterial disease surveillance in the United States, p. 529-535. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington. D.C.