Abstract
In many European countries, the level of pneumococcal resistance to macrolides has now passed the level of resistance to penicillin G. A total of 82 erythromycin A-resistant isolates of Streptococcus pneumoniae were collected by 11 laboratories in seven European countries. All of the isolates were tested for antimicrobial susceptibility, analyzed for clonal relatedness by multilocus sequence typing, and characterized for macrolide resistance genotypes. The prevalence of the macrolide resistance genotypes varied substantially between countries. In France (87.5% of all strains), Spain (77.3%), Switzerland (80%), and Poland (100%), strains were predominantly erm(B) positive, whereas higher levels of mef(A)-positive strains were reported from Greece (100%) and Germany (33.3%). Macrolide resistance was caused by the oligoclonal spread of some multilocus sequence types, but significant differences in clonal distribution were noted between France and Spain, countries from which high levels of macrolide resistance have been reported. Overall, sequence type 81 (Spain23F-1 clone) was by far the most widespread. The mainly erm(B)-positive serotype 14 clone (sequence type 143), first reported in Poland in the mid-1990s, is now widespread in France.
Streptococcus pneumoniae continues to be a significant cause of morbidity and mortality in humans (17). The worldwide increase in antibiotic resistance in this species has become a serious infectious disease problem within the last 20 years (1).
In many European countries, the rate of pneumococcal resistance to macrolides has passed the level of resistance to penicillin G. Prevalence rates of resistance to erythromycin A vary substantially among European countries. Erythromycin A resistance rates of >20% are reported from France, Spain, Poland, Greece, and Portugal, whereas significantly lower resistance rates are documented in Germany and Switzerland (5, 9).
Macrolide resistance in S. pneumoniae is usually caused by the presence of the erm(B) or the mefE [renamed mef(A)] resistance determinants. The erm(B) protein encodes a 23S rRNA methylase, and most pneumococcal strains harboring the gene are resistant to 14-, 15-, and 16-member ring macrolides, lincosamides, and streptogramin B (cMLSB phenotype). The mef(A) protein encodes an efflux pump that leads to resistance to only 14- and 15-member ring macrolides (M phenotype) (22, 31). Other mechanisms of macrolide resistance have been described in only a few clinical isolates of S. pneumoniae. Changes were clustered in a highly conserved region of domain V of 23S rRNA, which plays a key role in macrolide binding (2, 3, 28, 33), and in ribosomal proteins L4 and L22 (18, 28).
Multilocus sequence typing (MLST) is a recently developed technique that produces unambiguous molecular typing data that can be transmitted electronically via the Internet (4, 12). The method is highly portable, as any laboratory can compare the sequences of the seven loci in their isolates with those in a central database on the World Wide Web (http://www.mlst.net) and obtain the allelic profile of each isolate.
In the present study, this technique was used to analyze the genetic relatedness of clinical erythromycin A-resistant strains of S. pneumoniae isolated in seven European countries.
MATERIALS AND METHODS
Study design.
Eleven participating laboratories in seven European countries were requested to send consecutive clinical pneumococcal isolates to the Eijkman-Winkler Institute (the European reference center for the SENTRY Antimicrobial Surveillance Program) using Amies charcoal medium transport swabs (Difco), together with relevant information on the isolate. Isolates were cultured on blood agar and stored at −70°C using Microbank (Mast Diagnostics, Reinfeld, Germany) until they were further tested. The study has been described in detail elsewhere (9).
A total of 82 erythromycin A-resistant isolates of S. pneumoniae were sent to the German National Reference Centre for Streptococci, where these strains were further characterized. The strains came from the following countries: France, n = 37; Spain, n = 22; Greece, n = 4; Portugal, n = 1; Germany, n = 3; Switzerland, n = 10; and Poland, n = 5. Centers from the following countries were included in the study: France (three), Germany (one), Poland (one), Switzerland (one), Spain (three), Greece (one), and Portugal (one).
Susceptibility testing.
MIC testing was performed using the broth microdilution method as recommended by the National Committee for Clinical Laboratory Standards (NCCLS) (19). Microtiter plates containing penicillin G, cefotaxime, amoxicillin, erythromycin A, clarithromycin, roxithromycin, azithromycin, clindamycin, spiramycin, telithromycin, quinupristin-dalfopristin, ciprofloxacin, tetracycline, teicoplanin, and vancomycin with cation-adjusted Mueller-Hinton broth (Oxoid, Wesel, Germany) plus 5% lysed horse blood (Oxoid) were used. The final inocculum was 5 × 105 CFU/ml. MICs were determined following incubation at 35°C for 24 h in ambient air. S. pneumoniae ATCC 49619 was used as a control strain. Current NCCLS interpretive criteria were used to define antimicrobial resistance (19). Isolates were stored at −70°C on porous beads (Microbank; Mast Diagnostics, Rheinfeld, Germany).
PCR experiments.
PCR was performed as described previously (22, 24, 27). For the detection of erm(A) and mef(E), the primers described by Trieu-Cuot et al. (34) and by Tait-Kamradt et al. (32) with the sequences (erm) 5′ CGA GTG AAA AAG TAC TCA ACC 3′ (positions 362 to 382) and 5′ GGC GTG TTT CAT TGC TTG ATG (positions 978 to 958) and the sequences (mef) 5′ AGT ATC ATT AAT CAC TAG TGC 3′(positions 57 to 77) and 5′ GTA ATA GAT GCA ATC ACA GC 3′ (positions 551 to 532) were chosen.
Serotyping.
Pneumococcal strains were serotyped by Neufeld's Quellung reaction using type and factor sera provided by the Statens Serum Institut, Copenhagen, Denmark.
MLST.
MLST of all strains (n = 82) was carried out as described previously. Briefly, internal fragments of the aroE, gdh, gki, recP, spi, xpt, and ddl genes were amplified by PCR from chromosomal DNA with the primer pairs described by Enright and Spratt (4). The alleles at each of the seven loci provide the allelic profile of each isolate and also define their sequence type (ST). Allelic profiles are shown as the alleles at each of the seven loci in the order aroE, gdh, gki, recP, spi, xpt, and ddl. The allelic profiles of the German isolates were compared with each other, as well as with other isolates in the pneumococcal MLST database, using software available at the MLST website. Additional MLST data for control strains were obtained from http://www.mlst.net.
Phylogenetic analysis.
The 82 erythromycin A-resistant strains in this study were assigned to 23 different MLS types. Nineteen of the STs were already known.
For phylogenetic analysis, the sequence fragments of the aroE, gdh, gki, recP, spi, and xpt genes were concatenated using the concatenation tool on http://www.mlst.net. As recommended, the ddl gene was left out of the analysis, since it appears to be highly variable in penicillin-resistant isolates.
The concatenated sequences (2,715 bp) were aligned with the multiple sequence alignment tool ClustalW. The multiple alignment was fed to the programs Distances and Grow Tree to create a phylogenetic tree, using the unweighted pair group method with arithmetic means (UPGMA). All programs were from the HUSAR Program Package of the Biocomputing Service Group (http://genius.dkfz-heidelberg.de).
In addition, the genetic relatedness of representative isolates of the present investigation (eight strains) were compared with macrolide-susceptible strains from the MLST database (four strains).
RESULTS
A total of 82 isolates were consecutively collected by 11 centers. The strains were isolated from the respiratory tract (n = 67; 81.7%) and from blood (n = 15; 18.3%).
Data on antibiotic resistance are presented in Table 1. Macrolide-resistant S. pneumoniae strains usually showed cross-resistance to other 14- and 15-member ring macrolides. All strains were inhibited by 0.5 μg of telithromycin/ml. The quinupristin-dalfopristin MIC for one strain was 2 μg/ml (intermediate). More than 70% of the strains were found to be nonsusceptible to penicillin G. The strains also often showed cross-resistance to tetracycline (75.6%). All strains were susceptible to glycopeptides. erm(B)-positive-strains (n = 64) were characterized by resistance to 14- and 15-member macrolides and clindamycin. Interestingly, the erythromycin A MICs for two strains (19B157 and 18B084) were relatively low (4 μg/ml). mef(A)-positive strains (n = 18) were characterized by generally lower erythromycin MICs (MIC range, 1 to 8 μg/ml), and all strains remained susceptible to clindamycin. mef(A)-positive strains were more often found to be cross-resistant to penicillin G [83.3% of strains (combined rate of penicillin intermediate and resistant) versus 75.1% in erm(B)-positive strains]. All erm(B)-positive strains were found to exhibit reduced susceptibility to tetracycline (tetracycline intermediate, 4.7% of strains; tetracycline resistant, 95.3% of strains), whereas 38.9% of the mef(A)-positive strains remained susceptible to tetracycline.
TABLE 1.
MIC ranges, MIC50sa, MIC90s, and resistance rates of 82 erythromycin A-resistant pneumococcal isolates from seven European countries
| Antibioticb | MIC range | MIC50 | MIC90 | Susceptible
|
Intermediate
|
Resistant
|
|||
|---|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | ||||
| All strains (n = 82) | |||||||||
| Erythromycin A | 1-≥32 | ≥32 | ≥32 | 0 | 0 | 0 | 0 | 82 | 100 |
| Roxithromycin | 0.5-≥32 | ≥32 | ≥32 | 0 | 0 | 1 | 1.2 | 81 | 98.8 |
| Clarithromycin | 1-≥32 | ≥32 | ≥32 | 0 | 0 | 0 | 0 | 82 | 100 |
| Azithromycin | 1-≥32 | ≥32 | ≥32 | 0 | 0 | 5 | 6.1 | 77 | 93.9 |
| Spiramycin | 0.03-≥32 | ≥32 | ≥32 | ND | ND | ND | ND | ND | ND |
| Clindamycin | 0.06-≥32 | 16 | ≥32 | 17 | 20.7 | 1 | 1.2 | 64 | 78.0 |
| Telithromycin | ≤0.03-0.5 | ≤0.03 | 0.25 | 82 | 100 | 0 | 0 | 0 | 0 |
| Q-D | 0.03-2 | 0.5 | 1 | 80 | 97.6 | 2 | 2.4 | 0 | 0 |
| Penicillin G | 0.016-2 | 1 | 1 | 19 | 23.2 | 57 | 69.5 | 6 | 7.3 |
| Cefotaxime | 0.016-2 | 1 | 1 | 78 | 95.1 | 4 | 4.9 | 0 | 0 |
| Amoxicillin | 0.016-2 | 1 | 1 | 82 | 100 | 0 | 0 | 0 | 0 |
| Tetracycline | 0.06-≥32 | 16 | ≥32 | 8 | 9.8 | 12 | 14.6 | 62 | 75.6 |
| Ciprofloxacin | 0.06-2 | 1 | 1 | ND | ND | ND | ND | ND | ND |
| Vancomycin | 0.125-0.5 | 0.25 | 0.25 | 82 | 100 | 0 | 0 | 0 | 0 |
| Teicoplanin | 0.06-0.25 | 0.06 | 0.125 | ND | ND | ND | ND | ND | ND |
| Erm(B)-positive strains (n = 64) | |||||||||
| Erythromycin A | 4-≥32 | ≥32 | ≥32 | 0 | 0 | 0 | 0 | 64 | 100 |
| Roxithromycin | 4-≥32 | ≥32 | ≥32 | 0 | 0 | 0 | 0 | 64 | 100 |
| Clarithromycin | 2-≥32 | ≥32 | ≥32 | 0 | 0 | 0 | 0 | 64 | 100 |
| Azithromycin | 1-≥32 | ≥32 | ≥32 | 0 | 0 | 1 | 1.6 | 63 | 98.4 |
| Spiramycin | 2-≥32 | ≥32 | ≥32 | ND | ND | ND | ND | ND | ND |
| Clindamycin | 2-≥32 | ≥32 | ≥32 | 0 | 0 | 0 | 0 | 64 | 100 |
| Telithromycin | ≤0.03-0.5 | ≤0.03 | 0.25 | 64 | 100 | 0 | 0 | 0 | 0 |
| Q-D | 0.03-2 | 0.5 | 1 | 62 | 96.9 | 2 | 3.1 | 0 | 0 |
| Penicillin G | 0.016-2 | 1 | 1 | 16 | 25.0 | 44 | 68.8 | 4 | 6.3 |
| Cefotaxime | 0.016-2 | 0.5 | 1 | 62 | 96.9 | 2 | 3.1 | 0 | 0 |
| Amoxicillin | 0.016-2 | 1 | 2 | 64 | 100 | 0 | 0 | 0 | 0 |
| Tetracycline | 0.125-≥32 | 16 | ≥32 | 0 | 0 | 3 | 4.7 | 61 | 95.3 |
| Ciprofloxacin | 0.06-2 | 1 | 1 | ND | ND | ND | ND | ND | ND |
| Vancomycin | 0.125-0.5 | 0.25 | 0.25 | 64 | 100 | 0 | 0 | 0 | 0 |
| Teicoplanin | 0.06-0.25 | 0.06 | 0.125 | ND | ND | ND | ND | ND | ND |
| mef(A)-positive strains (n = 18) | |||||||||
| Erythromycin A | 1-8 | 2 | 8 | 0 | 0 | 0 | 0 | 18 | 100 |
| Roxithromycin | 0.5-16 | 4 | 8 | 0 | 0 | 1 | 5.6 | 17 | 94.4 |
| Clarithromycin | 1-16 | 2 | 8 | 0 | 0 | 0 | 0 | 18 | 100 |
| Azithromycin | 1-16 | 2 | 16 | 0 | 0 | 4 | 22.2 | 14 | 77.8 |
| Spiramycin | 0.03-8 | 2 | 4 | ND | ND | ND | ND | ND | ND |
| Clindamycin | 0.06-0.5 | 0.06 | 0.25 | 17 | 94.4 | 1 | 5.6 | 0 | 0 |
| Telithromycin | ≤0.03-0.125 | 0.03 | 0.06 | 18 | 100 | 0 | 0 | 0 | 0 |
| Q-D | 0.25-1 | 0.5 | 1 | 18 | 100 | 0 | 0 | 0 | 0 |
| Penicillin G | 0.016-2 | 1 | 2 | 3 | 16.7 | 13 | 72.2 | 2 | 11.1 |
| Cefotaxime | 0.016-2 | 1 | 1 | 16 | 88.9 | 2 | 11.1 | 0 | 0 |
| Amoxicillin | 0.016-2 | 1 | 1 | 18 | 100 | 0 | 0 | 0 | 0 |
| Tetracycline | 0.06-32 | 8 | 32 | 7 | 38.9 | 9 | 50.0 | 2 | 11.1 |
| Ciprofloxacin | 0.25-2 | 0.5 | 2 | ND | ND | ND | ND | ND | ND |
| Vancomycin | 0.03-0.25 | 0.25 | 0.25 | 18 | 100 | 0 | 0 | 0 | 0 |
| Teicoplanin | 0.03-0.125 | 0.06 | 0.125 | ND | ND | ND | ND | ND | ND |
MIC50 and MIC90, MICs at which 50 and 90% of isolates, respectively, are inhibited.
Breakpoints (intermediate and resistant) are as follows according to NCCLS (19): erythromycin A, 0.5 μg/ml and ≥1 μg/ml; clarithromycin, 0.5 μg/ml and ≥1 μg/ml; azithromycin, 1 μg/ml and ≥2 μg/ml; clindamycin, 0.5 μg/ml and ≥1 μg/ml; telithromycin, 2 μg/ml and ≥4 μg/ml, quinupristin-dalfopristin (Q-D), 2 μg/ml and ≥4 μg/ml; penicillin G, 0.1 to 1 μg/ml and ≥2 μg/ml; cefotaxime (nonmeningitis), 2 μg/ml and ≥4 μg/ml; amoxicillin, 4 μg/ml and ≥8 μg/ml; tetracycline, 4 μg/ml and ≥8 μg/ml; vancomycin (susceptible), ≤1 μg/ml. Roxithromycin breakpoints are not NCCLS approved. The breakpoints ≤1 μg/ml and ≥2 μg/ml were used. Ciprofloxacin, spiramycin, and teicoplanin breakpoints are not available. ND, not determined.
Serotyping showed serotypes 23F and 14 (30.5% each) to be the leading serotypes among the collection of macrolide-resistant pneumococcal isolates. Macrolide resistance was also found among serotypes 6B, 19F, and 9V. Only single isolates of other serotypes were encountered (Table 2).
TABLE 2.
Serotype distribution of erythromycin A-resistant S. pneumoniae isolates in seven European countries
| Serotype | No. of strains (%) |
|---|---|
| 3 | 1 (1.2) |
| 6A | 1 (1.2) |
| 6B | 12 (14.6) |
| 9V | 4 (4.9) |
| 14 | 25 (30.5) |
| 15A | 1 (1.2) |
| 15B | 1 (1.2) |
| 15F | 1 (1.2) |
| 19A | 1 (1.2) |
| 19F | 9 (11.0) |
| 23F | 25 (30.5) |
| 38 | 1 (1.2) |
| Total | 82 (100.0) |
In total, 64 of the 82 isolates (78.0%) were erm(B) positive and 18 (22.0) were mef(A) positive. The prevalence of the macrolide resistance genotypes varied substantially among countries. In France (87.5% of all strains), Spain (77.3%), Switzerland (80%), and Poland (100%), strains were predominantly erm(B) positive, whereas high levels of mef(A)-positive strains were reported from Greece (100%) and Germany (33.3%) (Table 3).
TABLE 3.
Distribution of macrolide resistance genotypes of clinical pneumococcal isolates in 11 centers from seven European countries
| Country | Center | No. of strains |
mef(A) positive
|
erm(B) positive
|
||
|---|---|---|---|---|---|---|
| No. | % | No. | % | |||
| France | Paris | 20 | 4 | 20.0 | 16 | 80.0 |
| France | Lyon | 5 | 1 | 20.0 | 4 | 80.0 |
| France | Lille | 12 | 0 | 0.0 | 12 | 100.0 |
| France | Total | 37 | 5 | 13.5 | 32 | 87.5 |
| Spain | Madrid | 11 | 2 | 18.2 | 9 | 81.8 |
| Spain | Seville | 3 | 0 | 0.0 | 3 | 100.0 |
| Spain | Barcelona | 8 | 3 | 37.5 | 5 | 62.5 |
| Spain | Total | 22 | 5 | 22.7 | 17 | 77.3 |
| Greece | Athens | 4 | 4 | 100.0 | 0 | 0.0 |
| Portugal | Coimbra | 1 | 1 | 100.0 | 0 | 0.0 |
| Germany | Düsseldorf | 3 | 1 | 33.3 | 2 | 66.7 |
| Switzerland | Lausanne | 10 | 2 | 20.0 | 8 | 80.0 |
| Poland | Warsaw | 5 | 0 | 0.0 | 5 | 100.0 |
| Total | 82 | 18 | 22.0 | 64 | 78.0 | |
Macrolide resistance was caused by the oligoclonal spread of some MLS types. Among those, sequence type 81 (Spain23F-1 serotype) was by far the most important, followed by the penicillin-resistant Polish serotype 14 clone (ST 143) and the Spain9V-3 clone (ST 156) (Table 4). The ST 81 isolates (n = 22) were found predominantly in Spain (n = 14) and France (n = 7) and in a single case in Germany. Notably, most of the strains were erm(B) positive, but only one isolate from France and five isolates from Spain were M phenotypes. With the exception of the German strain (MIC, ≤0.016 μg/ml), the penicillin MICs for all isolates were ≥1 μg/ml (Table 4).
TABLE 4.
Distribution of MLS types among 82 macrolide-resistant pneumococcal isolates from 11 centers in seven European countries
| MLS typee | Clone designation | No. | % | Predominant countries |
|---|---|---|---|---|
| 81 | Spain23F-1a | 22 | 26.8 | France, Spain, Germany |
| 143 | Pen-R Polish 14 cloneb | 11 | 13.4 | France |
| 156 | Spain9V-3a | 5 | 6.0 | France, Switzerland |
| 658 | New French 14 cloneb | 5 | 6.0 | France |
| 236 | Taiwn19F-14 conea | 4 | 4.9 | Greece |
| 315 | Poland6B-20 clonea,d | 4 | 4.9 | Poland |
| 621 | New French 19F clonec | 4 | 4.9 | France, Switzerland |
| 179 | Spain 19F cloneb | 3 | 3.7 | Switzerland |
| 619 | New French 6B cloneb | 3 | 3.7 | France |
| 620 | New Spanish 6B cloneb | 3 | 3.7 | Spain |
| 15 | SLV of England14-9 clonea,b | 2 | 2.4 | Switzerland |
| 73 | Multiresistant Spanish 15F cloneb | 2 | 2.4 | Portugal, Spain |
| 90 | Spain6B-2 clonea | 2 | 2.4 | Spain |
| 670 | New Swiss 14 cloneb | 2 | 2.4 | Switzerland |
| 9 | England14-9a | 1 | 1.2 | Germany |
| 276 | Netherlands 19 cloneb | 1 | 1.2 | France |
| 564 | German Serotype 14 cloneb | 1 | 1.2 | Germany |
| 699 | New French 23F clone | 1 | 1.2 | France |
| 657 | New French 23F clone | 1 | 1.2 | France |
| Others | New MLST | 5f | 6.0 | Various |
| Total | 82 | 100 |
Clones defined by the pneumococcal molecular epidemiology network.
For clone definitions, see MLST home page (http://www.mlst.net).
Serotype 14 variant of the ST 156 clone, which is generally of serotype 9V.
One isolate (13C056) was a serotype 23F.
Boldface numbers indicate clones primarily described in the present study.
Two isolates (17B044 and 17B047) are atypical pneumococci.
Significantly, differences were noted between the clonal distributions in France and Spain, countries for which relatively high levels of macrolide resistance have been reported. In Spain, >60% of macrolide resistance (14 of 22 strains) is due to the spread of a single clone (ST 81; Spain23F-1 serotype), and macrolide resistance was only rarely seen among some strains of ST 73, ST 90, and ST 620. In contrast, macrolide resistance in France is caused by a large number of clones, predominantly ST81 and ST143, but more than half of the macrolide resistance is due to the spread of other STs. Furthermore, the predominantly erm(B)-positive penicillin G-resistant Polish serotype 14 clone has spread to France but was found to carry the mef(E) gene. Notably, macrolide resistance has now spread to some serotypes, such as serotypes 3 and 38 and some serogroup 15 strains, not primarily involved in the clonal spread of resistance to date.
In addition, seven new macrolide-resistant clones were described for the first time in this investigation, including the French serotype 14 clone (ST 658), the French-Swiss 19F clone (ST 621), a French 6B clone (ST 619), a Spanish 6B clone (ST 620), a Swiss serotype 14 clone (ST 670), a French serotype 14 clone (ST 658), and a French 23F clone (ST 657).
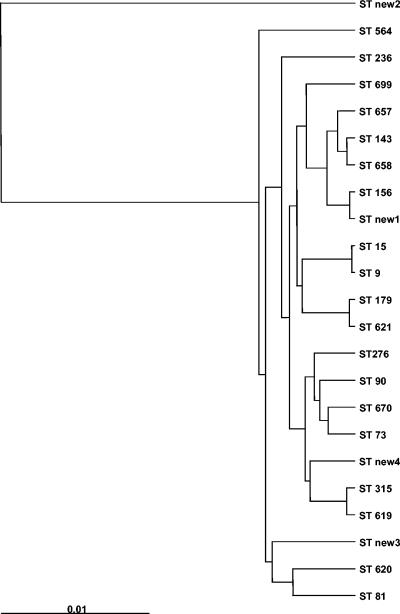
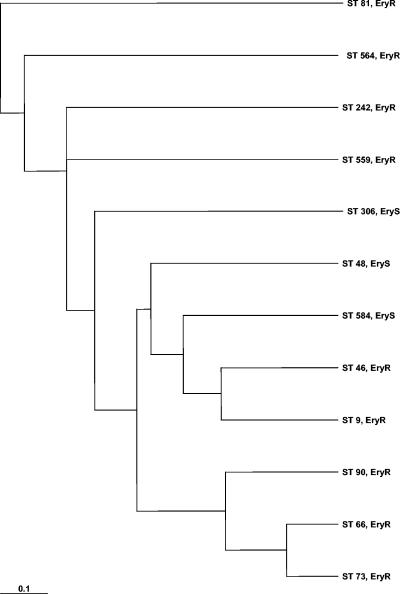
A dendrogram of the 23 MLS types included in the present study, constructed from the pairwise distance matrix using UPGMA, is presented in Fig. 1. The dendrogram shows that that ST new2 is only distantly related to the other MLS types. Within the other group of strains, ST new3, ST 81, and ST 620 are closely genetically related. Figure 2 shows the MLS types from the present study and representative strains of macrolide-susceptible and -resistant pneumococci from the MLST database. Analysis of the data indicated that the macrolide-resistant pneumococci are a genetically heterogeneous group within the pneumococci.
FIG. 1.
Dendrogram constructed from a pairwise distance matrix using UPGMA. Shown are the 23 MLSTs of the macrolide-resistant isolates from the present study.
FIG. 2.
Dendrogram constructed from a pairwise distance matrix using UPGMA. Shown are five MLSTs of strains of the present study (STs 9, 73, 81, 90, and 564) and four macrolide-resistant (STs 46, 66, 242, 559) and three macrolide-susceptible (STs 48, 306, 584) pneumococcal strains from the MLST database.
DISCUSSION
Within the last 10 years, macrolide resistance in S. pneumoniae has emerged on a dramatic scale. In Europe, it is now being increasingly reported from France and Spain. The present study shows that in France (87.5%), Spain (77.3%), Switzerland (80%), and Poland (100%), strains are predominantly erm(B) positive, whereas high levels of mef(A)-positive strains are reported from Greece (100%) and Germany (33.3%), confirming the findings of other investigators (6, 7, 20, 21, 25, 26). Notably, none of the strains was found to be mef(A)- and erm(B)-negative, suggesting the absence of one of the recently described new macrolide resistance mechanisms, such as mutations in the 23S rRNA or alterations in ribosomal proteins L4 and L22. Therefore, these mechanisms may at present be less important for the spread of macrolide resistance determinants in pneumococci.
Two serotypes (23F and 14) were identified by the present investigation as major contributors to the emergence of macrolide resistance in Europe. MLST, combined with recent developments in high-throughput sequencing and bioinformatics with established population genetics techniques, now provides a portable, reproducible, and scalable typing system that reflects the population and evolutionary biologies of bacterial pathogens (35) and permits analysis of the marked differences among pneumococcal strains belonging to one serotype. MLST has been increasingly used to analyze the clonal spread of antibiotic-resistant pneumococcal strains.
These studies include the identification of multidrug-resistant S. pneumoniae strains isolated in Poland (29), the spread of fluoroquinolone-resistant strains in the United States (11) and England (10), the genotypic characterization of two penicillin-susceptible serotype 6B S. pneumoniae clones circulating in Italy (8), and the characterization of erythromycin-resistant clinical isolates of the four major antimicrobial-resistant Spanish clones of S. pneumoniae (Spain23F-1, Spain6B-2, Spain9V-3, and Spain14-5) (13). For analysis of the epidemiologic relevance and the relative importance of individual MLS types, the evaluation of the MLST database may not be sufficient, as entries in the database do not all include consecutive isolates and therefore may not be representative of the clonal profiles of S. pneumoniae in individual countries. The present study is an approach to applying MLST to a large collection of macrolide-resistant pneumococci isolated in an international epidemiological study. A very small number of clones have been identified as being responsible for the spread of resistance, particularly highlighted by the worldwide spread of the serotype 23F pneumococcal clone (Spain23F-1), first identified in Spain in the early 1980s (16) and resistant to penicillin, chloramphenicol, tetracycline, and in some cases erythromycin A, as proved by the present investigation. This successful clone has now been found in the United States, South Africa, South America, and several European countries (14). The present study underscores the relevance of this clone for the spread of macrolide resistance, mainly in Spain. Furthermore, in Spain this clone was found to carry both widespread macrolide resistance mechanisms, mef(A) and erm(B), and to be nonsusceptible to penicillin. In contrast, in Germany, a country with a low level of penicillin G resistance (23), the only isolate of this clone was found to be penicillin susceptible.
The choice of empirical antibiotic therapy for the treatment of respiratory tract infections may have significant implications for the different frequencies of mef(A)- and erm(B)-mediated macrolide resistance in the several European countries. In France, we see the highest level of antibiotic and macrolide consumption in Europe (data available from the European Surveillance of Antibiotic Consumption [http://www.ua.ac.be/esac]). In addition, josamycin and spiramycin are widely used in France, which may contribute to the high rate of erm(B)-mediated macrolide resistance in that country. Furthermore, high macrolide consumption rates are reported from Italy and Greece. In Germany, the absolute consumption of all antibiotics is significantly lower than in Spain and France, but the consumption of macrolides is relatively high (36).
Notably, fluoroquinolone resistance was not observed in our collection of pneumococcal strains. It has also been observed by other investigators that macrolide and beta-lactam resistance is not associated with fluoroquinolone resistance in S. pneumoniae (5). Furthermore, high levels of macrolide consumption in many European countries may cause the selective antibiotic pressure responsible for this phenomenon.
In addition, this investigation describes some new clones, which may have the potential for further spread. Tetracycline resistance is frequently associated with erythromycin A resistance. In Europe, associations of >80% in erythromycin-resistant pneumococci isolated in Spain (30) and Italy (15) have recently been reported. This association may reflect the widespread presence in pneumococcal populations of transposons, typified by Tn1545, thought to result from the insertion over time of resistance determinants, such as erm(B) for erythromycin and aphA3 for kanamycin, into primitive gram-positive conjugative transposons carrying tet(M) and the integrase gene int-Tn, typified by Tn916.
Phylogenetic analysis showed all but one strain to be closely genetically related. Comparison with a representative group of macrolide-susceptible strains revealed no significant diversity between these two groups of strains (Fig. 1 and 2).
In summary, the present investigation demonstrates the genetic relatedness of macrolide-resistant strains in various European countries and underscores the high value of MLST in analyzing the genetic relatedness of antibiotic-resistant pneumococcal strains.
Acknowledgments
We thank the Eijkman-Winkler Institute (the European reference center for the SENTRY Antimicrobial Surveillance Program) for cooperation and for providing the isolates. We thank Nelli Neuberger, Maria Lemperle, and Claudia Cremer for excellent technical assistance and Susan Griesbach, Münster for copyediting.
The study was supported in part by grant RKI-415/1369235 from the German Ministry of Health (Bundesminister für Gesundheit) and by the German Ministry for Science and Technology (BMFT) (CAP net).
REFERENCES
- 1.Appelbaum, P. C. 2002. Resistance among Streptococcus pneumoniae: implications for drug selection. Clin. Infect. Dis. 34:1613-1620. [DOI] [PubMed] [Google Scholar]
- 2.Canu, A., B. Malbruny, M. Coquemont, T. A. Davies, P. C. Appelbaum, and R. Leclercq. 2002. Diversity of ribosomal mutations conferring resistance to macrolides, clindamycin, streptogramin, and telithromycin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Depardieu, F., and P. Courvalin. 2001. Mutation in 23S rRNA responsible for resistance to 16-membered macrolides and streptogramins in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:319-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 5.Felmingham, D., R. R. Reinert, Y. Hirakata, and A. Rodloff. 2002. Increasing prevalence of antimicrobial resistance among isolates of Streptococcus pneumoniae from the PROTEKT surveillance study, and comparative in vitro activity of the ketolide, telithromycin. J. Antimicrob. Chemother. 50(Suppl. 1):25-37. [DOI] [PubMed] [Google Scholar]
- 6.Fitoussi, F., C. Doit, P. Geslin, N. Brahimi, and E. Bingen. 2001. Mechanisms of macrolide resistance in clinical pneumococcal isolates in France. Antimicrob. Agents Chemother. 45:636-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fotopoulou, N., P. T. Tassios, D. V. Beste, S. Ioannidou, A. Efstratiou, E. R. Lawrence, J. Papaparaskevas, R. C. George, and N. J. Legakis. 2003. A common clone of erythromycin-resistant Streptococcus pneumoniae in Greece and the UK. Clin. Microbiol. Infect. 9:924-929. [DOI] [PubMed] [Google Scholar]
- 8.Gherardi, G., M. Del Grosso, A. Scotto D'Abusco, F. D'Ambrosio, G. Dicuonzo, and A. Pantosti. 2003. Phenotypic and genotypic characterization of two penicillin-susceptible serotype 6B Streptococcus pneumoniae clones circulating in Italy. J. Clin. Microbiol. 41:2855-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordon, K. A., D. J. Biedenbach, and R. N. Jones. 2003. Comparison of Streptococcus pneumoniae and Haemophilus influenzae susceptibilities from community-acquired respiratory tract infections and hospitalized patients with pneumonia: five-year results for the SENTRY Antimicrobial Surveillance Program. Diagn. Microbiol. Infect. Dis. 46:285-289. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, A. P., C. L. Sheppard, S. J. Harnett, A. Birtles, T. G. Harrison, N. P. Brenwald, M. J. Gill, R. A. Walker, D. M. Livermore, and R. C. George. 2003. Emergence of a fluoroquinolone-resistant strain of Streptococcus pneumoniae in England. J. Antimicrob. Chemother. 52:953-960. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, C. N., W. H. Benjamin, Jr., S. A. Moser, S. K. Hollingshead, X. Zheng, M. J. Crain, M. H. Nahm, and K. B. Waites. 2003. Genetic relatedness of levofloxacin-nonsusceptible Streptococcus pneumoniae isolates from North America. J. Clin. Microbiol. 41:2458-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marimon, J. M., L. Iglesias, D. Vicente, and E. Perez-Trallero. 2003. Molecular characterization of erythromycin-resistant clinical isolates of the four major antimicrobial-resistant Spanish clones of Streptococcus pneumoniae (Spain23F-1, Spain6B-2, Spain9V-3, and Spain14-5). Microb. Drug Resist. 9:133-137. [DOI] [PubMed] [Google Scholar]
- 14.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefevre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montanari, M. P., M. Mingoia, I. Cochetti, and P. E. Varaldo. 2003. Phenotypes and genotypes of erythromycin-resistant pneumococci in Italy. J. Clin. Microbiol. 41:428-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munoz, R., T. J. Coffey, M. Daniels, C. G. Dowson, G. Laible, J. Casal, R. Hakenbeck, M. Jacobs, J. M. Musser, and B. G. Spratt. 1991. Intercontinental spread of a multiresistant clone of serotype 23F Streptococcus pneumoniae. J. Infect. Dis. 164:302-306. [DOI] [PubMed] [Google Scholar]
- 17.Musher, D. M. 1992. Infections caused by Streptococcus pneumoniae: clinical spectrum, pathogenesis, immunity, and treatment. Clin. Infect. Dis. 14:801-807. [DOI] [PubMed] [Google Scholar]
- 18.Musher, D. M., M. E. Dowell, V. D. Shortridge, R. K. Flamm, J. H. Jorgensen, P. Le Magueres, and K. L. Krause. 2002. Emergence of macrolide resistance during treatment of pneumococcal pneumonia. N. Engl. J. Med. 346:630-631. [DOI] [PubMed] [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards. 2004. Performance standards for antimicrobial susceptibility testing. Fourteenth informational supplement M100-S14. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 20.Perez-Trallero, E., C. Fernandez-Mazarrasa, C. Garcia-Rey, E. Bouza, L. Aguilar, J. Garcia-de-Lomas, and F. Baquero. 2001. Antimicrobial susceptibilities of 1,684 Streptococcus pneumoniae and 2,039 Streptococcus pyogenes isolates and their ecological relationships: results of a 1-year (1998-1999) multicenter surveillance study in Spain. Antimicrob. Agents Chemother. 45:3334-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez-Trallero, E., J. M. Marimon, M. Montes, B. Orden, and M. de Pablos. 1999. Clonal differences among erythromycin-resistant Streptococcus pyogenes in Spain. Emerg. Infect. Dis. 5:235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reinert, R. R. 2004. Clinical efficacy of ketolides in the treatment of respiratory tract infections. J. Antimicrob. Chemother. 53:918-927. [DOI] [PubMed] [Google Scholar]
- 23.Reinert, R. R., A. Al-Lahham, M. Lemperle, C. Tenholte, C. Briefs, S. Haupts, H. H. Gerards, and R. Lütticken. 2002. Emergence of macrolide and penicillin resistance among invasive pneumococcal isolates in Germany. J. Antimicrob. Chemother. 49:61-68. [DOI] [PubMed] [Google Scholar]
- 24.Reinert, R. R., C. Franken, M. van der Linden, R. Lutticken, M. Cil, and A. Al-Lahham. 2004. Molecular characterisation of macrolide resistance mechanisms of Streptococcus pneumoniae and Streptococcus pyogenes isolated in Germany, 2002-2003. Int. J. Antimicrob. Agents 24:43-47. [DOI] [PubMed] [Google Scholar]
- 25.Reinert, R. R., R. Lütticken, A. Bryskier, and A. Al-Lahham. 2003. Macrolide-resistant Streptococcus pneumoniae and Streptococcus pyogenes in the pediatric population in Germany during 2000-2001. Antimicrob. Agents Chemother. 47:489-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reinert, R. R., S. Muckel, A. Al-Lahham, B. G. Spratt, A. B. Brueggemann, R. Hakenbeck, and R. Lütticken. 2003. Characterization of German penicillin non-susceptible serotype 23F pneumococci using multilocus sequence typing. J. Med. Microbiol. 52:981-987. [DOI] [PubMed] [Google Scholar]
- 27.Reinert, R. R., S. Simic, A. Al-Lahham, S. Reinert, M. Lemperle, and R. Lütticken. 2001. Antimicrobial resistance of Streptococcus pneumoniae recovered from outpatients with respiratory tract infections in Germany from 1998 to 1999: results of a national surveillance study. J. Clin. Microbiol. 39:1187-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reinert, R. R., A. Wild, P. Appelbaum, R. Lütticken, M. Y. Cil, and A. Al-Lahham. 2003. Ribosomal mutations conferring resistance to macrolides in Streptococcus pneumoniae clinical strains isolated in Germany. Antimicrob. Agents Chemother. 47:2319-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sadowy, E., J. Zhou, E. Meats, M. Gniadkowski, B. G. Spratt, and W. Hryniewicz. 2003. Identification of multidrug-resistant Streptococcus pneumoniae strains isolated in Poland by multilocus sequence typing. Microb. Drug Resist. 9:81-86. [DOI] [PubMed] [Google Scholar]
- 30.Seral, C., F. J. Castillo, M. C. Rubio-Calvo, A. Fenoll, C. Garcia, and R. Gomez-Lus. 2001. Distribution of resistance genes tet(M), aph3′-III, catpC194 and the integrase gene of Tn1545 in clinical Streptococcus pneumoniae harbouring erm(B) and mef(A) genes in Spain. J. Antimicrob. Chemother. 47:863-866. [DOI] [PubMed] [Google Scholar]
- 31.Sutcliffe, J., A. Tait Kamradt, and L. Wondrack. 1996. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob. Agents Chemother. 40:1817-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tait-Kamradt, A., T. Davies, P. C. Appelbaum, F. Depardieu, P. Courvalin, J. Petitpas, L. Wondrack, A. Walker, M. R. Jacobs, and J. Sutcliffe. 2000. Two new mechanisms of macrolide resistance in clinical strains of Streptococcus pneumoniae from Eastern Europe and North America. Antimicrob. Agents Chemother. 44:3395-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tait-Kamradt, A., T. Davies, M. Cronan, M. R. Jacobs, P. C. Appelbaum, and J. Sutcliffe. 2000. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 44:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trieu-Cuot, P., C. Poyart-Salmeron, C. Carlier, and P. Courvalin. 1990. Nucleotide sequence of the erythromycin resistance gene of the conjugative transposon Tn1545. Nucleic Acids Res. 18:3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Urwin, R., and M. C. Maiden. 2003. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 11:479-487. [DOI] [PubMed] [Google Scholar]
- 36.Vander Stichele, R. H., M. M. Elseviers, M. Ferech, S. Blot, and H. Goossens. 2004. European Surveillance of Antimicrobial Consumption (ESAC): data collection performance and methodological approach. Br. J. Clin. Pharmacol. 58:419-428. [DOI] [PMC free article] [PubMed] [Google Scholar]