Abstract
In the winter of 2002, an outbreak of mycoplasma infection in Vaal rhebok (Pelea capreolus) originating from South Africa occurred 15 weeks after their arrival in San Diego, Calif. Three rhebok developed inappetence, weight loss, lethargy, signs related to pulmonary or arthral dysfunction, and sepsis. All three rhebok died or were euthanized. Primary postmortem findings were erosive tracheitis, pleuropneumonia, regional cellulitis, and necrotizing lymphadenitis. Mycoplasmas were detected in numerous tissues by electron microscopy, immunohistochemistry, and PCR. The three deceased rhebok were coinfected with ovine herpesvirus-2, and two animals additionally had a novel gammaherpesvirus. However, no lesions indicative of herpesvirus were seen microscopically in any animal. The rheboks' mycoplasmas were characterized at the level of the 16S rRNA gene, the 16S-23S intergenic spacer region, and the fructose biphosphate aldolase gene. Denaturing gradient gel electrophoresis was carried out to address the possibility of infection with multiple strains. Two of the deceased rhebok were infected with a single strain of Mycoplasma capricolum subsp. capricolum, and the third animal had a single, unique strain most closely related to Mycoplasma mycoides subsp. mycoides large-colony. A PCR survey of DNA samples from 46 other ruminant species demonstrated the presence of several species of mycoplasmas in the mycoides cluster, including a strain of M. capricolum subsp. capricolum identical to that found in two of the rhebok. These findings demonstrate the pervasiveness of mycoplasmas in the mycoides cluster in small ruminants and the potential for interspecies transmission and disease when different animal taxa come in contact.
Mycoplasmas of the mycoides cluster are pathogens of ruminants and comprise six closely related species that are subdivided into two subgroups based on genetic similarity. The capricolum subgroup includes Mycoplasma capricolum subsp. capricolum, Mycoplasma capricolum subsp. capripneumoniae, and Mycoplasma subsp. bovine group 7 (BG7). The mycoides subgroup consists of Mycoplasma mycoides subsp. capri, Mycoplasma mycoides subsp. mycoides small-colony, and Mycoplasma mycoides subsp. mycoides large-colony (45). All six species of the mycoides cluster can cause respiratory, arthral, genitourinary, or mammary disease, although significant host- and strain-related variation in virulence exists. M. mycoides subsp. mycoides small-colony and M. capricolum subsp. capripneumoniae, the etiologies of contagious bovine pleuropneumonia (CBPP) and contagious caprine pleuropneumonia (CCPP), respectively, are the most virulent and typically induce fatal systemic disease in their hosts. Other members of the mycoides cluster, while pathogenic, do not usually cause life-threatening illness and, instead, establish protracted infections that result in low levels of chronic inflammation (8, 37).
Infection of sheep and goats with mycoplasmas in the mycoides cluster occurs worldwide. The insidious nature of many of these infections allows for the development of asymptomatic carriers able to harbor strains for prolonged periods (8). These reservoirs are extensive in some populations and can pose problems when different ruminant species come in contact or when naive animals are mixed with conspecific carriers (8, 37). Despite being widespread, however, mycoplasmas in the mycoides cluster have not been found in species of small ruminants other than sheep and goats. This may be, in part, because few studies have utilized sensitive molecular techniques, such as PCR, to survey nondomestic ruminants (8, 37). In this study, we report an outbreak of severe systemic mycoplasma infection in Vaal rhebok (Pelea capreolus) caused by two strains from the mycoides cluster and provide molecular epidemiologic data on possible reservoirs based on a PCR survey of 46 ruminant species.
MATERIALS AND METHODS
Animals and samples.
Thirteen (nine males, four females) Vaal rhebok antelope (Pelea capreolus, family Bovidae) were brought from South Africa to San Diego, Calif., in October 2002. All received complete physical examinations and routine diagnostic analyses for quarantined ruminants at the San Diego Zoo (SDZ). Fifteen weeks after arrival, three male rhebok developed clinical illness and were designated cases 1, 2, and 3 based on the chronology of their illness. Additional diagnostic tests were done on the three rhebok, which included complete blood counts (cases 1, 2, and 3), serum biochemical profiles (cases 1, 2, and 3), urinanalysis (case 3), blood cultures (cases 2 and 3), thoracic radiographs (cases 1, 2, and 3), and tracheal wash cytologies (cases 1 and 2). Nasal swabs and blood samples taken from four healthy rhebok housed in a distant location and three other ruminant species housed near the rhebok at the time of the outbreak, (eight Southern steenbok [Raphicerus campestris campestris], nine Cretan wild goats [Capra aegagrus cretica], and a Sichuan takin [Budorcas taxicolor tibetana]) were used in a PCR survey to identify possible reservoirs. Archived samples from one animal each of 43 ruminant species from the San Diego Zoo and San Diego Zoo's Wild Animal Park were also used in the PCR survey to identify reservoirs of mycoplasma. Samples consisted of lymph node, nasal swab, peripheral blood, liver, lung, oral mucosa, or intestine, and all samples were collected before the rhebok arrived in San Diego.
Necropsy.
Complete necropsies were performed on the three deceased rhebok (cases 1, 2, and 3). Complete sets of tissues were immersion fixed in 10% neutral buffered formalin for histology. Fresh tissue samples of liver, lung, spleen, retropharyngeal lymph node, and small intestine were taken by use of sterile technique from all three deceased rhebok in addition to oral mucosa (cases 2 and 3), kidney and heart (cases 1 and 3), pleura (case 2), and conjunctiva, tracheobronchial lymph node, and blood (case 3). Samples were frozen at −80°C for DNA extraction as described below. Samples were also sent to the Foreign Animal Disease Diagnostic Laboratory (Animal and Plant Health Inspection Service, United States Department of Agriculture, Orient Point, N.Y.) to exclude foreign animal diseases (CBPP, CCPP, and contagious agalactia).
Histopathology.
Formalin-fixed tissues were routinely processed, sectioned at 5 μm, and stained with hematoxylin and eosin (HE) for microscopic evaluation. Additional sections of pulmonary, splenic, and lymph node lesions were stained with Gram, Steiner's, Gimenez, periodic acid-Schiff, Ziehl-Neelsen, and Grocott's methenamine silver stains for bacteria, acid-fast organisms, and fungi.
DNA extractions.
DNA was extracted from tissues and nasal swabs with a QIAGEN (Valencia, Calif.) tissue kit according to the manufacturer's tissue sample protocol, except that the recommended amounts of sample were first placed with the lysis buffer in a 1.5-ml screw-cap FastPrep vial containing ceramic beads and were then lysed by agitation in a FastPrep shaker (Q-BIOgene, Carlsbad, Calif.) at a speed of 4 to 5.5 for 40 s. Proteinase K (20 mg/ml) (QIAGEN) was added to each sample and incubated overnight at 55°C. After the overnight incubation, another 20 μl of proteinase K was added and samples were incubated at 55°C for another hour. The lysates were then transferred to a clean Eppendorf tube for continuation of the QIAGEN tissue kit protocol.
Immunohistochemistry.
Five-micrometer sections of formalin-fixed, paraffin-embedded tissues on Platinum slides (Mercedes Medical, Sarasota, Fla.) were deparaffinized, hydrated, blocked with Peroxo-Block (Zymed Laboratories, South San Francisco, Calif.), washed in phosphate-buffered saline (PBS), and incubated with 0.1% protease (Sigma, St. Louis, Mo.) for 5 min at room temperature. After protease digestion, the sections were washed with PBS and blocked with CAS Block (Zymed Laboratories) for 30 min at room temperature. Sections were then incubated with a 1:500 dilution of the primary antibody, 5E5 (gift from H. J. Ball, Department of Agriculture for Northern Ireland, Stormont, Belfast, United Kingdom) overnight at 4°C. After the overnight incubation, the sections were washed in PBS and incubated with the secondary antibody, anti-mouse polymer horseradish peroxidase (Dako Corporation, Carpinteria, Calif.) for 1 h at room temperature. Sections were then washed in PBS and treated with diaminobenzidine tetrahydrochloride (DAB) (Vector Laboratories, Burlingame, Calif.) at room temperature for 1 to 5 min, washed in distilled water, counterstained with Gill's hematoxylin, dehydrated, and mounted.
Electron microscopy.
Lymph node and lung tissue initially preserved in 10% neutral buffered formalin were retrimmed and transferred to modified (half-strength) Karnovsky's (15) electron microscope fixative overnight before being rinsed in two changes of 0.2 M sodium cacodylate and postfixed in 2% osmium tetroxide reduced with 2.5% potassium ferrocyanide (34). Following osmium postfixation, tissues were washed again in 0.2 M sodium cacodylate and dehydrated through a graded ethanol series into propylene oxide before infiltration and embedment in Spurr's epoxy formulation (39). Thick sections were cut, mounted on glass slides, stained with Toluidine Blue O, and examined by light microscopy. Thin sections were cut and mounted on 300-mesh copper grids, stained briefly in 6% methanolic uranyl acetate, poststained with Reynold's lead citrate (29), and examined in a LEO 906E (LEO Electron Microscopy; Carl Zeiss electron microscopes, Thornwood, N.Y.) transmission electron microscope at a 60-kV accelerating voltage.
PCR.
Eight PCR assays were performed. Forty to 400 ng of DNA extracted from frozen tissues or nasal swabs were added to a 25-μl reaction mixture containing 15 mM Tris (pH 8.0), 50 mM KCl, 2.5 mM MgCl2, and 200 μM each of dATP, dCTP, dTTP, and dGTP (Promega, Madison, Wis.), 25 pmol of each primer, and 0.05 U of AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, Calif.)/μl. The eight PCR assays were as follows. (i) Universal bacteria: PCR targeting a segment of the 16S rRNA gene with primers Bactuniv F (5′-AGACTGCTACGGGAGGCAGCAGT-3′) and Bactuniv R (5′-GTTGCGCTCGTTGCGGGACTTAA-3′) was done as previously described (43). (ii) Mycoplasma 16S-23S rRNA intergenic spacer region (ITS): PCR targeting the 16S-23S rRNA ITS of mycoplasma with primers MycopIGSF (5′-CCCGTCACACCATGAGAGTT-3′) and MycopIGSR (5′-TCGGCTCCATTTTCCAAGGC-3′) was done as previously described (10, 11). (iii) Mycoplasma fructose biphosphate aldolase gene (fba): PCR specific for the putative membrane protein gene, fba, of mycoplasmas in the mycoides cluster was performed with primers Mycald P (5′-GCAATTGTTGGTATTGTTGTTGG-3′) and Capald M (5′-TATGTGCATCACTTACCATTTGTTTAG-3′) as previously described (41). (iv) Herpesvirus DNA polymerase: PCR using degenerate primers DFA (5′-GAYTTYGCNAGYTYITAYCC-3′), ILK (5′TCCTGGACAAGCAGCRNYSGCNMTNAA3′), and KG1 (3′-GTCCTTGCTCACCAGITCIACICCYTT3′) in a primary reaction and degenerate primers NY2 (5′-TGYAAYTCRGTDTAYGGITTYACIGGNGT3′) and NYGD (5′-CACRGAGTCCGTRTCNCCRTADAT-3′) in a secondary reaction that target consensus regions of herpesvirus DNA polymerase gene for amplification of all herpesviruses was performed as previously described (21, 33, 42). (v) Alcelaphine herpesvirus 1 (AlHV-1) and Alcelaphine herpesvirus 2 (AlHV-2): PCR that amplifies a homologous region partially within the major capsid protein of AlHV-1 and AlHV-2 was performed with primers H3D-F (5′-ATACATGTCATTTAAGACACCCACGCACCA-3′) and H3D-B (5′-CTGGTGCAGGATGACCACAATTTTACTATC-3′) as previously described (20, 25, 26). (vi) Ovine herpesvirus 2 (OvHV-2): PCR specific for a segment of the 140-kDa tegument protein was performed with primers OHV2-F (5′-GTCTGGGGTATATGAATCCAGATGGCTCTC-3′) and OHV2-B (5′-AAGATAAGCACCAGTTATGCATCTGATAAA-3′) as previously described (2). (vii) Adenovirus: PCR targeting a domain of the hexon protein for members of Adenoviridae was performed with primers AdenoKISSR (5′-CAGCRYRCCGCGGATGTCAAART-3′) and AdenoKISSF (5′-GCCGCARTGGTCTACATGCACATC-3′) as previously described (17). (viii) Caprine herpesvirus 2 (CaHV-2): PCR specific for a region of the DNA polymerase gene of CaHV-2 was performed with primers CaHV-2F (5′-CCTGCCTCACCATTGCGGAAA-3′) and CaHV-2R (5′-GGCATAGCTCCCTCAGGTGCT-3′), using touchdown PCR, as previously described (16, 22). PCR products were analyzed by electrophoresis on a 1% agarose gel, and relevant fragments were purified by using an Amicon Ultrafree-DA spin column (Millipore, Billerica, Mass.).
Denaturing gradient gel electrophoresis.
PCR products for the ITS and fba gene, as described above, were further analyzed by denaturing gradient gel electrophoresis (DGGE) by loading samples onto 10% polyacrylamide-bis (37.5:1) gels with a denaturing gradient from 0 to 100% (where 100% is 7 M urea and 40% deionized formamide) in 1× Tris-acetate-EDTA electrophoresis buffer. Electrophoresis was performed at 130 V at 56°C for 4 h on a DCode Universal mutation detection system (Bio-Rad, Hercules, Calif.). Gels were stained with a 1:20,000 dilution of SYBR Green (Molecular Probes, Eugene, Ore.) in 1× Tris-acetate-EDTA for 30 min at room temperature. Bands were visualized under UV illumination. Relevant bands were purified from the gel for cloning.
DNA manipulation and sequencing.
Purified fragments from agarose gels or the DGGE gels were cloned by using the TOPO TA or TOPO blunt cloning kits (Invitrogen, Carlsbad, Calif.). Sequencing reactions were performed by using the CEQ DTCS (dye terminator cycle sequencing) Quick-Start kit (Beckman Coulter, Fullerton, Calif.). Sequences were acquired by using a CEQ 2000XL capillary sequencer (Beckman Coulter).
Nucleotide sequence analyses.
Sequence analyses and alignments were conducted by using the Sequencher 4.1 (Gene Codes Corporation, Ann Arbor, Mich.) and the MacVector 6.5.3 (Accelrys, San Diego, Calif.) software packages. Sequence data were compared to the GenBank database with the basic local alignment search tool. For phylogenetic analyses, sequences were aligned with the MacVector ClustalW program (Accelrys). Phylogenetic relationships were estimated by using the program Paup 4.0 (40) with neighbor-joining and maximum-parsimony methods with the Kimura-2 parameter. Bootstrap values were counted as percentages over 1,000 replicates. A single majority-based prototype sequence and several minority variant sequences for three loci (16S rRNA, ITS, and fba segments) were obtained for mycoplasmas from the three rhebok after agarose and DGGE analyses of multiple PCR assays from different tissues of each animal.
Nucleotide sequence accession numbers.
GenBank accession numbers for the 16S rRNA gene segment prototype are AY625085 (cases 1 and 2) and A0Y625088 (case 3). GenBank accession numbers for the 16S rRNA gene segment minority variants are AY625086 (case 1); AY625087 (case 2); and AY625089 (case 3). GenBank accession numbers for the ITS prototypes are AY625090 (cases 1 and 2 and Dall's sheep); AY625082 (case 3); and AY625094 and AY625095 (blue sheep). GenBank accession numbers for the ITS minority variants are AY625092, AY625076, AY625077, and AY625078 (case 1); AY625091, AY625079, AY625080, and AY625081 (case 2); and AY625083 and AY625084 (case 3). GenBank accession numbers for the fba segment prototypes are AY625097 (case 1 and 2 and Dall's sheep); AY625098 (case 3); and AY625096 (blue sheep). GenBank accession number for the gammaherpesvirus found in two rhebok is AY685146.
RESULTS
History and clinical findings.
Thirteen (nine males and four females) rhebok antelope (Pelea capreolus, family Bovidae) from South Africa served quarantine periods in Warsaw, Poland, and Newburg, N.Y., before being transported to San Diego, Calif., on 11 October 2002. Upon their initial arrival in San Diego, the 13 rhebok were divided into six groups: four large adult males were housed individually; four small adult males (cases 1, 2, and 3 and the fourth rhebok) were housed together in a single pen; and four adult females were housed together with a 4-week-old male calf. Physical examinations and standard diagnostic screening evaluations for ruminants performed by veterinarians at SDZ detected intestinal infection of several rhebok with coccidia, but the animals were otherwise found to be healthy.
When moved from quarantine housing on 21 November 2002, the four small adult males were housed in groups of two individuals (case 1 and fourth rhebok together; cases 2 and 3 together) in a row of pens adjacent to a group of Cretan wild goats (Capra aegagrus cretica) and also near other animals. On 27 January 2003, a rhebok (case 1) became ill; a second rhebok (case 2) became ill on 11 February 2003, and a third (case 3) became ill on 23 February 2003. Cases 2 and 3 were separated on 4 February 2003, so that all rhebok were individually housed in the row of pens thereafter. The fourth rhebok was removed from the pen on 23 January 2003, before the first rhebok was ill, and was returned to the pen on 19 February 2003, just before the third rhebok became ill, and subsequently did not become ill. During this period (2 January 2003 to 25 February 2003), ambient temperatures ranged between 46°F and 83°F, winds varied from 10 to 38 miles per h, and six episodes of rain occurred.
The three ill rhebok showed subdued behavior, lack of appetite, and weight loss. Also present were ataxia (case 1), excessive salivation (cases 2 and 3), and head tremor (case 2). Physical examination findings included increased bronchovesicular lung sounds (cases 1 and 2) and pyrexia (cases 1 and 3). Case 1 subsequently developed generalized facial and oropharyngeal swelling, audible respirations, diarrhea, and recumbency. Radiographic changes included a diffuse bronchointerstitial pattern, infiltrative pneumonia, or bullae formation. Tracheal wash cytologies for cases 1 and 2 were indicative of inflammation.
In cases 1, 2, and 3, complete blood count analyses showed elevated fibrinogen, leukopenia, and a left shift with toxic changes in leukocytes. One rhebok (case 1) was euthanized, and two rhebok (cases 2 and 3) died within 3 days of the onset of clinical signs. None of the nine rhebok housed at distant locations were ill at any time during the outbreak.
Necropsy findings.
From necropsy, it was found that inflammation of skin, subcutis, and other tissues (salivary gland, skeletal muscle, oral cavity, nasal cavity, and pharynx) caused swollen heads in two animals (cases 1 and 3). All three animals had acute necrotizing lymphadenitis (Fig. 1A). Erosive and ulcerative tracheitis were present in two animals (cases 1 and 2). One rhebok had fibrinous pleuropneumonia and epicarditis (case 2) (Fig. 1B). Two rhebok (cases 1 and 3) had minimal multifocal vascular necrosis and neutrophilic vasculitis considered to be secondary to the surrounding inflammation. Special stains (Gram, Steiner's, Gimenez, Grocott's methenamine silver, periodic acid-Schiff, and acid-fast stains) performed on selected tissues did not clearly reveal infectious agents. However, granular, basophilic material suggestive of bacteria was seen in HE sections of tissues with lesions (Fig. 1C).
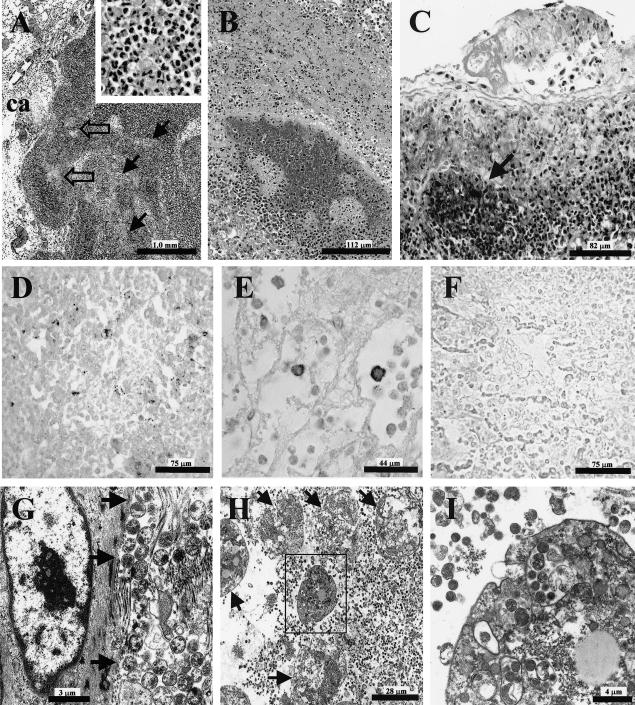
FIG. 1.
Photomicrographs of tissue lesions taken from affected rhebok at necropsy. (A) Tracheobronchial lymph node with marked necrosis and inflammation comprising karyorrhexis with accumulation of amorphous eosinophilic material and infiltrates of predominantly macrophages and neutrophils (inset); inflammatory cells fill sinuses (solid black arrows) and accompany necrosis in the cortex (clear arrows); also affected are the capsule (ca) and adjacent connective tissue, HE. (B) Effacement of pulmonary parenchyma, including bronchiole and surrounding alveoli, with severe inflammation and necrosis, HE. (C) Pleuropneumonia with fine basophilic material representing mycoplasma organisms (arrow), HE. (D) Immunohistochemistry on lung with antibody 5E5 showing extracellular staining of mycoplasma organisms around an area of necrosis, DAB with Gill's hematoxylin counterstain. (E) Intracellular immunostaining with antibody 5E5 of inflammatory cells in lung, DAB with Gill's hematoxylin counterstain. (F) negative control of lung tissue for immunohistochemistry, DAB, Gill's hematoxylin counterstain. (G) Electron micrograph from lymph node with numerous round-to-oval extracellular mycoplasma organisms (arrows) adjacent to collagen fibrils. (H) Lung, with extracellular and intracellular mycoplasma organisms and exudative leukocytes in various states of degeneration (arrows). (I) higher power of box inset from H demonstrating inflammatory cell with cytoplasmic mycoplasma organisms.
Antibody detection of mycoplasma in rhebok tissue sections.
Immunohistochemistry using the antibody, 5E5, which previously has been shown to react with antigens on all mycoplasma of the mycoides cluster (30-32), demonstrated mycoplasma organisms in lesions from each of the three deceased rhebok (Fig. 1D, E, and F). Staining was localized extracellularly around necrotic foci (Fig. 1D) and inside macrophages or neutrophils in areas of inflammation (Fig. 1E). Immunostaining for mycoplasma was not seen in normal tissues of the rhebok.
Detection of mycoplasma in rhebok tissue by electron microscopy.
Transmission electron microscopy on lymph node and lung tissue from cases 2 and 3 showed numerous extracellular organisms compatible with mycoplasma species. The lung had abundant, pleomorphic 0.15- to 0.46-μm-diameter alveolar organisms that were lying free within the interstitium and alveolar lumen. Lymph nodes had interstitial organisms, especially abundant in areas of connective tissue reticulum (Fig. 1G). Additional organisms were present within macrophage phagolysosomes (Fig. 1H and I). The organisms were limited by a single membrane. Smaller organisms had diffusely condensed cytoplasm, while in the larger bacteria, the condensed-granular cytoplasm was marginated.
PCR amplification of mycoplasma DNA in rhebok tissues.
The universal bacteria PCR for a segment of the 16S rRNA gene yielded 766-bp amplicons from multiple tissues of all three deceased rhebok. Phylogenetic analyses grouped the rhebok mycoplasma sequences in the mycoides cluster (Fig. 2). Amplicons from two rhebok (cases 1 and 2) had identical nucleotide sequences in the majority of clones. These sequences had 100% identity to M. capricolum subsp. capricolum (GenBank accession U26047) and MBG7 (GenBank accession AF261730). Amplicons from the majority of clones from the third rhebok (case 3) differed by a single base pair from the 16S rRNA gene segment sequences of the other two rhebok and had 100% identity to M. mycoides subsp. mycoides large-colony (GenBank accession no. U26050) and M. mycoides subsp. capri (GenBank accession no. U26036). In each rhebok, several clones of the 16S rRNA gene segment from different tissues differed by a single base pair from the majority of clones. These sequences were designated minority variants in the GenBank accessions. Excluding samples of intestine and oral cavity, no other bacteria were detected in any tissues of the rhebok.
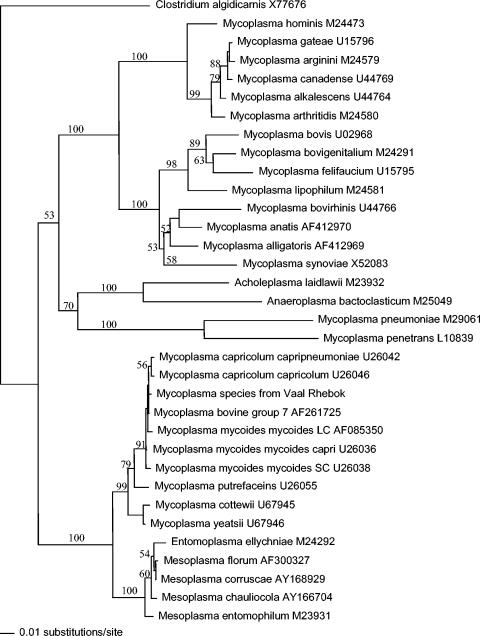
FIG. 2.
Phylogenetic tree analysis of 16S rRNA sequences from various bacterial organisms and the affected rhebok using the neighbor-joining method. Phylogram shows a clustering of the 16S rRNA sequence from the rhebok (case 1) DNA with the 16S rRNA sequences of mycoplasmas of the mycoides cluster. Numbers at the branch points represent percent probabilities obtained from bootstrap resampling. Clostridium algidicarnis (GenBank accession no. X77676) was the outgroup.
PCR for adenovirus was negative in all tissues of the rhebok. PCR for regions of the DNA polymerase gene conserved in all herpesviruses was positive in spleen samples from cases 1 and 3. The amplicons generated were identical in both rhebok and were similar to herpesviruses genetically unrelated to the malignant catarrhal fever group. PCR assays specific for the malignant catarrhal fever viruses, AlHV-1, AlHV-2, and CaHV, were negative with all rhebok samples, but PCR for OvHV-2 was positive with kidney (case 1), pleura (case 2), and oral mucosa (case 3). The amplicons generated from each rhebok sample were identical and had 100% identity to OvHV-2 (GenBank accession S64565).
Identification of the infectious mycoplasma species by PCR methods.
To further characterize the species of mycoplasma in the rhebok, PCR for the ITS and fba gene segment was done. PCR for the ITS generated 418-bp fragments from all tissues in cases 1 and 2 and from the majority of the tissues from case 3 (Fig. 3A). Case 1 and 2 had identical ITS sequences which had 100% identity to a strain of M. capricolum subsp. capricolum (GenBank accession no. AB031584), whereas case 3 had a unique sequence that was 98.7% similar to a strain of M. mycoides subsp. mycoides large-colony (GenBank accession no. AB031586) (Fig. 3B). PCR for the fba gene segment generated 302-bp amplicons from all tissues from cases 1 and 2 and the majority of tissues from case 3 (Fig. 4A). The fba sequences obtained from cases 1 and 2 were identical and had 100% identity to several strains of M. capricolum subsp. capricolum (GenBank accession no. AF162970, AF162952, AF162951, and AF162950). The fba sequences obtained from case 3 was unique and had 99.2% nucleotide identity to a strain of M. mycoides subsp. mycoides large-colony (GenBank accession no. AF162943) (Fig. 4B). The Foreign Animal Disease Diagnostic Laboratory performed PCR analysis on tissue samples to test for the presence of mycoplasmas exotic to the United States, which cause CCPP and CBPP. They were able to confirm our results that none of the rhebok were infected with the mycoplasma species, M. capricolum subsp. capripneumoniae or M. mycoides subsp. mycoides small-colony, that cause these diseases.
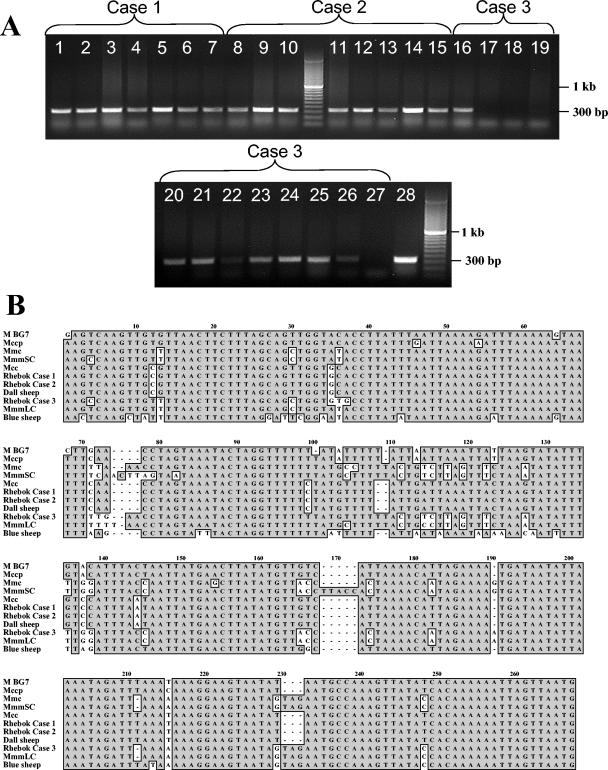
FIG. 3.
(A) Ethidium bromide-stained agarose gel of PCR amplicons of mycoplasma ITS from various tissues of the three deceased rhebok. Lanes 1 to 7, various tissues from rhebok case 1: lane 1, liver; lane 2, lung; lane 3, spleen; lane 4, small intestine; lane 5, retropharyngeal lymph node; lane 6, kidney; lane 7, heart. Lanes 8 to 15, various tissues from rhebok case 2: lane 8, liver; lane 9, lung; lane 10, spleen; lane 11, small intestine; lane 12, retropharyngeal lymph node; lane 13, nasal mucosa; lane 14, lung pleura; lane 15, oral mucosa. Lanes 16 to 27, various tissues from rhebok case 3: lane 16, liver; lane 17, lung; lane 18, spleen; lane 19, small intestine; lane 20, retropharygeal lymph node; lane 21, nasal mucosa; lane 22, kidney; lane 23, oral mucosa; lane 24, conjunctiva; lane 25, heart; lane 26, tracheobronchial lymph node; lane 27, blood. (B) Alignment of mycoplasma ITS DNA from the prototype sequence from the three deceased rhebok, Dall's sheep, and blue sheep and representative mycoplasma species of the mycoides cluster. Majority conserved nucleotides are enclosed in shaded boxes. For rhebok case 2, there were two sequences that were equally represented in the clones.
FIG. 4.
(A) Ethidium bromide-stained agarose gel of PCR amplicons of a segment of the fba gene of mycoplasma of the mycoides cluster from DNA extracted from various tissues of the three deceased rhebok. Lanes 1 to 7, various tissues from rhebok case 1: lane 1, liver; lane 2, lung; lane 3, spleen; lane 4, small intestine; lane 5, retropharyngeal lymph node; lane 6, kidney; lane 7, heart. Lanes 8 to 15, various tissues from rhebok case 2: lane 8, liver; lane 9, lung; lane 10, spleen; lane 11, small intestine; lane 12, retropharyngeal lymph node; lane 13, nasal mucosa; lane 14, lung pleura; lane 15, oral mucosa. Lanes 16 to 27, various tissues from rhebok case 3: lane 16, liver; lane 17, lung; lane 18, spleen; lane 19, small intestine; lane 20, retropharyngeal lymph node; lane 21, nasal mucosa; lane 22, kidney; lane 23, oral mucosa; lane 24, conjunctiva; lane 25, heart; lane 26, tracheobronchial lymph node; lane 27, blood; lane 28, Dall’s sheep, retropharyngeal lymph node. (B) Alignment of fba DNA prototype sequences from the three deceased rhebok, Dall's sheep, and blue sheep and sequences from representative mycoplasma species of the mycoides cluster. Majority conserved nucleotides for a given position are enclosed in shaded boxes.
Analysis of mycoplasma PCR products by denaturing gradient gel electrophoresis.
To assess the possibility that individual rhebok were infected with more than one species of mycoplasma, PCR amplicons from the ITS and the fba gene segment were analyzed using DGGE (data not shown). For all deceased rhebok (case 1, 2, and 3), the same ITS and fba sequences obtained from agarose gel electrophoresis were also detected in all the same tissues by DGGE and are designated prototype sequences in our GenBank accessions. For case 1, four other ITS sequences that varied by 1 to 5 bp from the prototype sequence were detected in several tissues. All of these sequences were most similar to those of a strain of M. capricolum subsp. capricolum (GenBank accession no. AB031584). For cases 2 and 3, other ITS sequences that varied by 1 to 6 bp from the prototype sequence were detected in several tissues, and all of these sequences were also most similar to a strain of M. capricolum subsp. capricolum (GenBank accession no. AB031584). For case 3, two other ITS sequences that varied by 1 to 4 bp from the prototype sequence were found in some tissues. These sequences were most similar to those of a strain of M. mycoides subsp. mycoides large-colony (GenBank accession no. AB031586). All alternative ITS sequences from the three deceased rhebok were designated minority sequence variants in our GenBank entries.
In cases 1 and 3, only a single fba sequence was found on DGGE analyses. These sequences were the same as those found after agarose gel electrophoresis. In case 2, one additional fba sequence was detected in several tissues by DGGE. This sequence differed by 2 bp from the prototype sequence, was most similar to sequence of strains of M. capricolum subsp. capricolum (GenBank accession no. AF162970, AF162952, AF162951, and AF162950), and was designated a minority sequence variant in our GenBank entry.
Analysis of possible reservoirs of mycoplasma.
Analyses of DNA from blood samples and nasal swabs taken subsequent to the outbreak from four healthy male rhebok that were shipped to San Diego with the three ill rhebok and housed at a distant location since the SDZ quarantine period, as well as from three other ruminant species (Southern steenbok, Cretan wild goats, and Sichuan takin), which were housed near the rhebok at the time of the outbreak, were negative by PCR for mycoplasmas of the mycoides cluster. Two of the archived DNA samples that were extracted from 43 ruminant species before the rhebok arrived in San Diego were positive by PCR for the ITS and fba gene segments. One positive sample was from a retropharyngeal lymph node of a Dall's sheep, and the other was from a retropharyngeal lymph node of a blue sheep. Both the ITS sequences and the fba sequences from the Dall's sheep were identical to the prototype sequences in two (cases 1 and 2) of the ill rhebok (Fig. 3B and 4B). Two unique ITS sequences were obtained from the blue sheep. The sequences were 93.2% similar to each other, 90.6 to 94.2% similar to the rhebok prototypes and minority variant sequences, and overall most similar to a strain of BG7 (GenBank accession no. AB031589) at 95.1 and 93.7% nucleotide identity, respectively (Fig. 3B). One fba sequence was detected in the blue sheep. This sequence had 82.7 to 83.5% sequence identity to the rhebok fba prototypes and minority variant sequences and 98.0% identity to sequence of a strain of M. mycoides subsp. mycoides large-colony (GenBank accession no. AF162949) (Fig. 4B).
DISCUSSION
Mycoplasma infections of animals are common, with more than 200 species having been described in a variety of mammals, birds, reptiles, fish, and insects (8, 37). In small ruminants, mycoplasmas have been reported in domestic goats and sheep, mountain goats, ibex, chamois, Dall's sheep, and gazelles (4, 8, 9, 41, 44). In most of these cases, infection was associated with disease. However, the vast majority of animal mycoplasmas are nonpathogenic commensals (8, 37). Notable exceptions are the six species of mycoplasmas in the mycoides cluster. These agents can produce disease that ranges in severity from mild transient illness to chronic localized inflammation or fatal septicemia with multiple organ damage (8). In our study, disease in rhebok was caused by a strain of M. capricolum subsp. capricolum and a novel mycoplasma closely related to M. mycoides subsp. mycoides large-colony. In all three cases, illness was severe and infection was systemic.
Mycoplasma was determined to be the most significant etiologic component of clinical disease in the three deceased rhebok based on findings from PCR assays, immunohistochemistry, electron microscopy, and histology. The finding of exclusively mycoplasma in all tissues from the three deceased rhebok indicated that mycoplasma was a significant contributing factor in disease. Detection of mycoplasma only within areas of inflammation and necrosis by immunohistochemistry and electron microscopy further supported a causal relationship between the organism and disease. The nature and pattern of inflammation in trachea, lung, lymph nodes, and skin as well as the development of septicemia were characteristic of M. capricolum subsp. capricolum and M. mycoides subsp. mycoides large-colony infection in domestic goats, especially young kids (3, 8, 32). The severity of necrosis and presence of neutrophilic vasculitis in rhebok, however, more resembled infection with M. mycoides subsp. mycoides small-colony and M. capricolum subsp. capripneumoniae, which are the most virulent of the mycoides cluster species and are often fatal (8). Lesions of malignant catarrhal fever, such as lymphocytic vasculitis and perivasculitis, ulceration, lymphoproliferation, and chronic vasculopathy were not seen in any animals, nor were findings indicative of an alternative etiology.
Multilocus genetic analyses were necessary to speciate the mycoplasmas. Characterization of coding (16S rRNA and fba) and noncoding (ITS) regions have been used to classify the six species and numerous strains of mycoplasmas in the mycoides cluster (1, 10-12, 23, 24, 27, 28, 41). PCR is an effective diagnostic method for rapidly evaluating clinical samples in human and veterinary medicine. In our study, the assay also proved to be valuable, since all body fluids and tissues derived from the rhebok were restricted from leaving zoo grounds by animal health-related governmental ordinances and the samples could be quickly analyzed in an on-site molecular laboratory. Two rhebok had a mycoplasma identical to several isolates of M. capricolum subsp. capricolum. Interestingly, the third rhebok in our study was infected with a mycoplasma closely related to a strain of M. mycoides subsp. mycoides large-colony. Based on the range of intraspecies genetic heterogeneity among M. mycoides subsp. mycoides large-colony strains, the mycoplasma from this rhebok is probably a new strain of M. mycoides subsp. mycoides large-colony (18, 24, 45). However, phenotypic and serologic data are needed to confirm this. M. mycoides subsp. mycoides large-colony has been reported as a pathogen of sheep and goats, whereas M. capricolum subsp. capricolum has been associated with disease in sheep, goats, and cattle (8, 41). Findings from our study indicate that both M. capricolum subsp. capricolum and M. mycoides subsp. mycoides large-colony are pathogenic in rhebok. The degree of pathogenicity is not clear, but it appears to be marked.
The course of mycoplasma infection in animals is often influenced by environmental and physiologic factors, including coincidental infection with other microbial agents (6, 13, 32, 37). Handling and transport of animals, overcrowding, inadequate ventilation, and inclement weather are some environmental factors that have been shown to increase the risk of mycoplasmal pneumonia in sheep (5). In zoo animals, stress caused by movement of animals to a new location was reported to be a significant contributing factor in the development of pneumonia and death caused by Mycoplasma ovipneumoniae in Dall's sheep (Ovis dalli dalli) (4). In our study, the three rhebok that became ill were housed together for various periods of time and were briefly exposed to inclement weather. The immunologic status of the rhebok with regard to mycoplasmas of the mycoides cluster was unknown. If the animals came from a naive herd, the lack of previous exposure could have increased their vulnerability to the development of severe disease. Herpesvirus infection is an additional factor to consider. While OvH-2 did not appear to contribute to clinical signs and lesions in the rhebok, the possibility that it influenced the onset of mycoplasmosis in some way cannot be completely excluded. OvH-2 has been shown under experimental conditions to modulate host immune responses, although it is not thought to be an immunosuppressive agent (26, 36). The significance of the other gammaherpesvirus in two of the rhebok is unclear.
DGGE analyses indicated that each rhebok was infected with only one species of mycoplasma. The minor variations in nucleotide sequences of coding and noncoding loci seen within individual animals were likely reflective of genetic heterogeneity typical of mycoplasmas (7, 18, 24). Alternatively, sequence differences could have been due to taq polymerase-induced errors. This possibility was considered improbable, since replicate PCR samples were analyzed. Genome plasticity is widely recognized in mycoplasmas of veterinary importance (19, 46, 49). In people, genetic variation of Mycoplasma hominis within individual patients has been shown during chronic infections (38). Complete sequencing of the M. mycoides subsp. mycoides small-colony genome and extensive portions of the Mycoplasma capricolum chromosome have shown that hypermutation is a favored mechanism of mycoplasmas for adjusting virulence in the face of host selection pressures (47). Given that, it is not surprising to find some degree of genotypic variance in mycoplasmas after protracted infections. In the rhebok, infection and disease appeared to be acute. However, it is likely that the animal or animals acting as reservoirs for the mycoplasmas were chronically infected and provided an environment allowing genetic diversification before infection of the rhebok.
The finding of two distinct mycoplasma species suggested that infection did not pass laterally from rhebok to rhebok in each case. Most likely, the rhebok acquired the mycoplasmas after arriving in San Diego. These animals were at the zoo for at least 15 weeks before the first clinical signs of disease, and molecular testing of in-contact animals, including healthy rhebok, showed no evidence of infection with mycoplasmas of the mycoides cluster. The Dall's sheep that had a strain of M. capricolum subsp. capricolum identical to the organism in two of the rhebok may or may not have been part of a group of animals that acted as long-term reservoirs. The Dall's sheep died several months before the rhebok came to the SDZ, and none of its cohorts were housed near the rhebok. However, the finding of an M. capricolum subsp. capricolum strain that was identical at three genetic loci to the mycoplasmas in two of the rhebok clearly demonstrated that the organism was present on the premises before the rhebok arrived. The blue sheep, which had an occult infection with two unique species of mycoplasmas in the mycoides cluster, was not a putative source because the strains it harbored differed from those found in the rhebok. Yet the finding of two mycoplasmas in the mycoides cluster in this animal emphasized the pervasiveness of these organisms in different species of small ruminants and drew attention to the likelihood that several reservoirs existed at the SDZ before the outbreak in rhebok. The Cretan wild goats were considered a possible reservoir and source of infection for the rhebok. The goats were housed adjacent to the three affected rhebok. It is not surprising that a species of goat would harbor M. capricolum subsp. capricolum and M. mycoides subsp. mycoides large-colony, since both of these organisms have frequently been isolated from domestic goats (41).
Our study illustrates how translocated species may be susceptible to infectious agents which are endemic but cryptic in other related taxa. Mycoplasmas within the mycoides cluster, while pathogenic, can be frequently found in ruminants without clinical signs of disease (6). Furthermore, mycoplasmas are notoriously difficult to isolate by culture and difficult to visualize histologically (35, 37). Consequently, it is likely that many mycoplasmal infections are never detected with use of conventional diagnostic techniques. Our PCR survey results from various ruminants indicated that organisms of the mycoides cluster may be more widespread than previously thought. We found that different species of mycoplasmas within the mycoides cluster were able to infect rhebok, Dall's sheep, and blue sheep. This capacity for interspecies infectivity poses significant problems in settings where different animal taxa may have direct or indirect contact and should be considered in advance when moving or managing animals in zoological gardens or when investigating disease outbreaks in the wild. M. capricolum subsp. capricolum and M. mycoides subsp. mycoides large-colony are exceptional because strains of each have been shown to infect goats, sheep, and cattle, and titers of antibody against M. mycoides subsp. mycoides large-colony have been found in other animals (8, 14, 37). The high degree of host fidelity previously ascribed to other members of the mycoides cluster may have been due to a lack of extensive sampling from different ruminant species or the use of relatively insensitive culture techniques (41).
In our three deceased rhebok, a definitive diagnosis of mycoplasmosis required molecular analyses despite thorough antemortem quarantine and diagnostic evaluations and complete necropsy examinations. Findings from this study demonstrate the potential vulnerability of Vaal rhebok to mycoplasmas of the mycoides cluster and suggest that these and other like animals should be tested, handled, and housed accordingly to prevent future outbreaks. In particular, the use of PCR is valuable for detecting carriers of mycoplasmas among exotic or rare ruminant species in captivity.
Acknowledgments
This work was supported by the Zoological Society of San Diego.
We thank Charles and Shirley Sykes of San Diego for financial support of our laboratory. We thank Katie Lyons, April Gorow, Julie Concha, Pam McGlynn, and Yvonne Cates for excellent technical assistance. We thank the Beckman Coulter Corporation for donation of the automated capillary sequencer and high-speed centrifuge used in these studies. We also thank H. J. Ball (Department of Agriculture for Northern Ireland, Stormont, Belfast, Northern Ireland) for the generous gift of the 5E5 antibody used in this study.
REFERENCES
- 1.Bascunana, C. R., J. G. Mattsson, G. Bolske, and K. E. Johansson. 1994. Characterization of the 16S rRNA genes from Mycoplasma sp. strain F38 and development of an identification system based on PCR. J. Bacteriol. 176:2577-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baxter, S. I., I. Pow, A. Bridgen, and H. W. Reid. 1993. PCR detection of the sheep-associated agent of malignant catarrhal fever. Arch. Virol. 132:145-159. [DOI] [PubMed] [Google Scholar]
- 3.Bergonier, D., X. Berthelot, and F. Poumarat. 1997. Contagious agalactia of small ruminants: current knowledge concerning epidemiology, diagnosis and control. Rev. Sci. Tech. 16:848-873. [DOI] [PubMed] [Google Scholar]
- 4.Black, S. R., I. K. Barker, K. G. Mehren, G. J. Crawshaw, S. Rosendal, L. Ruhnke, J. Thorsen, and P. S. Carman. 1988. An epizootic of Mycoplasma ovipneumoniae infection in captive Dall's sheep (Ovis dalli dalli). J. Wildl. Dis. 24:627-635. [DOI] [PubMed] [Google Scholar]
- 5.Brogden, K. A., D. Rose, R. C. Cutlip, H. D. Lehmkuhl, and J. G. Tully. 1988. Isolation and identification of mycoplasmas from the nasal cavity of sheep. Am. J. Vet. Res. 49:1669-1672. [PubMed] [Google Scholar]
- 6.DaMassa, A. J., P. S. Wakenell, and D. L. Brooks. 1992. Mycoplasmas of goats and sheep. J. Vet. Diagn. Investig. 4:101-113. [DOI] [PubMed] [Google Scholar]
- 7.Djordjevic, S. R., W. A. Forbes, J. Forbes-Faulkner, P. Kuhnert, S. Hum, M. A. Hornitzky, E. M. Vilei, and J. Frey. 2001. Genetic diversity among Mycoplasma species bovine group 7: clonal isolates from an outbreak of polyarthritis, mastitis, and abortion in dairy cattle. Electrophoresis 22:3551-3561. [DOI] [PubMed] [Google Scholar]
- 8.Frey, J. 2002. Mycoplasmas of animals. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 9.Giacometti, M., M. Janovsky, L. Belloy, and J. Frey. 2002. Infectious keratoconjunctivitis of ibex, chamois and other Caprinae. Rev. Sci. Tech. 21:335-345. [DOI] [PubMed] [Google Scholar]
- 10.Harasawa, R. 1999. Genetic relationships among mycoplasmas based on the 16S-23S rRNA spacer sequence. Microbiol. Immunol. 43:127-132. [DOI] [PubMed] [Google Scholar]
- 11.Harasawa, R., H. Hotzel, and K. Sachse. 2000. Comparison of the 16S-23S rRNA intergenic spacer regions among strains of the Mycoplasma mycoides cluster, and reassessment of the taxonomic position of Mycoplasma sp. bovine group 7. Int. J. Syst. Evol. Microbiol. 50:1325-1329. [DOI] [PubMed] [Google Scholar]
- 12.Heldtander, M., H. Wesonga, G. Bolske, B. Pettersson, and K. E. Johansson. 2001. Genetic diversity and evolution of Mycoplasma capricolum subsp. capripneumoniae strains from eastern Africa assessed by 16S rDNA sequence analysis. Vet. Microbiol. 78:13-28. [DOI] [PubMed] [Google Scholar]
- 13.Howard, C. J., and G. Taylor. 1985. Immune responses to mycoplasma infections of the respiratory tract. Vet. Immunol. Immunopathol. 10:3-32. [DOI] [PubMed] [Google Scholar]
- 14.Hung, A. L., A. Alvarado, T. Lopez, R. Perales, O. Li, and E. Garcia. 1991. Detection of antibodies to mycoplasmas in South American camelids. Res. Vet. Sci. 51:250-253. [DOI] [PubMed] [Google Scholar]
- 15.Karnovsky, M. J. 1965. A formaldehyde-glutaraldehyde fixative of high osmolarity for use in electron microscopy. J. Cell Biol. 27:137A. [Google Scholar]
- 16.Keel, M. K., J. G. Patterson, T. H. Noon, G. A. Bradley, and J. K. Collins. 2003. Caprine herpesvirus-2 in association with naturally occurring malignant catarrhal fever in captive sika deer (Cervus nippon). J. Vet. Diagn. Investig. 15:179-183. [DOI] [PubMed] [Google Scholar]
- 17.Kiss, I., K. Matiz, A. Allard, G. Wadell, and M. Benko. 1996. Detection of homologous DNA sequences in animal adenoviruses by polymerase chain reaction. Acta Vet. Hung. 44:243-251. [PubMed] [Google Scholar]
- 18.Kokotovic, B., N. F. Friis, and P. Ahrens. 2002. Characterization of Mycoplasma hyosynoviae strains by amplified fragment length polymorphism analysis, pulsed-field gel electrophoresis and 16S ribosomal DNA sequencing. J. Vet. Med. B Infect. Dis. Vet. Public Health 49:245-252. [DOI] [PubMed] [Google Scholar]
- 19.Kusiluka, L. J., B. Kokotovic, B. Ojeniyi, N. F. Friis, and P. Ahrens. 2000. Genetic variations among Mycoplasma bovis strains isolated from Danish cattle. FEMS Microbiol. Lett. 192:113-118. [DOI] [PubMed] [Google Scholar]
- 20.Lahijani, R. S., S. M. Sutton, R. B. Klieforth, M. F. Murphy, and W. P. Heuschele. 1994. Application of polymerase chain reaction to detect animals latently infected with agents of malignant catarrhal fever. J. Vet. Diagn. Investig. 6:403-409. [DOI] [PubMed] [Google Scholar]
- 21.Li, H., N. Dyer, J. Keller, and T. B. Crawford. 2000. Newly recognized herpesvirus causing malignant catarrhal fever in white-tailed deer (Odocoileus virginianus). J. Clin. Microbiol. 38:1313-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, H., J. Keller, D. P. Knowles, and T. B. Crawford. 2001. Recognition of another member of the malignant catarrhal fever virus group: an endemic gammaherpesvirus in domestic goats. J. Gen. Virol. 82:227-232. [DOI] [PubMed] [Google Scholar]
- 23.Lorenzon, S., H. Wesonga, L. Ygesu, T. Tekleghiorgis, Y. Maikano, M. Angaya, P. Hendrikx, and F. Thiaucourt. 2002. Genetic evolution of Mycoplasma capricolum subsp. capripneumoniae strains and molecular epidemiology of contagious caprine pleuropneumonia by sequencing of locus H2. Vet. Microbiol. 85:111-123. [DOI] [PubMed] [Google Scholar]
- 24.Maniloff, J., R. N. McElhaney, L. R. Finch, and J. B. Baseman (ed.). 1992. Mycoplasmas: molecular biology and pathogenesis, p. 561-573. American Society of Microbiology, Washington D.C.
- 25.Murphy, M. F., R. B. Klieforth, R. S. Lahijani, and W. P. Heuschele. 1994. Diagnosis of malignant catarrhal fever by polymerase chain reaction amplification of alcelaphine herpesvirus 1 sequence. J. Wildl. Dis. 30:377-382. [DOI] [PubMed] [Google Scholar]
- 26.O'Toole, D., H. Li, D. Miller, W. R. Williams, and T. B. Crawford. 1997. Chronic and recovered cases of sheep-associated malignant catarrhal fever in cattle. Vet. Rec. 140:519-524. [DOI] [PubMed] [Google Scholar]
- 27.Pettersson, B., G. Bolske, F. Thiaucourt, M. Uhlen, and K. E. Johansson. 1998. Molecular evolution of Mycoplasma capricolum subsp. capripneumoniae strains, based on polymorphisms in the 16S rRNA genes. J. Bacteriol. 180:2350-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pettersson, B., T. Leitner, M. Ronaghi, G. Bolske, M. Uhlen, and K. E. Johansson. 1996. Phylogeny of the Mycoplasma mycoides cluster as determined by sequence analysis of the 16S rRNA genes from the two rRNA operons. J. Bacteriol. 178:4131-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reynolds, E. S. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17:208-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez, F., H. J. Ball, D. Finlay, D. Campbell, and D. P. Mackie. 1996. Detection of Mycoplasma mycoides subspecies mycoides by monoclonal antibody-based sandwich ELISA. Vet. Microbiol. 51:69-76. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez, F., S. Kennedy, T. D. Bryson, A. Fernandez, J. L. Rodriguez, and H. J. Ball. 1996. An immunohistochemical method of detecting Mycoplasma species antigens by use of monoclonal antibodies on paraffin sections of pneumonic bovine and caprine lungs. Zentbl. Vetmed. B 43:429-438. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez, J. L., C. Gutierrez, D. L. Brooks, A. J. Damassa, J. Oros, and A. Fernandez. 1998. A pathological and immunohistochemical study of goat kids undergoing septicaemic disease caused by Mycoplasma capricolum subsp. capricolum, Mycoplasma mycoides subsp. capri and Mycoplasma mycoides subsp. mycoides (large colony type). Zentbl. Vet. B 45:141-149. [DOI] [PubMed] [Google Scholar]
- 33.Rovnak, J., S. L. Quackenbush, R. A. Reyes, J. D. Baines, C. R. Parrish, and J. W. Casey. 1998. Detection of a novel bovine lymphotrophic herpesvirus. J. Virol. 72:4237-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russell, L. D., and S. Burquet. 1977. Ultrastructure of Leydig cells as revealed by secondary tissue treatment with ferrocyanide:osmium mixture. Tissue Cell 9:751-766. [DOI] [PubMed] [Google Scholar]
- 35.Sanchez, S., K. Tyler, N. Rozengurt, and J. C. Lida. 1994. Comparison of a PCR-based diagnostic assay for Mycoplasma pulmonis with traditional detection techniques. Lab. Anim. 28:249-256. [DOI] [PubMed] [Google Scholar]
- 36.Schock, A., R. A. Collins, and H. W. Reid. 1998. Phenotype, growth regulation and cytokine transcription in ovine herpesvirus-2 (OHV-2)-infected bovine T-cell lines. Vet. Immunol. Immunopathol. 66:67-81. [DOI] [PubMed] [Google Scholar]
- 37.Simecka, J. W., J. K. Davis, M. K. Davidson, S. E. Ross, C. T. K.-H. Stadtlander, and G. H. Cassell. 1992. Mycoplasma diseases of animals. American Society for Microbiology, Washington, D.C.
- 38.Soroka, A. E., K. T. Momynaliev, A. M. Taraskina, A. M. Savicheva, and V. M. Govorun. 2001. Genetic heterogeneity of Mycoplasma hominis clinical isolates detected during observation of patients with recurrent urogenital inflammation. Bull. Exp. Biol. Med. 132:663-665. [DOI] [PubMed] [Google Scholar]
- 39.Spurr, A. R. 1969. A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 26:31-43. [DOI] [PubMed] [Google Scholar]
- 40.Swofford, D. L. 1998. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Sinauer, Sunderland, Mass.
- 41.Thiaucourt, F., S. Lorenzon, A. David, and A. Breard. 2000. Phylogeny of the Mycoplasma mycoides cluster as shown by sequencing of a putative membrane protein gene. Vet. Microbiol. 72:251-268. [DOI] [PubMed] [Google Scholar]
- 42.VanDevanter, D. R., P. Warrener, L. Bennett, E. R. Schultz, S. Coulter, R. L. Garber, and T. L. Rose. 1996. Detection and analysis of diverse herpesviral species by consensus primer PCR. J. Clin. Microbiol. 34:1666-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Villalobo, E., and A. Torres. 1998. PCR for detection of Shigella spp. in mayonnaise. Appl. Environ. Microbiol. 64:1242-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watson, G. L., and R. F. Slocombe. 1986. Mycoplasmosis in a Thomson's gazelle. Vet. Pathol. 23:329-331. [DOI] [PubMed] [Google Scholar]
- 45.Weisberg, W. G., J. G. Tully, D. L. Rose, J. P. Petzel, H. Oyaizu, and D. Yang. 1989. A phylogenetic analysis of the mycoplasmas: basis for their classification. J. Bacteriol. 171:6455-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wellehan, J. F., M. Calsamiglia, D. H. Ley, M. S. Zens, A. Amonsin, and V. Kapur. 2001. Mycoplasmosis in captive crows and robins from Minnesota. J. Wildl. Dis. 37:547-555. [DOI] [PubMed] [Google Scholar]
- 47.Westberg, J., A. Persson, A. Holmberg, A. Goesmann, J. Lundeberg, K. E. Johansson, B. Pettersson, and M. Uhlen. 2004. The genome sequence of Mycoplasma mycoides subsp. mycoides SC type strain PG1t, the causative agent of contagious bovine pleuropneumonia (CBPP). Genome Res. 14:221-227. [DOI] [PMC free article] [PubMed] [Google Scholar]