The emergence in recent times in the United Kingdom of extended-spectrum beta-lactamase (ESBL)-producing pathogenic enterobacteria poses a serious antibiotic management problem, as these genes are easily transferred from one organism to the other via plasmids. It is thus not surprising that these ESBL-containing organisms, which were hitherto found mainly in hospitals, are now becoming fairly common in community-acquired infections, especially those of the urinary tract.
This new development should alert the clinical microbiologist to devise ways and means of readily identifying these ESBL-producing organisms and instituting appropriate therapy. This is all the more important because resistance to one of the extended-spectrum cephalosporins (ceftazidime, cefotaxime, or ceftriaxone), when mediated by an ESBL, means therapeutic resistance to all even when sensitivity test results may indicate otherwise.
The Department of Microbiology of the Worcestershire Royal Hospital serves as the referral diagnostic center for the hospital, community hospitals, and General Practitioners' surgeries in the county of South Worcestershire, United Kingdom.
ESBLs are hydrolytic enzymes that mediate resistance to extended-spectrum cephalosporins, namely, ceftazidime, cefotaxime, and ceftriaxone. These enzymes are also active against monobactams such as aztreonam. However, the cephamycins—cefoxitin and cefotetan—are resistant to the hydrolytic effect of these enzymes, this stability being afforded by their methoxy group. The carbapenems—imipenem and meropenem—are also not affected.
Many ESBL-producing bacteria are also resistant to other antimicrobial agents, namely, aminoglycosides, trimethoprim, and the quinolones.
ESBLs are thought to have evolved by mutation of the TEM (named after the first patient from whom the pathogen was isolated) and SHV (sulfhydryl-variable) genes. These are members of the class A beta-lactamases, which are the commonest plasmid-mediated beta-lactamases found in gram-negative bacilli of the Enterobacteriaceae family. More than 100 different sequence variants of SHV and TEM genes with various levels of activity against ceftazidime, cefotaxime, and ceftriaxone have so far been demonstrated. However the most commonly encountered ones are TEM-3, which confers broad resistance to ceftazidime, cefotaxime, and ceftriaxone, and TEM-10, which confers high-level resistance to ceftazidime and appears to be sensitive in vitro to cefotaxime and ceftriaxone. Most ESBLs found in the United Kingdom are derived from the TEM and SHV genes.
In recent times, however, the CTX-M-derived ESBLs, which are the predominant types in China, Poland, Spain, and South America, have begun to appear in the United Kingdom (2). These usually confer resistance to cefotaxime but spare ceftazidime. These genes are thought to have evolved by mutation of the chromosomal beta-lactamase of Kluyvera spp.; the mutated forms have now escaped to plasmids and are being distributed among other enterobacteria.
In practical terms, detection of an ESBL in a clinically significant isolate, whether mediated by TEM, SHV, or CTX-M genes, should mean therapeutic resistance to all extended-spectrum cephalosporins (indeed, to all cephalosporins, aztreonam, and penicillins). The treatment of choice in such circumstances should be the use of the carbapenems or cephamycins, depending on the sensitivity result.
The aims and objectives of this study were to identify and characterize ESBLs in pathogenic enterobacteria, to determine what fraction of potential ESBL producers go on to be confirmed (and thus, whether it is worthwhile to report all potential ESBL producers as resistant to all cephalosporins), and to determine what fraction of the isolates are hospital or community acquired.
All clinically significant isolates of enterobacteria from clinical samples received at the microbiology department of the Worcestershire Royal Hospital in September 2003 were screened for evidence of potential extended-spectrum beta-lactamase production according to NCCLS (National Committee for Clinical Laboratory Standards) protocols. These included isolates from urine, blood, fluids (including cerebrospinal fluids), and swabs.
For isolates from blood, cerebrospinal fluids and other fluids, and swabs, sensitivity testing was done using the Stokes comparative disk diffusion method (this was the method of sensitivity testing used in the department at the time this study was done). The isolates were emulsified in saline to a 0.5 McFarland opacity standard and inoculated to the periphery of the test sensitivity agar plate (Biomerieux, Basingstoke, United Kingdom) by the use of a rotary plater. The control organism for the strain being tested was similarly treated and was inoculated to the center of the plate. Disks of ceftazidime (30 μg) and cefotaxime (Oxoid, Basingstoke, United Kingdom) (30 μg) were added to the routine panel of antibiotics for the organism after the plates were allowed to dry. The plates were incubated aerobically overnight. Isolates which showed evidence of resistance to ceftazidime or cefotaxime were retested using the standardized method as described by the NCCLS, and organisms with zones of inhibition less than 22 mm in diameter in response to the presence of ceftazidime or less than 27 mm in diameter in response to the presence of cefotaxime or both were presumed to be potential ESBL producers (Table 1).
TABLE 1.
Result of ESBL screening test (NCCLS)
| Lab | Organism | Cefotaxime zone diam (mm) | Ceftazidime zone diam (mm) |
|---|---|---|---|
| 535 | Enterobacter sakazaki | 6 | 6 |
| 892 | E. coli | 6 | 6 |
| 348 | K. pneumonia | 21 | 9 |
| 302 | E. coli | 6 | 8 |
| 605 | Enterobacter spp. | 6 | 6 |
| 001 | E. coli | 6 | 6 |
| 957 | E. coli | 12 | 10 |
| 376 | E. cloacae | 11 | 14 |
| 893 | E. aerogenes | 18 | 16 |
| 679 | Enterobacter spp. | 26 | 20 |
For urine isolates, the breakpoint sensitivity test method was employed. All isolates were emulsified in saline solution to a 0.5 McFarland opacity standard. Using the multipoint inoculators, approximately 1 to 2 μl of the broth was inoculated onto two test sensitivity agar plates (poured in house; materials supplied by Oxoid); one of the plates was impregnated with 2 mg of ceftazidime/liter, and the other was impregnated with 2 mg of cefotaxime/liter (this was the method of sensitivity testing of urine specimens used in the department at the time this study was done). For ease of interpretation of results, 2 mg of the antibiotics/liter was chosen for subsequent testing. The plates were incubated aerobically overnight, and isolates which grew on the antibiotic plates—i.e., on the ceftazidime plates or the cefotaxime plates or both—were presumed to be potential ESBL producers (Table 1).
The double-disk synergy method recommended by the British Society for Antimicrobial Chemotherapy (BSAC) was used to confirm ESBL production (3, 4).
Isolates presumed to be ESBL producers on the basis of screening test results as described above were picked up and emulsified in saline to a 0.5 McFarland turbidity standard. By the use of a cotton wool swab and the rotary platter, the broth was evenly spread on test sensitivity agar and allowed to dry. Disks of ceftazidime (30 μg), cefotaxime (30 μg), and co-amoxyclav (20 + 10 μg) were placed 2 cm apart on the plate in a straight line, with the co-amoxyclav disk in the middle. The plates were incubated aerobically overnight, and the results were read the following day. Isolates which showed an enlargement of the zone of inhibition greater than 5 mm on the co-amoxyclav side of the disk compared to the results seen on the side without co-amoxyclav were confirmed as ESBL producers (Table 2).
TABLE 2.
ESBL phenotype confirmatory test result (BSAC double-disc synergy method)a
| Lab or strain | Isolate | CTX zone diam (mm) | CTX + AMC zone diam (mm) | CAZ zone diam (mm) | CAZ + AMC zone diam (mm) |
|---|---|---|---|---|---|
| 892 | E. coli | 0 | 10 | 2 | 10 |
| 302 | E. coli | 0 | 10 | 2 | 10 |
| 957 | E. coli | 1 | 10 | 3 | 10 |
| 001 | E. coli | 0 | 8 | 3 | 10 |
| Positive control | UZA 1789 | 4 | 10 | 3 | 10 |
| Negative control | NCTC 10418 | 20 | 10 | 15 | 10 |
CTX, cefotaxime; CAZ, ceftazidime; AMC, amoxicillin-clavulanic acid.
This ESBL phenotypic confirmatory test method was subjected to quality control using an Escherichia coli strain (UZA 1789), a Pan-European Antimicrobial Resistance Surveillance System strain, as a positive control, and another E. coli strain (NCTC 10418) as a negative control.
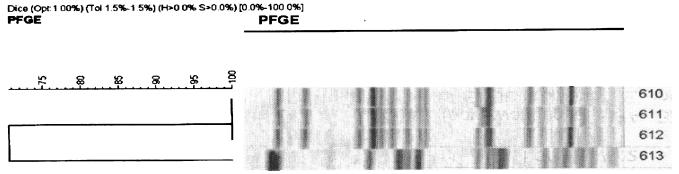
A total of 1,041 isolates of clinically significant enterobacteria were obtained during the study period. A total of 10 multiresistant organisms—5 Enterobacter species, 4 E. coli strains, and 1 Klebsiella pneumonia strains—were found. Of the five Enterobacter species, two were from anal swabs of hematology in-patients, two were from endotracheal secretions from the same patient in the neonatal unit, and one was from a urine specimen from a general practitioner. The five Enterobacter species were not considered any further, because this test method has not been validated for Enterobacter spp. and because cefepime and cefpirome—the antibiotic disks used in confirming the ESBL phenotype for Enterobacter spp.—were not routinely used in the laboratory. The only isolate of K. pneumonia (from a hip wound swab from an in-patient) gave mixed results in repeated subculturing and was not considered suitable for genotyping. The four pure isolates of E. coli (all from a urine specimen) were sent to the Antibiotic Resistance Monitoring and Reference Laboratory of the Health Protection Agency, Colindale, London, United Kingdom. All four isolates were found positive by PCR for the genes encoding the CTX-M enzyme. Further analysis by pulsed-field gel electrophoresis (PFGE) showed that these cefotaximases were clearly distinct from the five major CTX-M-15-producing E. coli strains previously described in the United Kingdom. The isolates showed >15% difference in their PFGE genotypes from the strains previously described. The two isolates (isolates 610 and 612) from the same patient had the same PFGE pattern (Fig. 1).
FIG. 1.
PFGE pattern of the isolates.
Of the four isolates of E. coli that were confirmed as harboring cefotaximases, two were from urine samples of patients obtained on admission at the Worcestershire Royal Hospital. The remaining two (isolates 610 and 612; Fig. 1) were from catheter specimens of urine samples taken 9 days apart from the same patient and sent from a general practitioner.
The use of both ceftazidime and cefotaxime in screening for ESBL in pathogenic enterobacteria as recommended by the NCCLS appears to be quite reliable, easy to perform, and cost effective. Previous studies using cefuroxime showed a sensitivity of 42.5% (6), and a similar study with ceftazidime alone was only able to detect ESBL in 73% of cases (5). The double-disk synergy method recommended by BSAC appears to be quite specific for phenotypic confirmation of ESBL production.
The need to screen isolates of pathogenic enterobacteria for evidence of ESBL production cannot be overemphasized. In district general hospitals, where technical resources and expertise may not be in abundant supply, routine testing of isolates of pathogenic enterobacteria for sensitivity to both ceftazidime and cefotaxime may represent a cost-effective, easy-to-perform, and reliable means of detecting ESBLs. The double-disk synergy method can easily be used to confirm the ESBL phenotype.
Acknowledgments
Special thanks to Jane Stockley (FRCPath) and Dr. Chris Catchpole (FRCPath), both consultant microbiologists at the Worcestershire Royal Hospital, for their support during the course of this study. Thanks also to all members of staff of the Medical Microbiology Department of Worcestershire Royal Hospital for graciously assisting me and supplying all the materials I needed during the course of this work.
REFERENCES
- 1.Division of Healthcare Quality Promotion, Centers for Disease Control and Prevention. 16. September 2004, access date. Laboratory detection of extended-spectrum β-lactamases. [Online.] www.cdc.gov/ncidod/hip/lab/factsheet/esbl.htm.
- 2.Health Protection Agency. 2003. Infection with organisms carrying extended-spectrum beta-lactamases in the community: First report. CDR Wkly. 13(32):3. [Google Scholar]
- 3.Livermore, D. 2002. Detection of β-lactamase mediated resistance, p. 1-18. In Standardized disc susceptibility method. User day report for November 2002 meeting held at Belfast City Hospital, Belfast. British Society for Antimicrobial Chemotherapy, Birmingham, United Kingdom.
- 4.Livermore, D. M., and D. F. J. Brown. 2001. Detection of β-lactamase-mediated resistance. J. Antimicrob. Chemother. 48(Suppl. 1):59-64. [DOI] [PubMed] [Google Scholar]
- 5.MacKenzie, F. M., C. A. Miller, and I. M. Gould. 2002. Comparison of screening methods for TEM and SHV derived ESBL production. Clin. Microbiol. Infect. 8:715-724. [DOI] [PubMed] [Google Scholar]
- 6.Navon-Venezia, S., O. Hammer-Munz, D. Schwartz, D. Turner, B. Kuzmenko, and Y. Carmeli. 2003. Occurrence and phenotypic characteristics of extended-spectrum β-lactamases among members of the family Enterobacteriaceae at the Tel-Aviv Medical Centre (Israel) and evaluation of diagnostic tests. J. Clin. Microbiol. 41:155-158. [DOI] [PMC free article] [PubMed] [Google Scholar]