Abstract
Cryptosporidiosis is an emerging protozoan disease associated with large waterborne outbreaks. Diagnosis relies on microscopic examination of stools, but this method cannot identify the infecting species of Cryptosporidium. We have developed a test based on nested PCR and restriction fragment length polymorphism (RFLP) that offers simple identification of Cryptosporidium hominis, Cryptosporidium parvum, and most other human infective species in stool samples. Purified C. parvum oocysts were used for PCR development. Extracted DNA was amplified by nested PCR targeting a 214-bp fragment of the 18S RNA gene. Enzymatic restriction sites were identified by bioinformatic analysis of all published Cryptosporidium 18S rRNA sequences. Experiments with spiked stool samples gave an estimated PCR detection limit of one oocyst. Specificity was assessed by testing 68 stool samples from patients with microscopically proven cryptosporidiosis and 31 Cryptosporidium-negative stools. Sixty-seven (98.5%) of the 68 stool samples from patients with microscopically proven cryptosporidiosis and 2 of the other stool samples were positive by PCR and could be genotyped. RFLP analysis identified 36 C. hominis, 19 C. parvum, 8 Cryptosporidium meleagridis, and 6 Cryptosporidium felis or Cryptosporidium canis samples. Species determination in 26 PCR-positive cases was in full agreement with DNA sequencing of the 18S rRNA hypervariable region. The excellent sensitivity of PCR, coupled with the accuracy of RFLP for species identification, make this method a suitable tool for routine diagnosis and genotyping of Cryptosporidium in stools.
Several Cryptosporidium species can cause severe acute diarrhea in humans and animals (5, 6, 13). Human cryptosporidiosis is usually self-resolving within a few days, but immunocompromised patients can develop life-threatening complications. Outbreaks due to drinking water contamination continue to occur (7, 10, 18, 32), and there is no effective treatment, making cryptosporidiosis a major public health issue and economic problem. Diagnosis is generally based on microscopic detection of oocysts in stools, but this offers no information on the infecting species and is not suited to epidemiological investigations.
The following 13 Cryptosporidium species are currently accepted, on the basis of host specificity, pathogenesis, morphology (9) and genotyping (8, 16, 21): Cryptosporidium hominis, Cryptosporidium parvum, Cryptosporidium wrairi, Cryptosporidium felis, Cryptosporidium canis, Cryptosporidium andersoni, and Cryptosporidium muris as infecting mammals; Cryptosporidium baileyi, Cryptosporidium meleagridis, and Cryptosporidium galli as infecting birds; Cryptosporidium serpentis and Cryptosporidium saurophilum as infecting reptiles; and Cryptosporidium molnari as infecting fish (36). Recent phylogenetic analyses based on sequencing of the small subunit rRNA gene (18S rRNA) (21, 22, 24, 33, 34), the hsp 70 gene (31), or other housekeeping or structural genes (24, 28-30) show a complex multispecies organization of the genus Cryptosporidium. The C. parvum complex includes subspecies that specifically infect cattle (former genotype 2), pigs, kangaroos, ferrets, or monkeys (23). The specific host of C. felis is the cat, but this species has also been isolated from a cow (2), while C. andersoni is morphologically close to C. muris but infects cattle rather than mice (17).
C. hominis and C. parvum are the main species causing cryptosporidiosis in humans, but other species, such as C. felis, C. meleagridis, C. muris, and C. canis, occasionally cause human diarrhea (11, 15, 19, 20, 25-27). As Cryptosporidium species identification is restricted to specialized laboratories, the role of species other than C. parvum species is probably underestimated.
The aim of this study was to develop a sensitive and specific method for the detection and species identification (genotyping) of Cryptosporidium oocysts in biological samples. Based on nested PCR of the 18S rRNA gene, combined with restriction fragment length polymorphism (RFLP) analysis, we propose a simple algorithm for species and subspecies identification of Cryptosporidium.
MATERIALS AND METHODS
Parasites.
Fresh C. parvum oocysts obtained from feces of a naturally infected calf were used to set up the PCR and RFLP protocols. Oocysts were concentrated from feces by using a one-step ethyl ether method. Briefly, 10 ml of dichromate feces suspension was vigorously mixed with 3 ml of diethyl ether and then centrifuged at 500 × g for 5 min at 4°C. The pellet was washed three times in phosphate-buffered saline (PBS) solution, pH 7.2, by centrifugation at 1,500 × g for 10 min at 4°C. The pellet was resuspended in 1 ml of PBS and then deposited on a two-layer saccharose gradient (dilution, 1.05/1.1) and centrifuged at 1,200 × g for 25 min at 4°C. Purified oocysts were collected from the interface between the sucrose layers, washed twice in PBS, and kept at +4°C until use.
Clinical specimens.
We studied 93 human fecal samples collected at our laboratory hospital and 6 samples from Dijon Hospital, France (Table 1). Sixty-eight samples were from patients with microscopically proven cryptosporidiosis, 21 samples were from patients with other intestinal protozoal or helminth infections (Giardia, 11 patients; microsporidia, 1 patient; intestinal amoeba, 8 patients; and Ancylostoma, 1 patient), and 10 samples were from patients with no diagnosed intestinal parasitosis.
TABLE 1.
Results of PCR-RFLP analysis of 99 stool samples
| Patient no. | Clinical statusc | Microscopic examination | PCR-RFLP result | Sequencing result |
|---|---|---|---|---|
| 1-16 | HIV | + | C. hominis | ND |
| 17-25a | HIV | + | C. hominis | C. hominis |
| 26-38 | HIV | + | C. parvum | ND |
| 39-41a | HIV | + | C. parvum | C. parvum |
| 42-46 | HIV | + | C. meleagridis | C. meleagridis |
| 47a | HIV | + | C. meleagridis | C. meleagridis |
| 48-49 | HIV | + | C. felis | C. felis |
| 50-52a | HIV | + | C. felis | C. felis |
| 53a | HM | + | C. hominis | C. hominis |
| 54 | HM | + | C. parvum | ND |
| 55-56a | HM | + | C. parvum | C. parvum |
| 57a | HM | + | C. felis | C. felis |
| 58 | HM | + | ||
| 59-62 | IC | + | C. hominis | ND |
| 63-68b | IC | + | C. hominis | C. hominis |
| 69-70 | IC | Other protozoa | C. meleagridis | C. meleagridis |
| 71-89 | ND | Other protozoa | ||
| 90-99 | ND | No parasites |
Cryptosporidiosis was diagnosed by microscopic demonstration of typical Cryptosporidium oocysts on a Ziehl-Neelsen-stained fecal smear after concentration by the one-step ethyl ether method. Selected stools were diluted in 2.5% dichromate solution or distilled water and kept at 4°C.
In 26 of the patients with Cryptosporidium infection (patients 17 to 25, 39 to 41, 47, 50 to 53, 55 to 57, and 63 to 68) (Table 1), the species and genotype had previously been determined by Guyot et al. (patients 17 to 25, 39 to 41, 47, 50 to 53, and 55 to 57) (12) and Dalle et al. (patients 63 to 68) (7), based on sequencing of the 18S rRNA gene hypervariable region (11).
PCR-RFLP experiments were done blindly to the results of microscopic examination and genotyping.
DNA extraction and purification.
DNA was harvested from purified oocysts or from fecal material by using alkaline lysis buffer containing 1 N NaOH, 0.5% sodium dodecyl sulfate, 25% Chelex-100 (Sigma, Saint-Quentin-Fallavier, France) and 1% polyvinylpyrrolidone-360K (PVP) (Sigma). At this step of oocyst lysis, we checked that the addition of PVP was as efficient as the use of Inhibitex tablets (QIAGEN, Courtaboeuf, France) for removing PCR inhibitors (data not shown).
In brief, 250 μl of oocyst concentrate or feces was mixed with 250 μl of lysis buffer and then incubated for 15 min at 70°C with vortexing at 900 rpm in a rotating heating block (Thermo-mixer; Eppendorf, Le Pecq, France). Then, 250 μl of 4 M NaH2PO4 was added. Chelex-100 was removed by centrifugation through a QIAshredder column (QIAGEN). DNA was recovered on a silica-activated column (QIAamp DNA Stool; QIAGEN) as recommended by Baeumner et al. (1). Eluted DNA was used immediately for PCR or kept at −20°C.
Bioinformatics.
Preliminary comparative analysis of the TRAP-C1, COWP, HSP70, and 18S rRNA gene sequences resulted in the selection of the Cryptosporidium 18S rRNA gene as the target for PCR and RFLP. For each Cryptosporidium species, we aligned all available sequences of the 18S rRNA gene by using ClustalW software and thereby identified species-specific consensus sequences. These sequences were aligned, and we then looked for specific restriction sites by using software available from http://www.infobiogen.fr. Primers were designed manually. Primer3 software was used as a decision tool. The HCNV4 human type sequence (accession number AF093489) was used as a reference for primer location.
PCR and RFLP.
All amplifications were performed by using a Gene TC Digital Thermal Cycler (Techne, Ozyme, Saint-Quentin en Yvelines, France). The primer pairs were designed to encompass the two first polymorphous regions located between nucleotides 179 and 271 of the 18S rRNA gene (see Results). Initial PCR amplification was performed in a 25-μl volume containing 5 μl of DNA template, 75 mM Tris-HCl, 20 mM (NH4)2SO4, 0.01% Tween 20, 2.5 mM MgCl2, a 0.15 mM concentration of each deoxynucleoside triphosphate (Amersham, Orsay, France), a 0.2 μM concentration of each primer, and 1.25 U of Taq DNA polymerase (Eurogentec, Seraing, Belgium). The second-round PCR mix was the same as the initial mix except that the final volume was 50 μl and the concentration of each primer was 0.4 μM. Five microliters of the initial amplification product was used as a template. The following PCR parameters were optimized (see Results): the MgCl2 concentration, the annealing temperature, and the annealing and elongation times. Each set of experiments included a positive PCR control consisting of 1 ng of specific DNA template and a negative PCR control (laboratory-grade distilled water). PCR specificity was checked by amplifying 2 ng of human, Giardia lamblia, Encephalitozoon intestinalis, Toxoplasma gondii, Trichophyton rubrum, and Escherichia coli DNA.
Aliquots of amplified fragments were visualized under UV light after 2% agarose gel electrophoresis and ethidium bromide staining.
Restriction assays were performed in a 30-μl volume with 2 units of restriction enzyme and 5 μl of PCR product per reaction. Mixes were incubated in a heating block. Digestion products were visualized under UV light after 2% agarose gel electrophoresis and ethidium bromide staining. The enzymes used were TaqI (Roche-Bohringer, Mannheim, Germany), AseI, MseI, BstUI, and SspI (New England Biolabs, Beverly, Mass.)
Cloning and sequencing.
In order to prepare reproducible PCR and RFLP controls, second-round PCR products of C. hominis, C. parvum, C. meleagridis, and C. felis were cloned into the pDrive vector (QIAGEN) as recommended by the supplier. EZ competent cells (QIAGEN) were then chemically transformed. Clones were selected on Luria-Bertani agar supplemented with 100 μg of ampicillin per ml and cultured overnight in Luria-Bertani broth supplemented with 100 μg of ampicillin per ml. The two strands of amplified DNA were sequenced by using the M13 Fwd and Rev primers and a Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems) and were analyzed in an ABI Prism 377XL sequencer (Applied Biosystems). Each insert was sequenced by the Centre d'Etude du Polymorphisme Humain (Paris, France) with the ABI Prism kit. Each sequence was compared with GenBank sequences by using the basic local alignment search tool (BLAST) algorithm. Plasmids were further used as templates to generate species-specific restriction controls.
Sequencing of 18S rRNA gene polymorphisms was also performed when rare Cryptosporidium species were found in human samples.
Sensitivity experiments.
Two sets of experiments were performed to assess the performance of our nested PCR method. We first examined sensitivity on serial dilutions of purified Cryptosporidium DNA. DNA was extracted from 108 purified C. parvum oocysts and then quantified by UV absorption at 260 nm. Serial 10-fold dilutions from 5 to 5 × 10−8 ng were prepared in distilled water and used as templates in the PCR mixture.
Second, suspensions of purified oocysts were prepared in distilled water or used to spike a stool suspension at concentrations of 100,000, 40,000, 4,000, 2,000, 800, 400, 200, 40, and 0 oocysts per ml. DNA extraction and purification were carried out as described above. Five microliters of each DNA stock preparation was used in PCR runs in order to obtain the equivalent of 2,500, 1,000, 100, 50, 20, 10, 5, 1, and 0 oocyst equivalents per PCR mixture.
RESULTS
Bioinformatics.
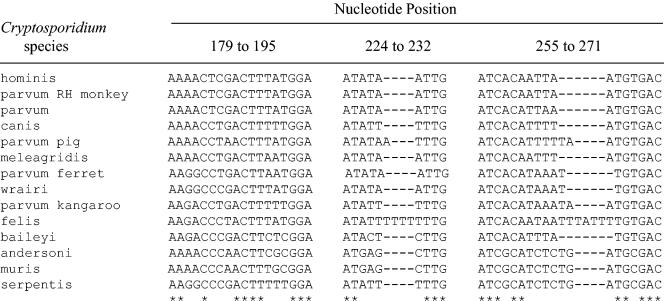
Compared to other Cryptosporidium gene sequences, such as those of TRAP-C1, COWP, and HSP70, the 18S rRNA gene is the most widely characterized and described sequence in known species of Cryptosporidium, with numerous available complete sequences. We thus collected all available sequences, clustered them according to the species, and aligned the species-clustered sequences. Apart from the hypervariable region, very low intragenic sequence variations were found within each species cluster. It was thus possible from the complete sequences to determine species consensus sequences. From the alignment of these sequences, we identified the previously described polymorphic regions (33), i.e., the hypervariable region encompassing nucleotides 615 to 850 and three polymorphous regions located between nucleotides 179 and 271.
We found that the region between nucleotides 179 and 271 was relevant and as informative as the hypervariable region, as each species consensus sequences was original (Fig. 1). Also, compared to the hypervariable region, where polymorphisms mainly involve poly(dA) and poly(dT) stretches, the region between nucleotides 179 and 271 has a higher GC percentage, and its polymorphisms were restricted to nucleotide mutations and to a few insertions or deletions (Fig. 2).
FIG. 1.
Alignments of the species-specific consensus polymorphous region. Except for C. parvum rhesus (RH) monkey, which is identical to C. hominis, the other species-specific sequences are original. As asterisk (*) indicates that the nucleotides in the corresponding column are identical in all sequences in the alignment, whereas a minus sign (−) indicates gaps.
FIG. 2.
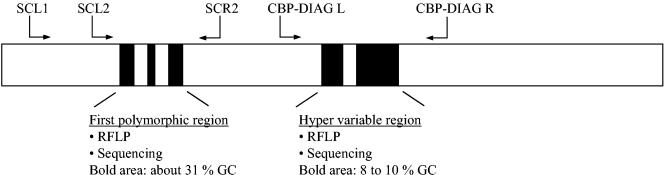
Schematic representation of the location of the polymorphous regions and the relative positions of the primers in the 18S rRNA gene. Black boxes correspond to the polymorphous regions used for genotyping. SCL2 and SCR2 amplify a 214-bp fragment containing the polymorphous region between nucleotides 179 and 271. CBP-DIAG L and CBP-DIAG R (described by Jonhson et al. [14]) amplify a 435-bp fragment containing the hypervariable region. SCL1 and CBP-DIAG R are used for the first round of nested PCR.
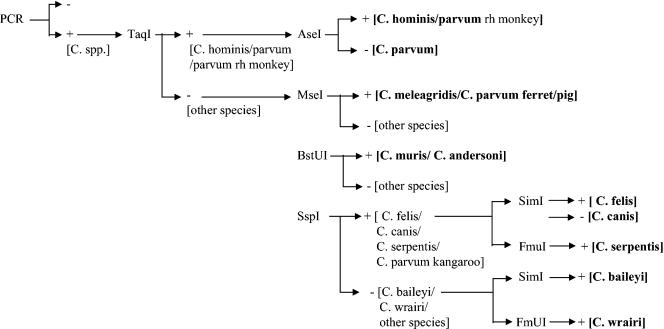
Specific enzymatic restriction sites for each species consensus sequence were sought. We found that sequential use of the restriction enzymes TaqI, AseI, MseI, BstUI, and SspI allowed us to discriminate C. hominis from C. parvum and to identify four clusters of other Cryptosporidium species (Fig. 3). As shown in the decision tree, RFLP analysis discriminated among the following species: C. hominis/C. parvum genotype rhesus monkey, C. parvum, C. meleagridis/C. parvum genotype ferret and pig, C. andersoni/C. muris, C. felis/C. canis/C. serpentis, and C. baileyi/C. wrairi. The restriction enzymes SimI and FmuI are not yet commercially available, but their use should permit specific identification of C. felis, C. canis, C. serpentis, C. baileyi, and C. wrairi.
FIG. 3.
Schematic representation of the sequential (or parallel) use of restriction enzymes for species and cluster identification.
Primer selection and PCR conditions.
When designing the primers for our nested PCR, we found few Cryptosporidium species-specific regions. The selected primer pairs encompass the polymorphous region of the 18S rRNA gene located between nucleotides 179 and 271. Initial amplification is performed with forward primer SCL1 (5′-CTGGTTGATCCTGCCAGTAG-3′), corresponding to nucleotides 4 to 23, and reverse primer CPB-DIAGR (5′-TAAGGTGCTGAAGGAGTAAGG-3′) (described in reference 14), corresponding to nucleotides 1016 to 1036. The optimized initial amplification conditions were as follows: 5 min at 94°C, initial denaturation for 30 s at 94°C, and 39 cycles of amplification (annealing for 45 s at 60°C, extension for 90 s at 72°C, and denaturation for 30 s at 94°C). The final extension step lasts 10 min, and the samples are then cooled to 4°C. The second-round PCR amplifies a 214-bp fragment. The forward primer (5′-CAGTTATAGTTTACTTGATAATC-3′; SCL2) corresponds to nucleotides 106 to 128, and the reverse primer (5′-CAATACCCTACCGTCTAAAG-3′; SCR2) corresponds to nucleotides 299 to 318. The optimized conditions for second-round PCR were the same as those for the first round, except that final volume was 50 μl, the primer concentrations were 0.4 μM, 5 μl of the initial amplification product was used as the template, and annealing lasted 45 s at 58°C and extension 60 s at 72°C.
Specificity and sensitivity.
The specificities of each primer were evaluated by amplifying human, G. lamblia, Encephalitozoon intestinalis, Toxoplasma gondii, Trichophyton rubrum, and E. coli DNA. No cross-amplification was detected with these DNAs or with DNA extracted from stools containing either no parasites or parasites other than Cryptosporidium.
PCR sensitivity was first tested on serial 10-fold dilutions of purified DNA. The detection limit was 5 × 10−5 ng of pure Cryptosporidium DNA per reaction mix, which corresponds approximately to one oocyst.
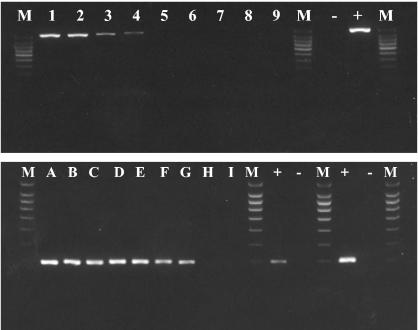
Sensitivity was then tested with fresh oocyst dilutions in water and human stools. Under both conditions the nested PCR procedure was able to detect the equivalent of one oocyst. According to the dilution and stool sample size used for PCR, 40 oocysts/ml of stools could be detected (Fig. 4).
FIG. 4.
Amplification by nested PCR of Cryptosporidium DNA extracted from an experimentally scale-charged Cryptosporidium-negative stool sample. The upper gel shows the amplification of a specific 1,032-bp fragment with primers SCL1 and CBP-DIAG R. The lower gel shows the amplification of a 214-bp species-specific fragment with primers SCL2 and SCR2. Lanes 1 and A, 2,500 oocysts; lanes 2 and B, 1,000 oocysts; lanes 3 and C, 100 oocysts; lanes 4 and D, 50 oocysts; lanes 5 and E, 20 oocysts; lanes 6 and F, 10 oocysts; lanes 7 and G, 5 oocysts; lanes 8 and H, 1 oocyst; lanes 9 and I, 0 oocyst; lane M, 100-bp molecular weight ladder. +, positive control; −, negative control. Detection of 1 oocyst per PCR mixture is equivalent to 40 oocysts in 1 ml of stool suspension.
RFLP and molecular characterization of Cryptosporidium in clinical samples.
The capacity for species discrimination and identification was first tested with plasmids containing C. parvum, C. hominis, C. meleagridis, and C. felis DNA fragments amplified with primers L2 and R2. In each case the restriction patterns were in total agreement with the bioinformatic analysis (Fig. 5). Oocysts or plasmids were not available for other species.
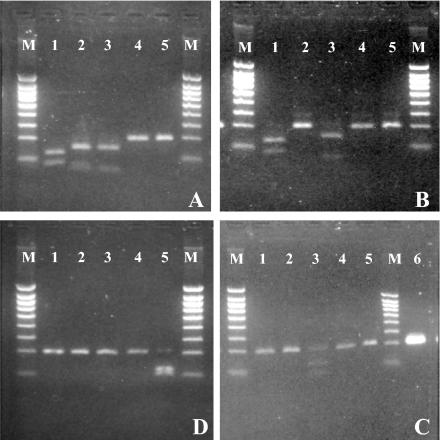
FIG. 5.
Comparative restriction patterns of C. hominis (A), C. parvum (B), C. felis (C), and C. meleagridis (D) obtained with the following enzymes: TaqI (lanes 1), AseI (lanes 2), MseI (lanes 3), BstUI (lanes 4), and SspI (lanes 5). Lanes 6, undigested C. hominis fragment; lanes M, 100-bp molecular weight ladder.
In order to further validate our RFLP typing method, we analyzed 26 human isolates of Cryptosporidium that had previously been genotyped (7, 12). When tested blindly to the results of previous DNA sequencing, RFLP analysis yielded typical restriction patterns for C. hominis in 16 cases (patients 17 to 25, 53, and 63 to 68) (Table 1), C. parvum in 5 cases (patients 39 to 41 and 55 to 56), C. meleagridis in 1 case (patient 47) and C. felis in 4 cases (patients 50 to 52 and 57). These results were in total agreement with those previously reported.
We then blind tested stool samples from 42 other patients with proven cryptosporidiosis (patients 1 to 16, 26 to 39, 43 to 46, 48 to 49, 54, and 57 to 62) (Table 1) and 31 patients with other parasitic diseases or with parasite-negative stool examination (patients 69 to 99). PCR-RFLP analysis yielded 44 positive samples, with the following species distribution: 21 C. hominis positives (47.7%), 15 C. parvum positives (34.1%), 6 C. meleagridis positives (13.6%), and 2 C. felis positives (4.5%). The identification of all C. felis and C. meleagridis samples was confirmed by sequencing the polymorphous regions between nucleotides 179 and 271 and the hypervariable region. One sample was PCR negative even though microscopy showed rare oocysts. This sample was found to contain a potent PCR inhibitor that could not be removed by PVP or Inhibitex tablets.
Twenty-nine of the 31 samples from Cryptosporidium-negative patients were PCR-negative. In two patients whose samples were microscopically negative and PCR positive, RFLP identified C. meleagridis. One patient presented an unexplained diarrhea. Species identification was confirmed by sequencing of the polymorphous regions between nucleotides 179 and 271 and the hypervariable region.
DISCUSSION
We propose a new simple, robust, and low-cost nested PCR-RFLP method for the detection and species identification of Cryptosporidium, with potential applications in human diagnostics and environmental surveillance.
Our method meets routine laboratory requirements for good sensitivity and for genotyping. The equivalent of a single oocyst can be detected in water or stools, and most Cryptosporidium species, including C. hominis and C. parvum, can be differentiated. This excellent detection limit is likely due to the use of nested PCR and to inclusion of PVP in the lysis buffer to remove PCR inhibitors.
We found that the 18S rRNA gene sequence was the most informative and practical. This gene is well known, and a large number of full sequences are available for most Cryptosporidium species. The gene contains four polymorphous regions of potential interest (33). We found that the region between nucleotides 179 and 271 was species- and subspecies-specific, with appropriate restriction sites. We were thus able to define a combination of restriction sites permitting the identification of most Cryptosporidium species with a small number of enzymes. For instance, the use of TaqI in combination with AseI identifies and separates C. parvum and C. hominis, the species most often responsible for human infection. This simple approach can be extended to other species, as shown by the decision tree in Fig. 3.
Our RFLP method did not clearly separate C. muris from C. andersoni or C. meleagridis from C. parvum genotype ferret and pig, although their respective 18S rRNA gene sequences showed certain specificities. Also, C. parvum rhesus monkey genotype (CPRM1; accession no. AF112569) could not be distinguished from C. hominis (HCNV4) as their sequences between primers L2 and R2 are identical.
Although this technique has some limitations, it does allow the identification of some rare species and has direct practical applications for the detection and characterization of Cryptosporidium species which account for at least 95% of infections in humans and farm animals. Also, RFLP yields restriction patterns that are more readily differentiated than those obtained with other targets in the 18S rRNA gene (34).
To our knowledge, this is the first time that the region between nucleotides 179 and 271 of the 18S rRNA gene has been used as the target for a genotyping and subgenotyping application. Blind analysis of 26 fecal samples that had previously been genotyped by DNA sequencing (7, 12) confirmed the reliability of our method. This result, and the use of bioinformatic-based sequence analysis, support the validity of this region, which appears to be less subject to sequence variations than other gene sequences used for genotyping (3, 13, 35).
When applied to routine clinical samples, PCR-RFLP identified Cryptosporidium species in 67 of 68 samples from patients with proven cryptosporidiosis and also in two samples from patients in whom oocysts had not been detected microscopically. Given the specificity of the PCR-RFLP method and confirmation by sequencing, we believe these results were not PCR false positives but, rather, reflect the higher sensitivity of PCR for low-level infection. Our genotyping study confirmed that C. hominis and C. parvum are the predominant species in both immunocompetent and immunocompromised patients; infection with other species was found in 18.5% (13 of 70) of cases of PCR-positive cryptosporidiosis and was not restricted to immunocompromised patients (4).
In conclusion, the proposed method is sensitive, thorough, and reliable, and the binary expression of the results avoids the risk of misinterpretation. It should find applications in routine clinical diagnosis and epidemiological investigations.
Acknowledgments
Stephane Coupe was supported in part by the Agence Nationale pour la Recherche Technologique (ANRT).
The authors thank C. Chartier (AFSSA, Niort, France) for providing calf oocysts of C. parvum, K. Guyot (Ecologie du Parasitisme, IFR 17, Institut Pasteur de Lille, Lille, France) for providing C. felis and C. meleagridis oocysts, F. Dalle (Laboratoire de Parasitologie Mycologie et Laboratoire de Microbiologie Medicale et Moleculaire [EA 562], CHU et Faculte de Medecine, Dijon, France) for providing DNA of Cryptosporidium from patients' stools from the Dracy-le-Fort Cryptosporidium epidemic, H. Bui (Centre d'Etude du Polymorphisme Humain, Paris, France) for sequence determinations, and David Young for editorial assistance.
REFERENCES
- 1.Baeumner, A. J., M. C. Humiston, R. A. Montagna, and R. A. Durst. 2001. Detection of viable oocysts of Cryptosporidium parvum following nucleic acid sequence based amplification. Anal. Chem. 73:1176-1180. [DOI] [PubMed] [Google Scholar]
- 2.Bornay-Llinares, F. J., A. J. da Silva, I. N. Moura, P. Myjak, and H. Pietkiewicz, W. Kruminis-Lozowska, T. K. Graczyk, N. J. Pieniazek. 1999. Identification of Cryptosporidium felis in a cow by morphologic and molecular methods. Appl. Environ. Microbiol. 65:1455-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caccio, S., W. Homan, R. Camilli, G. Traldi, T. Kortbeek, and E. Pozio. 2000. A microsatellite marker reveals population heterogeneity within human and animal genotypes of Cryptosporidium parvum. Parasitology 120:237-244. [DOI] [PubMed] [Google Scholar]
- 4.Chalmers, R. M., K. Elwin, A. L. Thomas, and D. H. Joynson. 2002. Infection with unusual types of Cryptosporidium is not restricted to immunocompromised patients. J. Infect. Dis. 185:270-271. [DOI] [PubMed] [Google Scholar]
- 5.Cordell, R. L., P. M. Thor, D. G. Addiss, J. Theurer, R. Lichterman, S. R. Ziliak, D. D. Juranek, and J. P. Davis. 1997. Impact of a massive waterborne cryptosporidiosis outbreak on child care facilities in metropolitan Milwaukee, Wisconsin. Pediatr. Infect. Dis. J. 16:639-644. [DOI] [PubMed] [Google Scholar]
- 6.Corso, P. S., M. H. Kramer, K. A. Blair, D. G. Addiss, J. P. Davis, and A. C. Haddix. 2003. Cost of illness in the 1993 waterborne Cryptosporidium outbreak, Milwaukee, Wisconsin. Emerg. Infect. Dis. 9:426-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalle, F., P. Roz, G. Dautin, M. Di-Palma, E. Kohli, C. Sire-Bidault, M. G. Fleischmann, A. Gallay, S. Carbonel, F. Bon, C. Tillier, P. Beaudeau, and A. Bonnin. 2003. Molecular characterization of isolates of waterborne Cryptosporidium spp. collected during an outbreak of gastroenteritis in South Burgundy, France. J. Clin. Microbiol. 41:2690-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fall, A., R. C. Thompson, R. P. Hobbs, and U. Morgan-Ryan. 2003. Morphology is not a reliable tool for delineating species within Cryptosporidium. J. Parasitol. 89:399-402. [DOI] [PubMed] [Google Scholar]
- 9.Fayer R, C. A. Speer, and J. P. Dubey. 1997. The general biology of Cryptosporidium, p. 1-41. In R. E. Fayer (ed.), Cryptosporidium and cryptosporidiosis. CRC Press, Inc., Boca Raton, Fla.
- 10.Furtado, C., G. K. Adak, J. M. Stuart, P. G. Wall, H. S. Evans, and D. P. Casemore. 1998. Outbreaks of waterborne infectious intestinal disease in England and Wales, 1992-5. Epidemiol. Infect. 121:109-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gatei, W., J. Greensill, R. W. Ashford, L. E. Cuevas, C. M. Parry, N. A. Cunliffe, N. J. Beeching, and C. A. Hart. 2003. Molecular analysis of the 18S rRNA gene of Cryptosporidium parasites from patients with or without human immunodeficiency virus infections living in Kenya, Malawi, Brazil, the United Kingdom, and Vietnam. J. Clin. Microbiol. 41:1458-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guyot, K., A. Follet-Dumoulin, E. Lelievre, C. Sarfati, M. Rabodonirina, G. Nevez, J. C. Cailliez, D. Camus, and E. Dei-Cas. 2001. Molecular characterization of Cryptosporidium isolates obtained from humans in France. J. Clin. Microbiol. 39:3472-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoxie, N. J., J. P. Davis, J. M. Vergeront, R. D. Nashold, and K. A. Blair. 1997. Cryptosporidiosis-associated mortality following a massive waterborne outbreak in Milwaukee, Wisconsin. Am. J. Public Health 87:2032-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, D. W., N. J. Pieniazek, D. W. Griffin, L. Misener, and J. B. Rose. 1995. Development of a PCR protocol for sensitive detection of Cryptosporidium oocysts in water samples. Appl. Environ. Microbiol. 61:3849-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katsumata, T., D. Hosea, I. G. Ranuh, S. Uga, T. Yanagi, and S. Kohno. 2000. Short report: possible Cryptosporidium muris infection in humans. Am. J. Trop. Med. Hyg. 62:70-72. [DOI] [PubMed] [Google Scholar]
- 16.Laxer, M. A., B. K. Timblin, and R. J. Patel. 1991. DNA sequences for the specific detection of Cryptosporidium parvum by the polymerase chain reaction. Am. J. Trop. Med. Hyg. 45:688-694. [DOI] [PubMed] [Google Scholar]
- 17.Lindsay, D. S., S. J. Upton, D. S. Owens, U. M. Morgan, J. R. Mead, and B. L. Blagburn. 2000. Cryptosporidium andersoni n. sp. (Apicomplexa: Cryptosporiidae) from cattle, Bos taurus. J. Eukaryot. Microbiol. 47:91-95. [DOI] [PubMed] [Google Scholar]
- 18.MacKenzie, W. R., N. J. Hoxie, M. E. Proctor, M. S. Gradus, K. A. Blair, D. E. Peterson, J. J. Kazmierczak, D. G. Addiss, K. R. Fox, J. B. Rose, and J. P. Davis. 1994. A massive outbreak in Milwaukee of Cryptosporidium infection transmitted through the public water supply. N. Engl. J. Med. 331:161-167. [DOI] [PubMed] [Google Scholar]
- 19.Morgan, U., R. Weber, L. Xiao, I. Sulaiman, R. C. Thompson, W. Ndiritu, A. Lal, A. Moore, and P. Deplazes. 2000. Molecular characterization of Cryptosporidium isolates obtained from human immunodeficiency virus-infected individuals living in Switzerland, Kenya, and the United States. J. Clin. Microbiol. 38:1180-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morgan, U., L. Xiao, I. Sulaiman, R. Weber, A. A. Lal, R. C. Thompson, and P. Deplazes. 1999. Which genotypes/species of Cryptosporidium are humans susceptible to? J. Eukaryot. Microbiol. 46:42S-43S. [PubMed] [Google Scholar]
- 21.Morgan, U. M., C. C. Constantine, D. A. Forbes, and R. C. Thompson. 1997. Differentiation between human and animal isolates of Cryptosporidium parvum using rDNA sequencing and direct PCR analysis. J. Parasitol. 83:825-830. [PubMed] [Google Scholar]
- 22.Morgan, U. M., P. T. Monis, R. Fayer, P. Deplazes, and R. C. Thompson. 1999. Phylogenetic relationships among isolates of Cryptosporidium: evidence for several new species. J. Parasitol. 85:1126-1133. [PubMed] [Google Scholar]
- 23.Morgan, U. M., L. Xiao, R. Fayer, A. A. Lal, and R. C. Thompson. 1999. Variation in Cryptosporidium: towards a taxonomic revision of the genus. Int. J. Parasitol. 29:1733-1751. [DOI] [PubMed] [Google Scholar]
- 24.Patel, S., S. Pedraza-Diaz, and J. McLauchlin. 1999. The identification of Cryptosporidium species and Cryptosporidium parvum directly from whole faeces by analysis of a multiplex PCR of the 18S rRNA gene and by PCR/RFLP of the Cryptosporidium outer wall protein (COWP) gene. Int. J. Parasitol. 29:1241-1247. [DOI] [PubMed] [Google Scholar]
- 25.Pedraza-Diaz, S., C. Amar, A. M. Iversen, P. J. Stanley, and J. McLauchlin. 2001. Unusual Cryptosporidium species recovered from human faeces: first description of Cryptosporidium felis and Cryptosporidium “dog type” from patients in England. J. Med. Microbiol. 50:293-296. [DOI] [PubMed] [Google Scholar]
- 26.Pedraza-Diaz, S., C. Amar, and J. McLauchlin. 2000. The identification and characterisation of an unusual genotype of Cryptosporidium from human faeces as Cryptosporidium meleagridis. FEMS Microbiol. Lett. 89:189-194. [DOI] [PubMed] [Google Scholar]
- 27.Pieniazek, N. J., F. J. Bornay-Llinares, S. B. Slemenda, A. J. da Silva, I. N. Moura, M. J. Arrowood, O. Ditrich, and D. G. Addiss. 1999. New Cryptosporidium genotypes in HIV-infected persons. Emerg. Infect. Dis. 5:444-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spano, F., L. Putignani, S. Guida, and A. Crisanti. 1998. Cryptosporidium parvum: PCR-RFLP analysis of the TRAP-C1 (thrombospondin-related adhesive protein of Cryptosporidium-1) gene discriminates between two alleles differentially associated with parasite isolates of animal and human origin. Exp. Parasitol. 90:195-198. [DOI] [PubMed] [Google Scholar]
- 29.Sulaiman, I. M., A. A. Lal, M. J. Arrowood, and L. Xiao. 1999. Biallelic polymorphism in the intron region of beta-tubulin gene of Cryptosporidium parasites. J. Parasitol. 85:154-157. [PubMed] [Google Scholar]
- 30.Sulaiman, I. M., A. A. Lal, and L. Xiao. 2002. Molecular phylogeny and evolutionary relationships of Cryptosporidium parasites at the actin locus. J. Parasitol. 88:388-394. [DOI] [PubMed] [Google Scholar]
- 31.Sulaiman, I. M., U. M. Morgan, R. C. Thompson, A. A. Lal, and L. Xiao. 2000. Phylogenetic relationships of Cryptosporidium parasites based on the 70-kilodalton heat shock protein (HSP70) gene. Appl. Environ. Microbiol. 66:2385-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weinstein, P., M. Macaitis, C. Walker, and S. Cameron. 1993. Cryptosporidial diarrhoea in South Australia. An exploratory case-control study of risk factors for transmission. Med. J. Aust. 158:117-119. [PubMed] [Google Scholar]
- 33.Xiao, L., L. Escalante, C. Yang, I. Sulaiman, A. A. Escalante, R. J. Montali, R. Fayer, and A. A. Lal. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 65:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao, L., U. M. Morgan, J. Limor, A. Escalante, M. Arrowood, W. Shulaw, R. C. Thompson, R. Fayer, and A. A Lal. 1999. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol. 65:3386-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao, L., J. R. Limor, L. Li, U. Morgan, R. C. Thompson, and A. A Lal. 1999. Presence of heterogeneous copies of the small subunit rRNA gene in Cryptosporidium parvum human and marsupial genotypes and Cryptosporidium felis. J. Eukaryot. Microbiol. 46:44S-45S. [PubMed] [Google Scholar]
- 36.Xiao, L., R. Fayer, U. Ryan, and S. J. Upton. 2004. Cryptosporidium taxonomy: recent advances and implications for public health. Clin. Microbiol. Rev. 17:72-97. [DOI] [PMC free article] [PubMed] [Google Scholar]