Abstract
The accurate identification of Salmonella enterica subsp. enterica serovar Typhi variants that fail to express the capsular polysaccharide, Vi, is an important and much discussed issue for medical microbiology. We have tested a multiplex PCR method which shows the presence or absence of the genetic locus required for Vi expression. Of 2,222 Salmonella serovar Typhi clinical isolates collected from patients' blood over a 4-year period in a region of Pakistan where typhoid is endemic, 12 tested negative for Vi expression by serological agglutination. However, only 1 of these 12 was Vi negative by the multiplex PCR method. This result was confirmed by immunofluorescence, the most sensitive method for Vi characterization in Salmonella serovar Typhi. The multiplex PCR described therefore represents a simple and accurate method for surveillance for Vi-negative variants of Salmonella serovar Typhi in Pakistan. Testing of clinical isolates of Salmonella serovar Typhi, before subculture, from other regions where Vi-negative Salmonella serovar Typhi has been described should be carried out so that the impact of vaccination with purified Vi antigen on the levels of Vi-negative Salmonella serovar Typhi in bacterial populations can be assessed.
The causative organism of human typhoid fever, Salmonella enterica subsp. enterica serovar Typhi, unlike most of the other serovars of Salmonella, expresses a capsular polysaccharide antigen known as Vi. In vitro studies have shown that Vi is antiopsonic and antiphagocytic (20), reduces the level of secretion of Salmonella serovar Typhi-induced tumor necrosis factor alpha (a marker of activation) by human macrophages (10), and increases the level of resistance of the organism to oxidative killing (20). The presence of the Vi antigen also increases the infectivity (11) of Salmonella serovar Typhi and the severity of disease (13) in volunteers. The Vi capsule is, however, not essential for infection, as Vi-negative mutants of Salmonella serovar Typhi are able to establish infection and cause a typhoid fever-like illness in human volunteers (11, 13). Furthermore, there have been reports of outbreaks of typhoid fever caused by Vi-negative Salmonella serovar Typhi (5).
The reliance of serological (agglutination) testing for Vi expression has some limitations. The level of Vi expression on Salmonella serovar Typhi varies in response to changes in culture conditions and is associated with regulation by the OmpR and EnvZ two-component regulatory system, which is influenced by factors such as osmolarity (26). Also, Vi-negative mutants of Salmonella serovar Typhi are known to accumulate from previously Vi-positive Salmonella serovar Typhi isolates following laboratory passage. Fortunately, the genetic control of Vi expression is now relatively well understood, and so determination of the genetic conformation of truly Vi-negative Salmonella serovar Typhi is now possible. The Vi biosynthetic and export proteins are encoded within the viaB locus (9), a region of DNA located within the acquired DNA island known as Salmonella pathogenicity island 7 (SPI-7) (25, 27). Insertion and deletion mutations in the tviA gene, encoded within the viaB locus, that can lead to the loss of Vi expression and an increase in invasive properties have been reported (30). Furthermore, spontaneous loss of the whole of SPI-7 in Vi-negative mutants has been reported in stored isolates (23), and precise excision of SPI-7 has been demonstrated (6). Thus, Vi antigen expression is encoded on an unstable and possibly mobile region of the Salmonella serovar Typhi genome, which has been well described (27).
An effective vaccine based on purified Vi antigen has been developed and licensed for use as a parenteral vaccine against typhoid fever in humans. A single dose of the antigen induces a protective immune response against Salmonella serovar Typhi in adults and older children (7), and when it is conjugated to a protein carrier the vaccine is also effective in young children (19). Purified Vi antigen has been used in regions where typhoid is endemic (1, 5, 18), and there is a long-running debate concerning its efficacy in the presence of Vi-negative isolates (3, 4, 16, 22, 28). Experience with Streptococcus pneumoniae vaccines based on combination capsular antigens has indicated that the immunization of populations with such vaccines may favor the emergence of capsular types not present in the vaccine (21, 24). There is potential selective pressure in favor of Vi-negative Salmonella serovar Typhi from the extended use of Vi-based vaccines, and there is a paucity of surveillance data based on molecular biology-based tests. We have therefore developed and characterized tools which allow us to monitor the prevalence of Vi-negative Salmonella serovar Typhi in regions where the Vi vaccine is being introduced. Here we show that Vi-negative Salmonella serovar Typhi was very rare in a region of Pakistan where typhoid was highly endemic before vaccination with purified Vi antigen was introduced.
MATERIALS AND METHODS
Bacterial isolates.
All Salmonella serovar Typhi isolates investigated were isolated from blood in the Microbiology Laboratory of the Aga Khan University, Karachi, Pakistan, between 1999 and 2002. Control strains SARB63 and SARB64 (Salmonella Reference Collection, Calgary, Alberta, Canada), CT18 (25), and Ty2 (a gift from M. Popoff, Pasteur Institute, Paris, France) were used as negative and positive controls, as described for each method. Primary isolation of the organism from blood was carried out with commercial blood culture bottles (BACTEC 9240; Becton Dickinson, Oxford, United Kingdom) and a single round of growth on sheep blood agar (Oxoid, Basingstoke, United Kingdom). Identification and Vi agglutination (Murex, Dartford, United Kingdom) were carried out with isolates obtained directly from the blood agar. Isolates were identified as Salmonella serovar Typhi by identification of the following characteristics: glucose positive without gas formation, lactose negative and H2S positive on a Kliglar iron agar slant (Oxoid), citrate (Oxoid) negative, indole (Oxoid) negative, and positive serology with both O9 and Hd antigens (Murex, Dartford, United Kingdom). If an isolate was Vi positive but negative for O9, the organism was boiled for 10 min and the agglutination test was repeated. Agglutination was carried out according to the instructions of the manufacturer. API 20E strips (bioMérieux, Basingstoke, United Kingdom) and DNA sequencing of a variable region (the phoN-pheU intergenic region) and a conserved region (aroC) of the genome were used to confirm the identities of unusual organisms.
PCR amplification for presence of Vi genes.
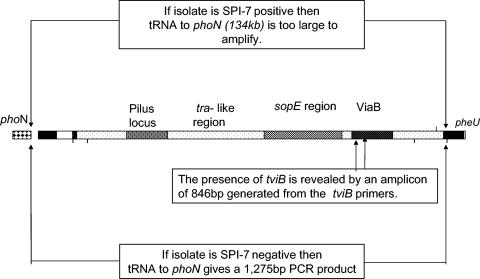
A multiplex PCR was developed to detect the deletion of the genes encoding Vi, including tviB. Three sets of primers were designed (Table 1) to specifically amplify (i) an internal region of the tviB gene in the viaB locus, a gene essential for the production of Vi capsular antigen (primers tviB-F and tviB-R); (ii) an intergenic region of 1,275 bp between the pheU (phenylalanine tRNA) and phoN genes, which produces a product only in the absence of SPI-7 (primers DE0032-F and DE0083-R); and (iii) a chromosomal gene, aroC, which acts as an internal PCR control (primers aroC-F and aroC-R) (Fig. 1). PCRs were performed with an Expand High Fidelity PCR kit (Roche, Basel, Switzerland). Each reaction mixture contained 300 nM primer, 100 ng of DNA template, 200 μM deoxynucleoside triphosphates, and 2.6 U of Taq polymerase in a total volume of 50 μl of reaction buffer. PCR conditions were 95.0°C for 3 min, followed by 28 cycles of 95.0°C for 30 s, 57.0°C for 1 min, and 72.0°C for 3 min. The PCR products were visualized by ethidium bromide staining after electrophoresis on a 1% agarose gel.
TABLE 1.
Primers used in the multiplex PCR for detection of tviB and SPI-7 in Vi agglutination-negative Salmonella isolates
| Interpretation if band detected | Primer | Sequence | Positive control strain(s) | Product size (bp [gene amplified]) |
|---|---|---|---|---|
| Genes for Vi production present | tviB-F | 5′-CGAGTGAAACCGTTGGTACA-3′ | SARB63, TY2, CT18 | 846 (tviB) |
| tviB-R | 5′-CAATGATCGCATCGTAGTGG-3′ | |||
| SPI-7 absent | DE0032-F | 5′-GCTCAGTCGGTAGAGCAGGGGATT-3′ | SARB 64 | 1,275 (tRNA-phoN) |
| DE0083-R | 5′-TCATCTTCAGGACGGCAGGTAGAATG-3′ | |||
| Salmonella DNA present | aroC-F | 5′-GGCACCAGTATTGGCCTGCT-3′ | CT18 | 639 (aroC) |
| aroC-R | 5′-CATATGCGCCACAATGTGTTG-3′ |
FIG. 1.
Schematic representation of the multiplex PCR for the detection of SPI-7 and tviB.
Sequencing of aroC and tRNA PCR products.
The PCR products were purified with DNA purification kits (Qiagen, Crawley, United Kingdom). Sequencing of the products obtained with the specific primers was performed with BigDye Chain Terminators on an ABI 3700 capillary machine. The sequence was edited and aligned by using Bioedit software (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). It is known that the aroC sequence varies among S. enterica isolates and is conserved within particular Salmonella serotypes (17). Consequently, the precise DNA sequence of aroC provides an accurate signature of a particular S. enterica serovar, and so this approach was used to independently confirm the identities of selected isolates. Experience with the intergenic region phoN to pheU of different Salmonella serovars (27) and aroC of Salmonella serovar Typhi (17) allowed us to use sequence data from these regions to support the identification.
Immunofluorescence detection of Vi.
All clinical isolates which were Vi negative by agglutination were investigated for Vi expression by immunofluorescence. Bacteria were grown at 37°C in Luria-Bertani (LB) broth (0.17 M NaCl) for 8 h with aeration, subcultured at a 1/100 dilution into fresh LB broth (0.17 M NaCl), and cultured for a further 16 h at 37°C without aeration. Bacteria from 500 μl of culture were harvested by centrifugation in a benchtop microcentrifuge (Eppendorf 5415D) (13,000 rpm, 2 min) and resuspended in 500 μl of phosphate-buffered saline (PBS). The bacteria were diluted 1/100 in PBS, and a 50-μl drop was placed in the wells of a glass chamber slide (Falcon, Poole, United Kingdom). The slides were incubated at room temperature for 30 min. Excess bacteria and PBS were removed, and the slides were fixed in 4% paraformaldehyde in PBS for 30 min at room temperature. The slides were washed twice with PBS and then placed in PBS containing 20 mM ammonium chloride and stored at 4°C until they were used. Before the slides were stained, they were washed twice with PBS. Primary antibodies (goat anti-Salmonella antibody and rabbit anti-Vi antibody; Murex) diluted 1/200 in PBS containing 10% horse serum were applied to the bacteria (40 μl per well) for 30 min at room temperature in a humid atmosphere. The slides were washed twice in PBS, and secondary antibodies (donkey anti-goat antibody conjugated to Cy2 and donkey anti-rabbit antibody conjugated to Rhodamine Redex, diluted 1/200 and 1/400, respectively) in PBS containing 10% horse serum were applied to the bacteria (40 μl per well) for 30 min at room temperature in a humid atmosphere. The slides were washed twice in PBS and once in water, dried, and mounted. The bacteria were visualized by epifluorescence microscopy. The bacteria were visible on the green channel, and the Vi antigen was visible on the red channel.
RESULTS
Vi agglutination.
In the routine diagnostic laboratory at the Aga Khan University during the period of study (1999 to 2002), 2,227 bacterial blood isolates were provisionally identified as Salmonella serovar Typhi on the basis of biochemistry test results and agglutination with O9 and Hd antisera. Of these, 2,210 (99.2%) isolates gave positive results for Vi by agglutination carried out at the same time as the identification tests. The 17 Vi agglutination-negative putative Salmonella serovar Typhi isolates identified from direct culture in this screening were subjected to a more detailed analysis after storage. Of these original 17 isolates, 5 were not Salmonella serovar Typhi. Three were positive for O9 but negative for Hd, which led to their classification as group D salmonellae but not Salmonella serovar Typhi. Interestingly these isolates gave a false-positive result for Hd agglutination when they tested by the slide agglutination format and were negative for citrate utilization when they were tested with 2.5 ml of citrate agar. Final confirmation of the identification was done with the API 20E system and sequencing of aroC. The two remaining isolates were identified as Salmonella serovar Paratyphi A, and we believe that these isolates were present as mixed cultures with Salmonella serovar Typhi on initial isolation. This is supported by the finding for one culture that contained organisms that were both SPI-7 negative, typical of Salmonella serovar Paratyphi A, and tviB positive, typical of Salmonella serovar Typhi (Fig. 2, lane 17). These classifications were subsequently confirmed by sequencing of the aroC gene of each isolate, which provided a genetic signature of the serovar (Fig. 3). Thus, although these bacteria were responsible for invasive salmonellosis in humans, they had been misclassified by standard laboratory methods. Therefore, of the 2,222 Salmonella serovar Typhi isolates recovered, 12 (0.5%) were agglutination negative for Vi on primary subculture. The reported rates of occurrence of Vi-negative Salmonella serovar Typhi by year are presented in Table 2.
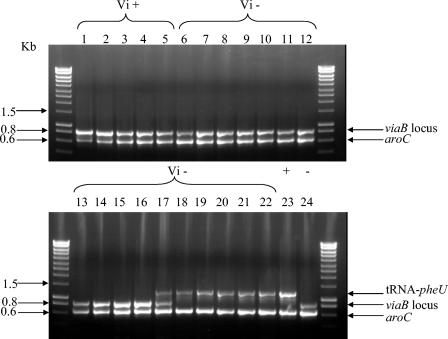
FIG. 2.
Multiplex PCR for detection of SPI-7 and tviB. The presence of the tviB gene in the viaB locus is shown by a band at about 0.8 kb, and the absence of the SPI-7 region is shown by a band at about 1.5 kb. The aroC products are at 0.6 kb. Lanes: Vi-positive controls Ty2 (lane 1), SARB63 (lane 16), and CT18 (lane 24); Vi-positive clinical isolates BL13807, BL15339, and BL13510 (lanes 2 to 4, respectively); Vi agglutination-negative Salmonella isolates BL9282 (lane 5), BL14907 (lane 6), BL13843 (lane 7), BL9283 (lane 8), BL12035 (lane 9), BL3081 (lane 10), BA788 (lane 11), BL4426 (lane 12), BL6886 (lane 13), BL6397 (lane 14), BL6583 (lane 15), BL2934 (lane 22), BL13994 (lane 17), BL9737 (lane 18), BL1838 (lane 20), and BL2580 (lane 21); Vi negative controls Salmonella serovar Paratyphi A BL11161 (lane 19) and SARB64 (lane 23).
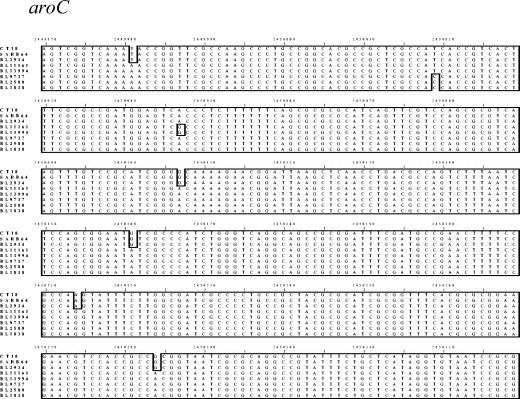
FIG. 3.
Sequence alignment of PCR product from the aroC gene of Vi agglutination-negative S. enterica. The sequences were compared to that of aroC of strain CT18. Boxed nucleotides represent variations from the consensus sequence.
TABLE 2.
Occurrence of Vi agglutination-negative Salmonella, by year
| Yr | Total no. of isolates | No. (%) of Vi agglutination-negative isolates |
|---|---|---|
| 2002 | 565 | 6 (1.06) |
| 2001 | 783 | 5 (0.06) |
| 2000 | 321 | 4 (1.24) |
| 1999 | 558 | 2 (0.358) |
| All yr | 2,227 | 17a (0.76) |
Five of the isolates were later shown to be non-Salmonella serovar Typhi salmonellae.
Multiplex PCR for ViaB and SPI-7.
To test if the phenotypically Vi-negative Salmonella serovar Typhi strains encoded the genes for Vi production, a multiplex PCR was developed to detect either the presence of tviB within the actual ViaB (Vi capsule biosynthesis)-coding region or the absence of the whole 134-kbp SPI-7 sequence (Fig. 1). This PCR was used to amplify all initial Salmonella isolates negative for Vi by agglutination (12 Salmonella serovar Typhi isolates, 2 Salmonella serovar Paratyphi A isolates, and 3 group D Salmonella isolates). Three Salmonella serovar Typhi clinical isolates and strains SARB63, CT18, and TY2, all freshly positive for Vi agglutination, acted as positive controls. Salmonella serovar Typhi SARB64 and three clinical isolates of Salmonella serovar Paratyphi A served as negative controls. The presence of Salmonella DNA was confirmed by including Salmonella aroC-specific primers in the multiplex PCR. All positive and negative controls routinely yielded the expected results: a PCR product of 846 bp for Vi-positive isolates and a PCR product of 1,275 bp, indicating the absence of SPI-7, for the negative controls (Fig. 1 and 2). Of the 12 agglutination-negative Salmonella serovar Typhi isolates, 11 gave products of 846 bp, indicating the presence of tviB (tviB+), and 1 gave a product of 1,275 bp, suggesting that this organism did not harbor SPI-7. The PCR results therefore show that 11 of 12 of the Vi agglutination-negative Salmonella serovar Typhi isolates possessed the tviA gene required for the synthesis and expression of the Vi antigen. Significantly, over the 4-year period only one isolate of Salmonella serovar Typhi had lost the whole gene sequence for SPI-7, as shown by the presence of a product between the pheU and phoN (Fig. 4).
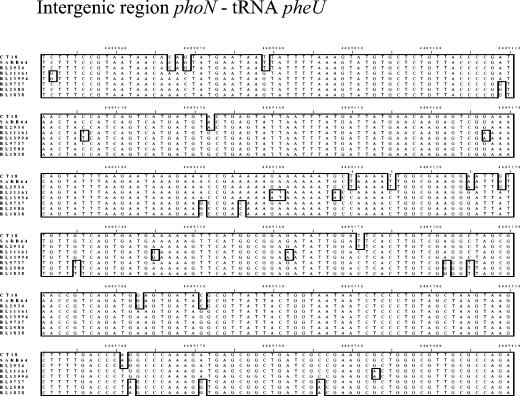
FIG. 4.
Sequence alignment of PCR product across the pheU tRNA gene of Vi agglutination-negative S. enterica. The sequence from Salmonella serovar Typhi CT18 with SPI-7 removed is presented for comparison. Boxed nucleotides represent variations from the consensus sequence.
Subculture on medium with low osmolarity.
There is evidence that Vi expression is influenced by osmolarity and that it is enhanced by the culture medium and conditions of low osmolarity, e.g., those obtained with 0.17 M NaCl (26). At the time of isolation, all of the Vi agglutination-negative Salmonella serovar Typhi isolates were routinely subcultured onto low-osmolarity LB agar (0.17 M NaCl; Difco, Detroit, Mich.). However, none of these isolates became detectably Vi agglutination positive following subculture. However, following storage on Protect Beads (Prolabs, Poole, United Kingdom) for various periods of several months to years at −70°C and subsequent subculture on LB agar containing 0.17 M NaCl, 9 of 11 previously Vi agglutination-negative tviB+ isolates grew as cultures that were positive for Vi by agglutination. Hence, the storage conditions used apparently favored the generation of Salmonella serovar Typhi capable of surface-associated Vi production. Interestingly, there were still two Salmonella serovar Typhi isolates which had the tviB gene but which could not be shown to express Vi by agglutination after storage.
Immunofluorescence detection of Vi.
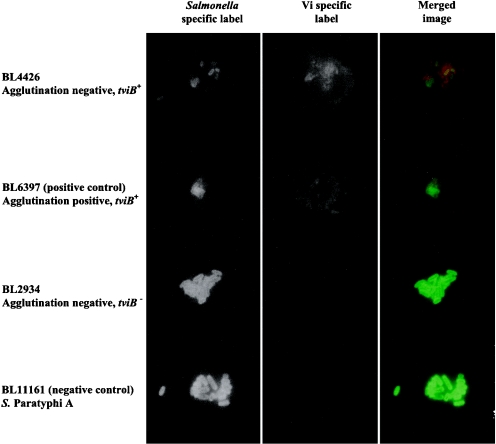
Immunofluorescence microscopy facilitates the direct detection of the Vi capsule on individual Salmonella serovar Typhi bacteria and was found to be a very sensitive method for the detection of Vi expression (our results, not shown). All of the original 11 Vi agglutination-negative Salmonella serovar Typhi isolates, which were positive for both SPI-7 and tviB, were tested for Vi expression by immunofluorescence microscopy with the same anti-Vi antiserum used for the agglutination tests. All were positive for Vi production by comparison with known negative controls. Interestingly, this included the two Salmonella serovar Typhi isolates that were negative by agglutination after storage (Fig. 5). The reason for this is likely to be the higher sensitivity of the immunofluorescence microscopy technique compared with that of agglutination.
FIG. 5.
Immunofluorescent staining of Vi agglutination-negative Salmonella serovar Typhi. Strain BL6397 is the Vi-positive control, and strain BL11161 is the Vi-negative control. First column, bacteria labeled with anti-Salmonella antibody (green channel); second column, bacteria labeled with anti-Vi antibody (red channel); third column, merged images of the first two columns. Original magnification, ×1,000.
Thus, of 17 Salmonella serovar Typhi isolates from clinical specimens originally reported to be Vi negative, only 1 was in fact a truly Vi-negative Salmonella serovar Typhi isolate. Eleven cases of false negativity were caused by a false-negative agglutination test result when the primary isolates were tested. The results of all the investigations are summarized in Table 3.
TABLE 3.
Summary of results for Salmonella isolates that were Vi agglutination negative on primary culture and three Salmonella serovar Typhi isolates that were Vi agglutination positive
| Isolate | Serovar | Agglutination result for Vi on:
|
PCR results for:
|
IFa result for Vi | ||
|---|---|---|---|---|---|---|
| First isolation | storage | tviB | pheUb | |||
| BL13807 | Typhi | + | NTc | + | − | NT |
| BL13539 | Typhi | + | NT | + | − | NT |
| BL13510 | Typhi | + | NT | + | − | NT |
| BL6583 | Typhi | − | + | + | − | + |
| BL12035 | Typhi | − | + | + | − | + |
| BL13843 | Typhi | − | + | + | − | + |
| BL14907 | Typhi | − | + | + | − | + |
| BL6397 | Typhi | − | + | + | − | + |
| BL6886 | Typhi | − | + | + | − | + |
| BL9283 | Typhi | − | + | + | − | + |
| BL9282 | Typhi | − | + | + | − | + |
| BL3081 | Typhi | − | − | + | − | + |
| BA788 | Typhi | − | + | + | − | + |
| BL4426 | Typhi | − | − | + | − | + |
| BL2934 | Typhi | − | − | − | + | − |
| BL11161 | Paratyphi A | − | − | − | + | − |
| BL13994d | Paratyphi A | + | − | + | + | − |
| BL1838 | O9 Hd− | − | − | − | + | − |
| BL2580 | O9 Hd− | − | − | − | + | − |
| BL9737 | O9 Hd− | − | − | − | + | − |
IF, immunofluorescence.
The presence of a pheU product means that SPI-7 is absent.
NT, not tested.
Probable mixed culture of Salmonella serovar Paratyphi A and Salmonella serovar Typhi.
DISCUSSION
We show here that more than 99% (2,221 of 2,222) of human blood isolates of Salmonella serovar Typhi in Pakistan were able to express the Vi capsular antigen. There were originally 17 non-Vi-agglutinable isolates, of which 5 were shown to be non-serovar Typhi Salmonella isolates. Of the remaining 12 isolates, 11 were shown to be capable of expressing Vi under some conditions. The one truly Vi-negative Salmonella serovar Typhi isolate had lost not only the viaB locus, which encodes proteins associated with Vi biosynthesis, but the whole of SPI-7, which contains other genes associated with virulence (25). Given the unstable nature of this region of the Salmonella serovar Typhi genome (27), we believe that this shows a remarkably high degree of conservation of Vi expression in primary isolates when they are characterized prior to storage and extensive laboratory passage. In contrast, reports from different regions of India have described both a high prevalence (22) and a low prevalence (14) of Vi-negative Salmonella serovar Typhi isolates when serovar Typhi isolates were tested.
Given that the use of the capsular polysaccharide-based vaccines for protection against other pathogens, such as S. pneumoniae, has led to concern about an increase in the proportion of strains expressing capsular types not incorporated into the vaccine (8), the proportion of Vi-negative Salmonella serovar Typhi isolates in natural bacterial populations potentially has major implications for the use of Vi antigen-based typhoid vaccines (4, 12, 16). We therefore believe that it will be critically important to establish robust methods for the analysis of Vi expression by Salmonella serovar Typhi in areas where Vi vaccines are used and recommend a serological approach combined with genetic analysis that incorporates PCR analysis. Attempts to identify Vi-negative Salmonella serovar Typhi isolates by purely agglutination-based approaches should be viewed with caution. Vi expression by Salmonella serovar Typhi has been shown to be subject to regulation involving two-component sensors-regulators, RcsB-RcsC (2) and OmpR-EnvZ (26), which respond to changes in environmental conditions, such as osmolarity. Vi expression is thus complex and is likely to be subject to regulation by other unknown factors. It is possible that Salmonella serovar Typhi isolates reported to be Vi agglutination negative from direct cultures in clinical microbiology laboratories are actually Vi positive but demonstrate a down regulation of Vi that could occur either in the patient's blood or during culture in the laboratory. Indeed, we report this to be the case with nearly all (11 of 12) of the Vi-negative isolates whose agglutination was determined in this study.
Vi-negative Salmonella serovar Typhi isolates accumulate at significant rates following laboratory passage of the organism, and so to investigate this, we have systematically tested all Salmonella serovar Typhi isolates recovered at the Aga Khan laboratories in Karachi over a 4-year period (1999 to 2002) for Vi expression using traditional microbiology identification techniques and then screened the Vi-negative Salmonella serovar Typhi isolates using specific immunofluorescence and genetic characterization. During primary isolation, 0.5% (12 of 2,222) of the Salmonella serovar Typhi isolates were found to be Vi agglutination negative. Eleven of these 12 Vi agglutination-negative isolates were positive for the tviB gene, which suggested that the genetic machinery required for Vi expression was present. Although none of the 11 tviB+ isolates became Vi agglutination positive after passage on low-osmolarity agar at the time of isolation, 9 of them became positive after they were frozen and recultured in low-osmolarity broth. This illustrates the difficulties in attempting to determine the Vi types of Salmonella serovar Typhi isolates purely by serological methods. By immunofluorescence, all 11 isolates were shown to be capable of expressing Vi after storage (Fig. 5). Of importance to this study is that all of the Salmonella serovar Typhi isolates which possessed tviB were able to express the Vi antigen, as shown by immunofluorescence, suggesting that the PCR used here for the detection of tviB has application for the detection of Vi-negative Salmonella serovar Typhi on primary culture. Further characterization of the relationship between the presence of tviB and the expression of Vi is needed, however, as an IS1 insertion sequence is present in this region in CT18, and the effects of this on Vi expression are unknown. Although we (27) and others (23) have shown the presence of SPI-7-negative Salmonella serovar Typhi strains in culture collections, these may accumulate because of the culture or storage conditions used. It is therefore important to test Salmonella serovar Typhi isolates for the presence of the Vi genes on primary isolation with as little subculturing as possible. It is possible that the particular strains of Salmonella serovar Typhi or the culture and isolation conditions used in Pakistan may favor the retention of the SPI-7 island and that the conditions and/or the strains encountered elsewhere may favor the loss of SPI-7 or Vi expression. Thus, it will be important to perform studies similar to ours in different geographical locations in laboratories that use different isolation techniques.
The genetics of Vi are well understood. In addition to regulatory genes that influence the levels of Vi expression, 11 genes within the ViaB operon are involved in Vi biosynthesis (9). Furthermore, ViaB is encoded on a potentially mobile pathogenicity island, SPI-7, which contains 134 kbp of DNA (Fig. 1). From sequence analysis it is predicted that SPI-7 is capable of excision from the Salmonella serovar Typhi chromosome (6, 27). The question remains whether the genes for Vi expression could be lost under the potentially overwhelming selective pressure against this capsule from Vi vaccination programs. The licensed parenteral typhoid vaccines are based on purified Vi antigen alone, and new vaccines in which the antibody response to this antigen is enhanced by conjugation of the antigen to carrier proteins are being developed (19). These vaccines will not provide protection against other Salmonella serovars, such as Salmonella serovar Paratyphi and nontyphoidal group D salmonellae, which may thus emerge as important causes of systemic salmonellosis. Furthermore, live oral typhoid vaccines which constitutively express Vi are also being developed and may have similar effects (29). Vi-negative Salmonella serovar Typhi can become clinically important only if the capsule is not essential for pathogenesis or transmission. At present, the Vi antigen is considered important in the pathogenesis of typhoid fever and possibly protects the bacterium from complement-mediated killing and phagocytosis. However, Vi-negative Salmonella serovar Typhi can cause a typhoid fever-like illness, as can Salmonella serovar Paratyphi A, which does not typically express the Vi antigen (15). Our data suggest that Vi may have a vital role in typhoid fever, but further study of the molecular epidemiology of the bacteria which cause invasive salmonellosis, particularly in regions where Vi vaccination is extensive, is imperative. The multiplex PCR and immunofluorescence assay developed for this study should prove invaluable for screening for Vi-negative Salmonella serovar Typhi postvaccination.
Acknowledgments
We acknowledge the work of the technical staff at the Aga Kahn University Microbiology Laboratory and thank Kate Unsworth at Imperial College and Ken Sanderson at University of Calgary for useful discussions.
G.D. was supported by funding from The Wellcome Trust, and C.K. was supported by a Wellcome Trust Ph.D. studentship. This project was funded by the Wellcome Trust of the United Kingdom and Aga Khan University.
REFERENCES
- 1.Acharya, I. L., C. U. Lowe, R. Thapa, V. L. Gurubacharya, M. B. Shrestha, M. Cadoz, D. Schulz, J. Armand, D. A. Bryla, B. Trollfors, et al. 1987. Prevention of typhoid fever in Nepal with the Vi capsular polysaccharide of Salmonella typhi. A preliminary report. N. Engl. J. Med. 317:1101-1104. [DOI] [PubMed] [Google Scholar]
- 2.Arricau, N., D. Hermant, H. Waxin, C. Ecobichon, P. S. Duffey, and M. Y. Popoff. 1998. The RcsB-RcsC regulatory system of Salmonella typhi differentially modulates the expression of invasion proteins, flagellin and Vi antigen in response to osmolarity. Mol. Microbiol. 29:835-850. [DOI] [PubMed] [Google Scholar]
- 3.Arya, S. C. 1997. Efficacy of Salmonella typhi Vi capsular polysaccharide vaccine in South Africa. Vaccine 15:244. (Letter.) [DOI] [PubMed] [Google Scholar]
- 4.Arya, S. C. 1999. Efficacy of Vi polysaccharide vaccine against Salmonella typhi. Vaccine 17:1015-1016. (Letter.) [DOI] [PubMed] [Google Scholar]
- 5.Arya, S. C. 2002. Field effectiveness of Vi polysaccharide typhoid vaccine in the People's Republic of China. J. Infect. Dis. 185:845. (Author's reply, 185:845-846.) [DOI] [PubMed] [Google Scholar]
- 6.Bueno, S. M., C. A. Santiviago, A. A. Murillo, J. A. Fuentes, A. N. Trombert, P. I. Rodas, P. Youderian, and G. C. Mora. 2004. Precise excision of the large pathogenicity island, SPI7, in Salmonella enterica serovar Typhi. J. Bacteriol. 186:3202-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engels, E. A., M. E. Falagas, J. Lau, and M. L. Bennish. 1998. Typhoid fever vaccines: a meta-analysis of studies on efficacy and toxicity. BMJ Clin. Res. Ed. 316:110-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feikin, D. R., and K. P. Klugman. 2002. Historical changes in pneumococcal serogroup distribution: implications for the era of pneumococcal conjugate vaccines. Clin. Infect. Dis. 35:547-555. [DOI] [PubMed] [Google Scholar]
- 9.Hashimoto, Y., T. Ezaki, N. Li, and H. Yamamoto. 1991. Molecular cloning of the ViaB region of Salmonella typhi. FEMS Microbiol. Lett. 69:53-56. [DOI] [PubMed] [Google Scholar]
- 10.Hirose, K., T. Ezaki, M. Miyake, T. Li, A. Q. Khan, Y. Kawamura, H. Yokoyama, and T. Takami. 1997. Survival of Vi-capsulated and Vi-deleted Salmonella typhi strains in cultured macrophage expressing different levels of CD14 antigen. FEMS Microbiol. Lett. 147:259-265. [DOI] [PubMed] [Google Scholar]
- 11.Hone, D. M., S. R. Attridge, B. Forrest, R. Morona, D. Daniels, J. T. LaBrooy, R. C. Bartholomeusz, D. J. Shearman, and J. Hackett. 1988. A galE via (Vi antigen-negative) mutant of Salmonella typhi Ty2 retains virulence in humans. Infect. Immun. 56:1326-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hormaeche, C. 1999. Letter to the editor. Vaccine 17:1016. [Google Scholar]
- 13.Hornick, R. B., S. E. Greisman, T. E. Woodward, H. L. DuPont, A. T. Dawkins, and M. J. Snyder. 1970. Typhoid fever: pathogenesis and immunologic control. Second of two parts. N. Engl. J. Med. 283:739-746. [DOI] [PubMed] [Google Scholar]
- 14.Jesudason, M. V., G. Sridharan, S. Mukundan, and T. J. John. 1994. Vi-specific latex agglutination for early and rapid detection of Salmonella serotype typhi in blood cultures. Diagn. Microbiol. Infect. Dis. 18:75-78. [DOI] [PubMed] [Google Scholar]
- 15.Kapil, A., S. Sood, V. Reddaiah, B. Das, and P. Seth. 1997. Paratyphoid fever due to Salmonella eneterica serotype Paratyphi A. Emerg. Infect. Dis. 3:407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keddy, K. H., K. P. Klugman, and J. B. Robbins. 1998. Efficacy of Vi polysaccharide vaccine against strains of Salmonella typhi: reply. Vaccine 16:871-872. (Letter.) [DOI] [PubMed] [Google Scholar]
- 17.Kidgell, C., U. Reichard, J. Wain, B. Linz, M. Torpdahl, G. Dougan, and M. Achtman. 2002. Salmonella typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infect. Genet. Evol. 2:39-45. [DOI] [PubMed] [Google Scholar]
- 18.Klugman, K. P., I. T. Gilbertson, H. J. Koornhof, J. B. Robbins, R. Schneerson, D. Schulz, M. Cadoz, and J. Armand. 1987. Protective activity of Vi capsular polysaccharide vaccine against typhoid fever. Lancet ii:1165-1169. [DOI] [PubMed] [Google Scholar]
- 19.Lin, F. Y., V. A. Ho, H. B. Khiem, D. D. Trach, P. V. Bay, T. C. Thanh, Z. Kossaczka, D. A. Bryla, J. Shiloach, J. B. Robbins, R. Schneerson, S. C. Szu, M. N. Lanh, S. Hunt, L. Trinh, and J. B. Kaufman. 2001. The efficacy of a Salmonella typhi Vi conjugate vaccine in two-to-five-year-old children. N. Engl. J. Med. 344:1263-1269. [DOI] [PubMed] [Google Scholar]
- 20.Looney, R. J., and R. T. Steigbigel. 1986. Role of the Vi antigen of Salmonella typhi in resistance to host defense in vitro. J. Lab. Clin. Med. 108:506-516. [PubMed] [Google Scholar]
- 21.Mbelle, N., R. E. Huebner, A. D. Wasas, A. Kimura, I. Chang, and K. P. Klugman. 1999. Immunogenicity and impact on nasopharyngeal carriage of a nonavalent pneumococcal conjugate vaccine. J. Infect. Dis. 180:1171-1176. [DOI] [PubMed] [Google Scholar]
- 22.Mehta, G., and S. C. Arya. 2002. Capsular Vi polysaccharide antigen in Salmonella enterica serovar Typhi isolates. J. Clin. Microbiol. 40:1127-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nair, S., S. Alokam, S. Kothapalli, S. Porwollik, E. Proctor, C. Choy, M. McClelland, S. L. Liu, and K. E. Sanderson. 2004. Salmonella enterica serovar Typhi strains from which SPI7, a 134-kilobase island with genes for Vi exopolysaccharide and other functions, has been deleted. J. Bacteriol. 186:3214-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Obaro, S. K., R. A. Adegbola, W. A. Banya, and B. M. Greenwood. 1996. Carriage of pneumococci after pneumococcal vaccination. Lancet 348:271-272. [DOI] [PubMed] [Google Scholar]
- 25.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 26.Pickard, D., J. Li, M. Roberts, D. Maskell, D. Hone, M. Levine, G. Dougan, and S. Chatfield. 1994. Characterization of defined ompR mutants of Salmonella typhi: ompR is involved in the regulation of Vi polysaccharide expression. Infect. Immun. 62:3984-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pickard, D., J. Wain, S. Baker, A. Line, S. Chohan, M. Fookes, A. Barron, P. O. Gaora, J. A. Chabalgoity, N. Thanky, C. Scholes, N. Thomson, M. Quail, J. Parkhill, and G. Dougan. 2003. Composition, acquisition, and distribution of the Vi exopolysaccharide-encoding Salmonella enterica pathogenicity island SPI-7. J. Bacteriol. 185:5055-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robbins, J. D., and J. B. Robbins. 1984. Reexamination of the protective role of the capsular polysaccharide (Vi antigen) of Salmonella typhi. J. Infect. Dis. 150:436-449. [DOI] [PubMed] [Google Scholar]
- 29.Wang, J. Y., F. R. Noriega, J. E. Galen, E. Barry, and M. M. Levine. 2000. Constitutive expression of the Vi polysaccharide capsular antigen in attenuated Salmonella enterica serovar Typhi oral vaccine strain CVD 909. Infect. Immun. 68:4647-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao, L., T. Ezak, Z. Y. Li, Y. Kawamura, K. Hirose, and H. Watanabe. 2001. Vi-suppressed wild strain Salmonella typhi cultured in high osmolarity is hyperinvasive toward epithelial cells and destructive of Peyer's patches. Microbiol. Immunol. 45:149-158. [DOI] [PubMed] [Google Scholar]