Abstract
A total of 254 serotype GH rotavirus strains were detected in Palermo, Italy, from 1985 to 2003. Out of 38 serotype G4 strains selected for genetic analysis, 14 were recognized by genotyping as type G9. Strains confirmed to belong to the G4 type showed temporal patterns of genetic evolution in their VP7 and VP4 gene sequences, and the latest Italian G4 strains were distantly related to the reference vaccinal ST3 strain.
Rotaviruses are the major agents of severe gastroenteritis in infants and young children throughout the world. On the basis of the two outer layer proteins, VP4 and VP7, that elicit the production of neutralizing antibodies, human rotaviruses are classified into 10 G (VP7-specific) and 11 P (VP4-specific) types (11). Strains with type G1P[8] are unanimously acknowledged as the most prevalent and ubiquitous type; strains with types G2P[4], G3P[8], and G4P[8] are ubiquitous, but their diffusion is temporal and regional (14). Since 1996 the circulation of strains with G9P[8] or G9P[6] type has been widely reported, while other G types and G/P combinations seem to be sporadic and only locally relevant (24).
In the whole of Europe, high frequency and marked fluctuation of serotype G4 strains have been reported (6, 13, 15, 19, 23, 28, 29, 32). In Palermo, Italy, serotyping of 1,084 rotavirus strains recovered from 1985 to 2003 showed an intermittent pattern of G4 type circulation with frequencies ranging from 33 to 71% for strains recovered from 1990 to 1993, 24 to 37% for strains recovered from 1999 to 2001, and 39.3% for strains recovered from 2003. In the intermediate periods, the frequency of detection ranged from 0 to 8.8% (1-4, 17).
It is known that antigenic variation within a serotype is a mechanism by which variants of rotavirus emerge to escape host immunity (33) and that antigenic and genetic variation can occur within the G4 serotype (29, 30, 31). In this study, we analyze the VP7- and VP4-encoding genes of 38 G4 rotavirus strains selected from a total of 254 strains exhibiting G4 reactivity to the ST3:1 monoclonal antibody (MAb). In particular, 28 specimens were representative of the years of higher frequency of detection of G4 rotaviruses and 10 samples were representative of the intermediate periods. All strains belonged to subgroup II and exhibited the long RNA pattern (2-4, 17).
To determine the G and P genotypes, specimens were analyzed by seminested reverse transcription-PCR strategy with type-specific primers by the methods of Gouvea et al. (20) and Gentsch et al. (16), respectively. Gene 9 (VP7) and gene 4 (VP4) were sequenced on reamplified PCR products (10, 16) (MWG Biotech, Ebersberg, Germany), and the sequences were aligned by using CLUSTAL W (34). Phylogenetic analysis of partial VP7 and VP4 sequences (nucleotides [nt] 63 to 880 and 27 to 430, respectively) and deduced amino acid sequences (amino acids [aa] 22 to 293 for VP7 and aa 10 to 143 for VP4) was performed by using MEGA version 3.0 software (26) using the Kimura two-parameter model as a substitution method and the neighbor-joining method to reconstruct the phylogenetic tree. The statistical significance of the phylogenies inferred was estimated by bootstrap analysis with 1,000 pseudoreplicate data sets.
Out of the 38 rotavirus strains serotyped as G4, 23 strains were confirmed by genotyping, while 14 strains recovered in 1999 to 2003 turned out to belong to the G9 serotype. Despite several attempts, PCR products could not be obtained from the only available strain from the 1980s, which was therefore excluded from further evaluation. Comparison of the nucleotide sequences of the VP7-encoding genes of the 14 G9 strains revealed more than 99% nucleotide identity among these strains and the G9 strains recovered in Palermo, Italy, since 1999 (5), which were not reactive to the ST3:1 MAb. The cross-reactivity of the G9 strains with the ST3:1 MAb has already been observed by other researchers (8, 25, 35), but they could not demonstrate the molecular reasons for such cross-reactivity. It has been hypothesized that the VP4 structure might influence the expression of VP7 epitopes (12). Interestingly, we observed that VP4 sequences of G4 and G9 cross-reacting strains shared >99% nucleotide and amino acid identity.
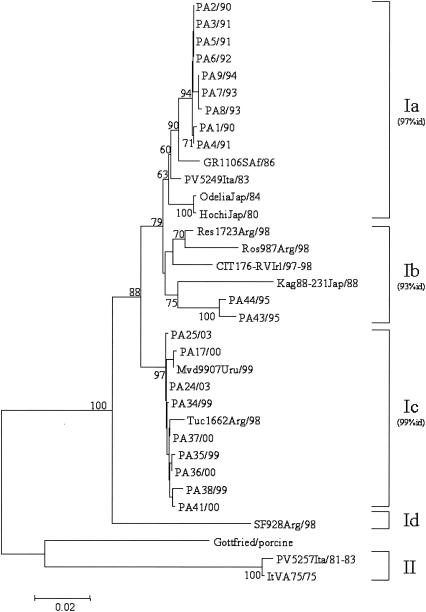
VP7 nucleotide sequence comparison and phylogenetic reconstruction, including the prototype ST3 strain and a selection of G4 rotaviruses, indicated that our 23 G4 viral strains could be easily separated into three well-defined groups: (i) strains collected from 1990 to 1994; (ii) strains collected in 1995; and (iii) strains collected from 1999 to 2003.
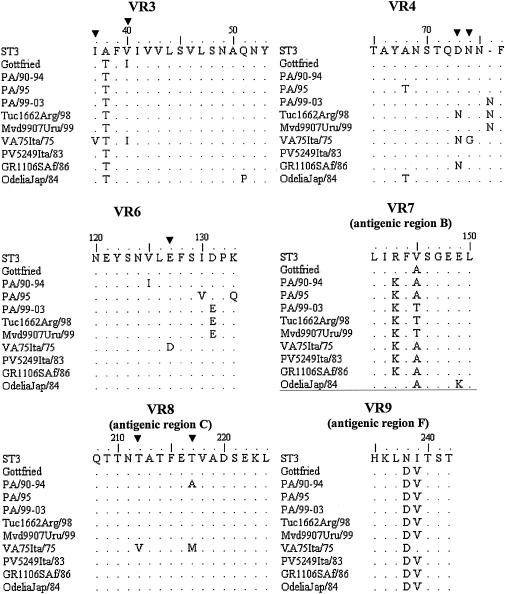
As shown in Fig. 1, all three groups of strains belonged to lineage I, and segregate into three different sublineages, namely, Ia, Ib, and Ic, as defined by Bok et al. (7). The overall nucleotide sequence divergence of the Italian strains within each sublineage was less than 1% for strains of sublineages Ia and Ic and 1.8% for strains of sublineage Ib. There was no correlation between electropherotype and sublineage, as strains virtually identical to each other (100% nucleotide identity) displayed a wide variety of electropherotypes. The analysis of the deduced amino acid sequences corresponding to eight of the nine variable regions (VRs 2 to 9) of VP7 (Fig. 2) showed distinctive amino acid substitutions in VRs 3, 4, 6, 7, 8, and 9 of our strains compared to the amino acids of the corresponding domains of reference G4 strain ST3 and other representative G4 strains. The amino acid sequence alignment revealed an important substitution in sublineage Ic strains, i.e., the insertion of Asn at position 76, which could increase the hydrophilicity of the domain. Bok et al. (7) demonstrated that the increased frequency of G4 rotavirus strains in Argentina in 1998 was due to the emergence of this variant. Interestingly, 1 year later, this emerging strain was also detected in Uruguay. Note that the same insertion was found in a Finnish strain (G484) isolated in 1984 (29). The sequences of the Uruguayan, Argentinean, and Italian strains also exhibited the distinctive amino acid changes 131-Asp→Glu and 146-Val→Thr. Such findings support the hypothesis that sublineage Ic G4 strains were introduced into Italy at the end of the 1990s.
FIG. 1.
Phylogenetic analysis of partial VP7 nucleotide sequences (nt 63 to 880) of serotype G4 strains. The phylogenetic tree was constructed using the neighbor-joining method and Kimura two-parameter model. Percentage bootstrap values above 60% are given at branch nodes. Nucleotide identity (id) within lineages is indicated in parentheses to the right of the sublineage. The number of substitutions per site is indicated by the scale bar.
FIG. 2.
Comparison of amino acid sequences of VP7 VRs 3, 4, and 6 to 9. Amino acids different from those in the ST3 reference vaccinal strain are shown. Conserved amino acids are indicated by periods. The codons discriminating between subtype A (ST3-like) and B (VA75-like) (21) are indicated (▾).
In the past, serotype G4 rotaviruses were divided into two subtypes, namely, subtype A (ST3-like) and subtype B (VA70-like) (18, 21). Direct inspection of the amino acid sequence alignment allowed us to classify all our strains as subtype A. However, the strains collected from 1990 to 1994 showed a 217-Thr→Ala substitution which is atypical for both subtypes (subtypes A and B).
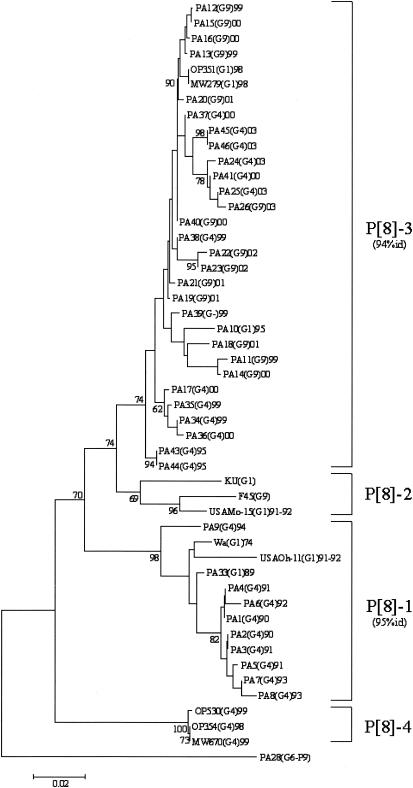
All G4 strains exhibited the P[8] genotype, and the sequences of their VP4-encoding genes clustered into two previously defined lineages (9, 22, 27) (Fig. 3). Chronological clustering was also observed: P[8] sequences obtained from 1990 to 1994 clustered into lineage P[8]-1 and those obtained from 1999 to 2003 clustered into lineage P[8]-3. It is also worth noting that G1 strains collected in Palermo, Italy, from 1989 to 1993 fell within the same lineage P[8]-1 and that G1 and G9 strains recovered from 1995 to 2003 clustered into the lineage P[8]-3. Nucleotide sequence divergence within each group ranged from 0 to 2.9% for the Italian strains of lineage P[8]-1, with the exception of strain PA9 (6.1% nucleotide variation), and from 0 to 5% for the Italian strains of lineage P[8]-3.
FIG. 3.
Phylogenetic analysis of partial VP4 nucleotide sequences (nt 29 to 449) of type P[8] strains. The phylogenetic tree was constructed using the neighbor-joining method and Kimura two-parameter model. Type P [9] PA28 sequence was used as an outgroup. Percentage bootstrap values above 60% are given at branch nodes. Nucleotide identity (id) within lineages is indicated in parentheses to the right of the lineage. The number of substitutions per site is indicated by the scale bar.
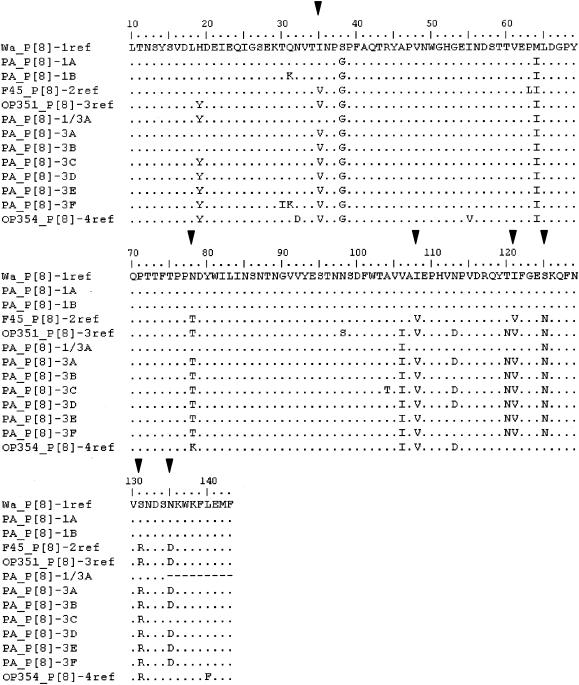
There was more than 94 and 95% amino acid identity among the strains in lineages P[8]-3 and P[8]-1, respectively (Fig. 4). However, compared to strain Wa, which is representative of lineage P[8]-1, two patterns of amino acid substitutions (1A and 1B) could be described. Six patterns (3A to 3F) were distinguished with regard to strain OP351 representative of lineage P[8]-3.
FIG. 4.
Comparison of conserved amino acids between residues 10 and 143 of VP4. Conserved amino acids are indicated by periods. Amino acids different from those in the Wa reference vaccinal strain are shown. Amino acids related to P[8] lineage specificity according to Maunula et al. (27) and Cunliffe et al. (9) are indicated (▾).
In the United Kingdom, Iturriza-Gòmara et al. (22) found that most of the G4 strains belonged to the P[8]-2 lineage and, with much lower frequency, to the P[8]-3 lineage, while only G1 strains were found in the P[8]-1 lineage. In Finland, G4 strains were identified belonging to the lineages P[8]-1 and P[8]-2, while no G4P[8]-3 strains were found (27). These data show that different G4P[8] strains have been circulating in northern and southern Europe in the past 10 years. Immigration into southern Europe has probably caused some of the variety in strains.
Over the last 13 years in Palermo, Italy, two different populations of G4P[8] rotavirus strains have been infecting the infant population. The detection of G1P[8]-3 lineage strains in Palermo, Italy, in 1995 suggests that the new G4P[8] strains may have emerged after the reassortment between the newly introduced G4P[8] strains and the more frequently cocirculating G1P[8] strains and that the VP4 gene of the P[8]-3 lineage might confer a replicative advantage to G4 strains over other P[8] lineages. However, in contrast to what we observed with VP7 gene sequences, a lower nucleotide identity was found within each P[8] lineage, determining different patterns of amino acid substitutions. Considering the role of anti-VP4 neutralizing antibodies in rotavirus immunity (36), such variability could influence their continuous circulation in the infant population and should be taken into consideration for vaccinal strategies.
In conclusion, we have evidence that the latest serotype G4 Italian rotavirus strains are more closely related to recent isolates from other countries than to the reference strains ST3, isolated in the 1970s and included in the vaccine, and VA70, subtype 4B, isolated in 1975 and apparently no longer circulating. Since vaccination can reduce the severity of rotavirus disease and several vaccine formulations are being developed to obtain specific protection, it is important to surveil the circulating strains to anticipate possible antigenic changes that might affect the effectiveness of the vaccine. However, epidemiological surveys based only on serological methods should take into consideration the risk of cross-reaction between G9 and G4 types.
REFERENCES
- 1.Arista, S., L. Giovannelli, N. Passarani, L. Titone, and G. Gerna. 1986. Electropherotyping of human rotaviruses: an epidemiological survey of rotavirus infections in Sicily. Eur. J. Epidemiol. 2:104-107. [DOI] [PubMed] [Google Scholar]
- 2.Arista, S., L. Giovannelli, D. Pistoia, A. Cascio, M. Parea, and G. Gerna. 1990. Electropherotypes, subgroups and serotypes of human rotavirus strains causing gastroenteritis in infants and young children in Palermo, Italy, from 1985 to 1989. Res. Virol. 141:435-448. [DOI] [PubMed] [Google Scholar]
- 3.Arista, S., E. Vizzi, D. Ferraro, A. Cascio, and R. Di Stefano. 1997. Distribution of VP7 serotypes and VP4 genotypes among rotavirus strains recovered from Italian children with diarrhea. Arch. Virol. 142:2065-2071. [DOI] [PubMed] [Google Scholar]
- 4.Arista, S., E. Vizzi, M. C. Migliore, E. Di Rosa, and A. Cascio. 2003. High incidence of G9P[8] rotavirus infections in Italian children during the winter season 1999-2000. Eur. J. Epidemiol. 18:711-714. [DOI] [PubMed] [Google Scholar]
- 5.Arista, S., G. M. Giammanco, S. De Grazia, M. C. Migliore, V. Martella, and A. Cascio. 2004. Molecular characterization of the genotype G9 human rotavirus strains recovered in Palermo, Italy, during the winter of 1999-2000. Epidemiol. Infect. 132:343-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bànyai, K., J. R. Gentsch, R. I. Glass, M. Uj, I. Mihàly, and G. Szücs. 2004. Eight-year survey of human rotavirus strains demonstrates circulation of unusual G and P types in Hungary. J. Clin. Microbiol. 42:393-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bok, K., D. O. Matson, and J. A. Gomez. 2002. Genetic variation of capsid protein VP7 in genotype G4 human rotavirus strains: simultaneous emergence and spread of different lineages in Argentina. J. Clin. Microbiol. 40:2016-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coulson, B. S., J. R. Gentsch, B. K. Das, M. K. Bhan, and R. I. Glass. 1999. Comparison of enzyme immunoassay and reverse transcriptase PCR for identification of serotype G9 rotaviruses. J. Clin. Microbiol. 37:3187-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunliffe, N. A., J. S. Gondwe, S. M. Graham, B. D. M. Thindwa, W. Dove, R. L. Broadhead, M. E. Molyneux, and C. A. Hart. 2001. Rotavirus strain diversity in Blantyre, Malawi, from 1997 to 1999. J. Clin. Microbiol. 39:836-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das, B. K., J. R. Gentsch, H. G. Cicirello, T. O. Woods, A. Gupta, M. Ramachandran, R. Kumar, M. K. Bhan, and R. I. Glass. 1994. Characterization of rotavirus strains from newborns in New Delhi, India. J. Clin. Microbiol. 32:1820-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desselberger, U., M. Iturriza-Gòmara, and J. Gray. 2001. Rotavirus epidemiology and surveillance. Novartis Symp. Ser. 238:125-147. [DOI] [PubMed] [Google Scholar]
- 12.Dunn, S. J., J. W. Burns, T. L. Cross, P. T. Vo, R. L. Ward, M. Bremont, and H. B. Greenberg. 1994. Comparison of VP4 and VP7 of five murine rotavirus strains. Virology 203:250-259. [DOI] [PubMed] [Google Scholar]
- 13.Ehlken, B., B. Laubereau, W. Karmaus, G. Petersen, A. Rohwedder, J. Forster, and RoMoD Study Group. 2002. Prospective population-based study on rotavirus disease in Germany. Acta Paediatr. 91:769-775. [DOI] [PubMed] [Google Scholar]
- 14.Estes, M. K. 2001. Rotaviruses and their replication, p. 1747-1785. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 15.Gault, E., R. Chikhi-Brachet, S. Delon, N. Schnepf, L. Alviges, E. Grimprel, J.-P. Girardet, P. Begue, and A. Garbang-Chenon. 1999. Distribution of human rotavirus G types circulating in Paris, France, during the 1997-1998 epidemic: high prevalence of type G4. J. Clin. Microbiol. 37:2373-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gentsch, J. R., R. I. Glass, P. Woods, V. Gouvea, M. Gorziglia, J. Flores, B. K. Das, and M. K. Bahn. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 30:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerna, G., S. Arista, N. Passarani, A. Saracini, and M. Battaglia. 1987. Electropherotype heterogeneity within serotypes of human rotavirus strains circulating in Italy. Arch. Virol. 95:129-135. [DOI] [PubMed] [Google Scholar]
- 18.Gerna, G., A. Sarasini, A. di Matteo, M. Parea, P. Orsolini, and M. Battaglia. 1988. Identification of two subtypes of serotype 4 human rotavirus by using VP7-specific neutralizing monoclonal antibodies. J. Clin. Microbiol. 26:1388-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ginevskaya, V. A., N. N. Amitina, T. P. Eremeeva, G. A. Shirman, L. S. Priimagi, and S. G. Drozdov. 1994. Electropherotypes and serotypes of human rotavirus in Estonia in 1989-1992. Arch. Virol. 137:199-207. [DOI] [PubMed] [Google Scholar]
- 20.Gouvea, V., R. I. Glass, P. Woods, K. Taniguchi, H. F. Clark, B. Forrester, and Z. Y. Fang. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28:276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green, K. Y., A. Sarasini, Y. Qian, and G. Gerna. 1992. Genetic variation in rotavirus serotype 4 subtypes. Virology 188:362-368. [DOI] [PubMed] [Google Scholar]
- 22.Iturriza-Gòmara, M., J. Green, D. W. G. Brown, U. Desselberger, and J. J. Gray. 2000. Diversity within the VP4 gene of rotavirus P[8] strains: implications for reverse transcription-PCR genotyping. J. Clin. Microbiol. 38:898-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iturriza-Gòmara, M., J. Green, D. W. G. Brown, M. Ramsay, U. Desselberger, and J. J. Gray. 2000. Molecular epidemiology of human group A rotavirus infection in the United Kingdom between 1995 and 1998. J. Clin. Microbiol. 38:4394-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kapikian, A. Z., Y. Hoshino, and R. M. Chanock. 2001. Rotaviruses, p. 1787-1833. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 25.Kirkwood, C., N. Bogdanovic-Sakran, E. Palombo, P. Masendycz, H. Bugg, G. Barnes, and R. Bishop. 2003. Genetic and antigenic characterization of rotavirus serotype G9 strains isolated in Australia between 1997 and 2001. J. Clin. Microbiol. 41:3649-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 27.Maunula, L., and C.-H. von Bonsdorff. 1998. Short sequences define genetic lineages: phylogenetic analysis of group A rotaviruses based on partial sequences of genome segments 4 and 9. J. Gen. Virol. 79:321-332. [DOI] [PubMed] [Google Scholar]
- 28.Maunula, L., and C.-H. von Bonsdorff. 2002. Frequent reassortments may explain the genetic heterogeneity of rotaviruses: analysis of Finnish rotavirus strains. J. Virol. 76:11793-11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Halloran, F., M. Lynch, B. Cryan, H. O'Shea, and S. Fanning. 2000. Molecular characterization of rotaviruses in Ireland: detection of novel strains circulating in the population. J. Clin. Microbiol. 38:3370-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palombo, E. A. 1999. Genetic and antigenic diversity of human rotaviruses: potential impact on the success of candidate vaccines. FEMS Microbiol. Lett. 181:1-8. [DOI] [PubMed] [Google Scholar]
- 31.Parra, G. I., K. Bok, M. Martinez, and J. A. Gomez. 2004. Evidence of rotavirus intragenic recombination between two sublineages of the same genotype. J. Gen. Virol. 85:1713-1716. [DOI] [PubMed] [Google Scholar]
- 32.Sanchez-Fauquier, A., I. Wilhelmi, J. Colomina, E. Cubero, and E. Roman. 2004. Diversity of group A human rotavirus types circulating over a 4-year period in Madrid, Spain. J. Clin. Microbiol. 42:1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taniguchi, K., Y. Hoshino, K. Nishikawa, K. Y. Green, W. L. Maloy, Y. Morita, S. Urasawa, A. Z. Kapikian, R. M. Chanock, and M. Gorziglia. 1988. Cross-reactive and serotype-specific neutralization epitopes on VP7 of human rotavirus: nucleotide sequence analysis of antigenic mutants selected with monoclonal antibodies. J. Virol. 62:1870-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Unicomb, L. E., G. Podder, J. R. Gentsch, P. A. Woods, K. Z. Hasan, A. S. Faruque, M. J. Albert, and R. I. Glass. 1999. Evidence of high-frequency genomic reassortment of group A rotavirus strains in Bangladesh: emergence of type G9 in 1995. J. Clin. Microbiol. 37:1885-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan, L., S. I. Ishida, S. Honma, J. T. Patton, D. Hodgins, A. L. Kapikian, and Y. Hoshino. 2004. Homotypic and heterotypic serum isotype-specific antibody responses to rotavirus nonstructural protein 4 and viral protein (VP) 4, VP6, and VP7 in infants who received selected live oral rotavirus vaccines. J. Infect. Dis. 189:1833-1845. [DOI] [PubMed] [Google Scholar]