Abstract
We describe a novel molecular method for the differentiation and identification of 29 mycobacterial species. The target is the secA1 gene that codes for the essential protein SecA1, a key component of the major pathway of protein secretion across the cytoplasmic membrane. A 700-bp region of the secA1 gene was amplified and sequenced from 47 American Type Culture Collection strains of 29 Mycobacterium species as well as from 59 clinical isolates. Sequence variability in the amplified segment of the secA1 gene allowed the differentiation of all species except for the members of the Mycobacterium tuberculosis (MTB) complex, which had identical sequences. A range of 83.3 to 100% interspecies similarity was observed. All species could also be differentiated by their amino acid sequences as deduced from the sequenced region of the secA1 gene, with the exception of the MTB complex. Partial sequences of secA1 from clinical isolates belonging to nine frequently isolated species of mycobacteria revealed a very high intraspecies similarity at the DNA level (typically >99%; range, 96.0 to 100%); all clinical isolates were correctly identified. Comparison of the deduced 233-amino-acid sequences among clinical isolates of the same species showed between 99.6 and 100% similarity. To our knowledge, this is the first time a secretion-related gene has been used for the identification of the species within a bacterial genus.
The global increase of infections caused by Mycobacterium tuberculosis and nontuberculous mycobacteria (NTM) has received attention worldwide. Traditionally, the definitive diagnosis of mycobacterial infections depends on the isolation and phenotypic identification of the causative agent, a procedure that is time-consuming and labor intensive.
The development of DNA probes for identification of mycobacteria greatly improved the identification speed and accuracy in the mycobacteriology laboratory. The Accuprobe system (Gen-Probe, San Diego, Calif.) is a rapid and sensitive method, but it offers only a limited number of species- or complex-specific probes (39). Recently, a new DNA probe kit, INNO-LiPA Mycobacteria (Innogenetics, Ghent, Belgium), targeting the 16S-23S rRNA spacer region, was developed for the identification of Mycobacterium spp. This assay can presently identify 16 mycobacterial species and was reported to be sensitive (100%) and specific (94.4 to 100%) (37). However, its complex procedure makes it difficult to implement in a clinical laboratory, and it is not available in the United States. The transcription-mediated amplification test (Amplified Mycobacterium Tuberculosis Direct [MTD] test) from Gen-Probe is the first approved commercial method for amplification of mycobacterial nucleic acids directly from respiratory specimens. This method is only useful for detecting members of the M. tuberculosis complex. Compared to culture, the sensitivity of the MTD test is variable, ranging from 65 to 97% (29).
PCR has been used to analyze various mycobacterial genes for diagnostic purposes, including the 16S rRNA gene (4, 26), the 16S-23S internal transcribed spacer (27), the 65-kDa heat shock protein (25, 32), recA (3), rpoB (16), and gyrB (15). The 16S rRNA gene-based methods are presently widely used for the identification and differentiation of mycobacteria (7, 13, 34, 36, 38). However, because the number of polymorphic sites in the 16S rRNA gene in the genus Mycobacterium is rather small, some species cannot be differentiated by their 16S sequences (e.g., M. kansasii and M. gastri), while others possess a very high degree of sequence similarity (e.g., M. marinum and M. ulcerans, M. abscessus and M. chelonae). The sequences of the 16S-23S rRNA internal transcribed spacer can distinguish between M. kansasii and M gastri; however, they fail to distinguish between M. marinum and M. ulcerans (27). The hsp65 gene-based PCR restriction pattern analysis (PRA) is also widely used for the identification of Mycobacterium spp.; however, species with similar patterns, and multiple patterns within a single species, do occur (12, 32, 34). Recently, McNabb et al. assessed the use of partial sequences of the hsp65 gene for routine identification of mycobacteria and reported an overall agreement of 85.2% with other identification methods; discrepancies were most frequently encountered with isolates of M. chelonae, M. fortuitum, M. gordonae, M. scrofulaceum, and M. terrae (18).
Protein export is an important aspect of bacterial pathogenesis, because a majority of bacterial virulence factors are extracytoplasmic proteins (11, 19). Although little is known about the protein export pathway in mycobacteria, the general secretory (Sec) pathway has been extensively studied in other bacteria, Escherichia coli in particular (9, 21). SecA1 is the mycobacterial homologue of the E. coli SecA protein, an essential preprotein translocase ATPase that provides the driving force for the export of proteins across the cytoplasmic membrane. The mycobacterial Sec pathway is unusual in that it has two SecA proteins: SecA1 is the essential housekeeping SecA protein, while SecA2 is a nonessential accessory secretion factor (5, 6, 22). The purpose of this study was to evaluate the use of secA1 gene sequences for the identification of Mycobacterium spp. The secA1 gene sequence of only five Mycobacterium species (M. tuberculosis [AE007144, Z95121], M. bovis [U66080.1], M. leprae [AL583919.1], M. smegmatis [U66081.1], and a partial sequence of M. avium [AF320124.1]) was available in GenBank at the time this study was initiated. We targeted a 700-bp fragment of secA1 coding for 233 amino acid residues located on the N-terminal region of the protein, which includes the substrate specificity domain (SSD) and contiguous sequences essential for protein translocation (2, 28). Because of the critical association between protein secretion and cell wall biosynthesis as well as the particular composition of the mycobacterial cell envelopes (24), we hypothesized that the SecA1 protein would be relatively conserved in the genus Mycobacterium, while exhibiting some amino acid differences among the different species. The present study shows the potential of secA1 gene sequences for the identification of Mycobacterium spp. by using 29 type strains as a foundation. This procedure has also been applied to 18 other reference strains, as well as 59 well-characterized clinical isolates, to demonstrate the feasibility of this identification method. As far as we know, this is the first time a secretion-related gene has been used for the species-level identification of congeneric bacteria.
MATERIALS AND METHODS
Mycobacterial reference strains and clinical isolates.
The 47 reference strains and 59 clinical isolates used in this study are listed in Table 1. Conventional identification of clinical isolates included an initial assessment of pigment production, growth rate, and colony characteristics. Slowly growing mycobacteria were identified using the AccuProbe assays (Gen-Probe), which are specific for M. avium complex (MAC), M. avium, M. intracellulare, M. tuberculosis (MTB) complex, M. gordonae, and M. kansasii. Additional biochemical testing of MTB complex isolates included niacin and nitrate tests. AccuProbe-negative slowly growing isolates were identified by 16S rRNA gene sequencing. Rapidly growing isolates were identified by hsp65 PRA (32) and 16S rRNA gene sequencing when necessary.
TABLE 1.
List of mycobacterial reference strains and clinical isolatesa
| Species | Reference strain(s) and source | No. of clinical isolates |
|---|---|---|
| M. abscessus | ATCC 19977 (T) | 6 |
| M. africanum | ATCC 25420 (T) | |
| ATCC 35711 | ||
| M. asiaticum | ATCC 25276 (T) | 1 |
| M. avium | ATCC 25291 (T) | 5 |
| M. bovis | ATCC 19210 (T) | |
| ATCC 35734 | ||
| M. celatum | ATCC 51131 (T) | |
| ATCC 51130 | ||
| M. chelonae | ATCC 35752 (T) | 2 |
| M. flavescens | ATCC 14474 (T) | |
| ATCC 23008 | ||
| ATCC 23033 | ||
| M. fortuitum | ATCC 6841 (T) | 5 |
| M. gastri | ATCC 15754 (T) | |
| ATCC 25157 | ||
| M. gordonae | ATCC 14470 (T) | 6 |
| M. haemophilum | ATCC 29548 (T) | |
| ATCC 33206 | ||
| M. intracellulare | ATCC 13950 (T) | 6 |
| M. kansasii | ATCC 12478 (T) | 6 |
| M. leprae | AL583919.1 | |
| M. malmoense | ATCC 29571 (T) | 1 |
| M. marinum | ATCC 927 (T) | 5 |
| M. mucogenicum | ATCC 49650 (T) | 5 |
| M. nonchromogenicum | ATCC 19530 (T) | |
| ATCC 25142 | ||
| M. peregrinum | ATCC 14467 (T) | 1 |
| M. scrofulaceum | ATCC 19981 (T) | 1 |
| M. shimoidei | ATCC 27962 (T) | |
| ATCC 49773 | ||
| M. simiae | ATCC 25275 (T) | 1 |
| M. smegmatis | ATCC 19420 (T) | |
| ATCC 14468 | ||
| M. szulgai | ATCC 35799 (T) | 1 |
| M. terrae | ATCC 15755 (T) | |
| ATCC 25267 | ||
| ATCC 25268 | ||
| M. triviale | ATCC 23292 (T) | |
| ATCC 23290 | ||
| ATCC 23291 | ||
| M. tuberculosis | ATCC 27294 (T) | 6 |
| ATCC 25177 | ||
| M. ulcerans | ATCC 19423 (T) | |
| ATCC 25900 | ||
| ATCC 25897 | ||
| ATCC 35839 | ||
| M. xenopi | ATCC 19250 (T) | 1 |
T, type strain.
Nonmycobacterial reference strains.
Seven American Type Culture Collection (ATCC) nonmycobacterial strains were also included in this study: Gordonia bronchialis [ATCC 25592 (T)], Gordonia terrae [ATCC 25594 (T)], Nocardia brasiliensis [ATCC 19296 (T)], Nocardia farcinica [ATCC 3318 (T)], Rhodococcus equi [ATCC 6939 (T)], Rhodococcus rhodochrous [ATCC 13808 (T)], and Tsukamurella paurometabola [ATCC 8368 (T)].
DNA extraction.
Mycobacterial cells were disrupted by an alkaline wash and heat lysis method (1), with a few modifications. One small loopful of organisms grown on Middlebrook 7H11 agar (Remel, Lenexa, Kans.) for 7 to 21 days was resuspended in 20 μl of lysis buffer (0.25% sodium dodecyl sulfate [QB Inc., Gaithersburg, Md.], 0.05 N NaOH [Sigma, St. Louis, Mo.]), mixed by vortexing, and heated at 95°C for 5 min. After the addition of 180 μl of water and mixing by vortexing, samples were incubated at 95°C for 25 min. Samples were then spun at 16,000 × g for 5 min to remove cell debris. Nucleic acid concentration in the supernatant was determined spectrophotometrically (A260). DNA extraction from the nonmycobacterial reference strains was performed by a phenol-chloroform-isoamyl alcohol method as described elsewhere (8).
Primer design and PCR.
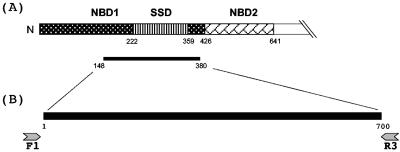
Two conserved regions were identified after aligning secA1 sequences from five Mycobacterium spp. which were available in the National Center for Biotechnology Information GenBank database at the time this study was initiated. These regions were used to design two primers (Mtu.Forward1 and Mtu.Reverse3), which correspond to M. tuberculosis secA1 gene positions 412 to 440 and 1141 to 1172, respectively (GenBank accession no. BX842582.1, nucleotides 150019 to 152868). A schematic representation of the primer design is shown in Fig. 1.
FIG. 1.
Schematic representation of the 649-amino-acid N-terminal region of the SecA1 protein and primer design for the amplification of the mycobacterial secA1 gene. (A) The SecA1 protein from M. tuberculosis contains two nucleotide-binding domains (NBD1 and NBD2) and a substrate specificity domain (SSD). The SSD is embedded in NBD1. The thin black bar shows the region of the protein coded by the secA1 gene region targeted in the assay. Numbers indicate amino acid residues. (B) Primers Mtu.Forward1 (F1) and Mtu.Reverse3 (R3) were used to generate a 700-bp fragment from the secA1 gene. Numbers indicate the nucleotide position in the amplified fragment.
Mtu.Forward1 (5′-GAC AGY GAG TGG ATG GGY CGS GTG CAC CG-3′) and Mtu.Reverse3 (5′-ACC ACG CCC AGC TTG TAG ATC TCG TGC AGC TC-3′) were commercially synthesized (Midland Certified Reagent Company, Midland, Tex.). Primers used for sequencing of the secA1 gene regions were tailed with the M13 sequencing primer sites M13 forward tail (5′-GTA AAA CGA CGG CCA G-3′) and M13 reverse tail (5′-CAG GAA ACA GC TAT GAC-3′). The PCR amplification using Mtu.Forward1 (M13 tailed) and Mtu.Reverse3 (M13 tailed) generated a product of 700 bp (excluding the primers; see Fig. 1). PCRs were performed in a Perkin-Elmer 9600 Thermocycler with a reaction mix containing 2.5 mM MgCl2, 1× LightCycler-Fast Start DNA Hybridization Master Probes (Roche, Mannheim, Germany), 1 pmol of forward primer, 1 pmol of reverse primer, 1 U of uracil N-glycosylase (UNG) enzyme (Roche), 5 μl of extracted DNA, and ultrapure water to a final volume of 25 μl. The PCR thermocycling program consisted of an initial step of 10 min of incubation at 30°C (UNG), followed by 10 min at 95°C and then 49 cycles of 1 min at 95°C, 1 min at 65°C, 1 min at 72°C, and a final incubation step of 10 min at 72°C. A negative control of ultrapure water was included with every amplification reaction mixture.
DNA samples of selected strains were also amplified and sequenced for the 16S rRNA gene by using the MicroSeq Full Gene 16S rRNA Bacterial Isolation Sequencing kit (Applied Biosystems, Foster City, Calif.) according to the manufacturer's protocol.
PCR product detection and purification.
PCR products were visualized by UV illumination of an ethidium bromide-stained 2% agarose gel following electrophoresis. The purification of the remaining PCR product was achieved with Microcon-100 microconcentrators (Millipore, Bedford, Mass.), following the manufacturer's instructions.
DNA sequencing.
The ABI Prism BigDye Terminator v1.1 Cycle Sequencing Ready Reaction kit (Applied Biosystems) was used for the sequencing of the PCR product. The sequencing reaction mixture contained 4 μl of Big Dye premix, 0.5× buffer, 3.2 pmol of sequencing primer, and approximately 150 ng of PCR product template in a total volume of 20 μl. The following M13 primers were used for sequencing: M13 Forward, 5′-GTA AAA CGA CGG CCA G-3′; M13 Reverse, 5′-CAG GAA ACA GCT ATG AC-3. The sequencing reaction and template preparation were performed in accordance with the instructions of the manufacturer. Sequencing products were purified with a CleanSEQ Sequencing Reaction Clean-Up system (Agencourt, Beverly, Mass.) and were analyzed with the 3100 Genetic Analyzer (Applied Biosystems), following the manufacturer's instructions.
Sequence and phylogenetic analysis.
The Lasergene program (version 5.51; DNASTAR, Inc. Madison, Wis.) was used for sequence assembly and alignment. Multiple-sequence alignment of secA1 700-bp sequences was done by the CLUSTAL W method (33). Phylogenetic analyses were performed with the PHYLIP, version 3.5c, package (10). Distance matrices based on Kimura's two-parameter model (17) were produced with the DNADIST program, and a neighbor-joining tree was constructed with the NEIGHBOR program. The resulting trees were depicted using the TreeView, version 1.4, package (23). The stability of the grouping was assessed by the bootstrap method using SEQBOOT, DNADIST, NEIGHBOR, and CONSENSE programs. A total of 1,000 bootstrapped trees were generated.
Assembled 16S rRNA sequences from selected isolates were compared with 16S rRNA sequences available in GenBank databases by using the standard nucleotide-nucleotide Basic Local Alignment Search Tool (BLAST) program (National Center for Biotechnology Information, Bethesda, Md.). The isolate was identified as most closely related to the reference species if its 16S rRNA sequence demonstrated the highest relatedness and had over 98.5% identical bases compared to the respective reference sequence, based on the data from a previous study (30).
Nucleotide sequence accession numbers.
Partial sequences of mycobacterial and nocardial secA1 genes were deposited in GenBank under accession numbers AY724701 to AY724734 and AY781799 to AY781800, respectively. 16S rRNA sequences were deposited in GenBank under accession numbers AY734991 to AY734996.
RESULTS
SecA1 partial sequences of the mycobacterial type strains.
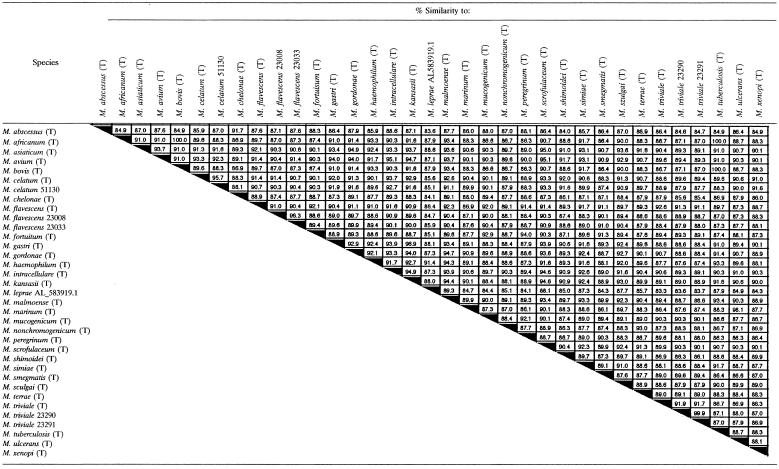
A 700-bp section of secA1 was amplified from 29 Mycobacterium type strains, and the nucleotide sequences were determined and compared pairwise. No insertions or deletions were detected. A range of 83.3 to 100% interspecies similarity was observed (Table 2). Sequence variability occurred throughout the 700-bp region targeted. However, a particularly hypervariable region was observed at the beginning of the 700-bp region, between nucleotides 7 and 70. The members of the MTB complex (M. tuberculosis, M. africanum, and M. bovis) had identical sequences. Members of the following closely related species could be differentiated: M. gastri and M. kansasii (96.9% similarity), M. abscessus and M. chelonae (91.7% similarity), and M. marinum and M. ulcerans (98.1%).
TABLE 2.
Interspecies similarity of partial secA1 gene sequences (700 bp)a
T, type strain; five-digit number, ATCC number; Genbank accession number AL583919.1 used for M. leprae.
No amplification of the secA1 gene was observed with the negative control, which was included with every amplification reaction.
Amino acid sequences of the mycobacterial type strains.
The deduced amino acid sequences of the amplified 700-bp fragment of secA1 comprised 233 amino acid residues (F148 to A375 according to M. tuberculosis numbering; protein accession no. CAE55574.1). Each type strain examined displayed unique amino acid sequences in this region, except for the members of the MTB complex, which shared the same protein sequence. Interspecies similarity at the amino acid level ranged from 87.6 to 100% (100% for the members of the MTB complex). High variability was observed in the first half of the 233-deduced-amino-acid sequence, with three particularly hypervariable regions between residues 5 to 23, 46 to 57, and 84 to 118. The following pairs of closely related species could also be differentiated from one another at the protein level: M. gastri and M. kansasii (99.1% similarity), M. abscessus and M. chelonae (96.6% similarity), and M. marinum and M. ulcerans (98.3% similarity).
Evaluation of the secA1 identification procedure on nonmycobacterial reference strains.
In addition to amplifying and sequencing the secA1 gene from mycobacteria, we tested seven nonmycobacterial isolates in genera considered related to the genus Mycobacterium. No amplification of secA1 was observed with the type strains of Gordonia bronchialis, Gordonia terrae, Rhodococcus equi, Rhodococcus rhodochrous, and Tsukamurella paurometabola. In contrast, amplification of a 700-bp region of secA1 was observed with the type strains of Nocardia brasiliensis and Nocardia farcinica. Sequencing of the amplified product allowed a clear differentiation of the Nocardia isolates from each other and from all Mycobacterium spp. strains tested. The ranges of similarity between N. brasiliensis and the type strains of Mycobacterium spp. were 82.6 to 87.6% and 82.4 to 88.8% for nucleotide and amino acid sequences, respectively. For N. farcinica these values were 82.0 to 89.4% and 83.7 to 89.7%, respectively.
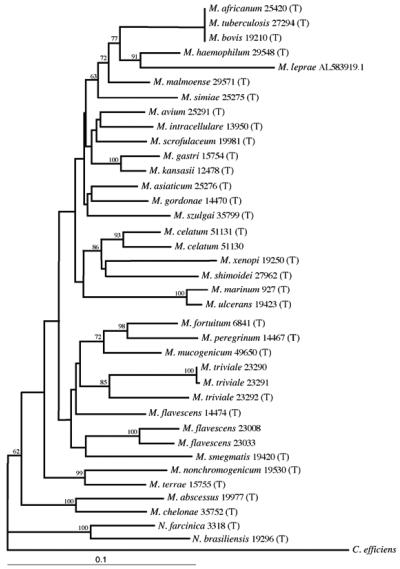
Phylogenetic tree.
A phylogenetic tree of 34 sequenced mycobacterial reference strains, as well as M. leprae, and two sequenced nocardial reference strains was constructed by the neighbor-joining method, using Corynebacterium efficiens as the outgroup (Fig. 2). All mycobacterial species studied showed good separation. Within the consensus tree, four major clusters of mycobacteria could be defined. Most of the slow-growing species grouped together in the first cluster. The rapid growers M. flavescens, M. fortuitum, M. mucogenicum, M. peregrinum, and M. smegmatis grouped together in the second cluster. Interestingly, M. triviale also appeared in that group. A third cluster included the closely related species M. terrae and M. nonchromogenicum. Finally, a long separate branch included M. abscessus and M. chelonae.
FIG. 2.
Phylogenetic tree derived from secA1 sequences from 34 mycobacterial reference strains and M. leprae (GenBank accession no. AL583919.1) and two nocardial reference strains. The tree was constructed by the neighbor-joining method, using Corynebacterium efficiens as the outgroup (GenBank accession no. AP005216.1). T, type strain; the 3- to 5-digit numbers are ATCC numbers. Numbers on the branches represent the percentage of 1,000 bootstrap samples supporting the branch; only values greater than 50% are shown.
The members of the M. tuberculosis complex were distantly separated from all other species. Slowly growing pathogenic M. kansasii and nonpathogenic M. gastri could be easily distinguished, although they cannot be separated by 16S rRNA. Two other pairs of species closely related by 16S rRNA sequences, M. abscessus-M. chelonae and M. marinum-M. ulcerans, were separated as well.
N. brasiliensis and N. farcinica clustered together as an independent branch, which was clearly separated from all the mycobacteria. In addition, the two Nocardia species could be easily differentiated from one another.
Correlation of additional mycobacterial isolates with the type strains.
In addition to the type strains, the 700-bp fragment of secA1 was sequenced from one or more reference strains or well-characterized clinical isolates (Table 1). Five or six clinical isolates were obtained and sequenced from each of nine commonly isolated species of mycobacteria (Table 1 and separately in Table 3).
TABLE 3.
Range of similarity to the type strain of 50 clinical isolates from nine commonly isolated Mycobacterium spp.
| Species | No. of clinical isolates | Range of % similarity (DNA) | Range of % similarity (amino acid) |
|---|---|---|---|
| M. abscessus | 6 | 100 | 100 |
| M. avium | 5 | 98.4-99.1 | 99.6-100 |
| M. fortuitum | 5 | 99.0-99.9 | 100 |
| M. gordonae | 6 | 96.0-99.0 | 99.6-100 |
| M. intracellulare | 6 | 99.4-100 | 100 |
| M. kansasii | 6 | 97.3-100 | 99.6-100 |
| M. marinum | 5 | 99.1-100 | 99.6-100 |
| M. mucogenicum | 5 | 99.4-100 | 100 |
| M. tuberculosis | 6 | 100 | 100 |
For species of mycobacteria listed in Table 1 only (not in Table 3), the level of similarity with respective type strains was typically higher than 99%. All isolates were correctly identified by the secA1 gene and also clustered with their respective type strains in the phylogenetic tree (data not shown), with the exception of two reference strains of M. flavescens (ATCC 23008 and 23033) that appeared more closely related to M. smegmatis (Fig. 2). These two M. flavescens reference strains are more closely related to each other (96.3% similarity) than to the type strain (91.0 and 90.4%) according to their secA1 gene sequences (Table 2, Fig. 2). To compare the results obtained by secA1 with 16S rRNA sequences, we performed sequencing of the full 16S rRNA gene on the three M. flavescens reference strains used in this study. 16S rRNA gene sequencing agreed with the sequences appearing in GenBank for those isolates. The percentage of similarity among the three species was lower than one would expect for 16S rRNA sequences of isolates of the same species (range, 98.1 to 98.4%). Similarly, although all three reference strains of M. triviale grouped together in a separate branch in the phylogenetic tree (Fig. 2), secA1 sequences of the M. triviale ATCC 23290 and 23291 isolates were almost identical to each other (99.9% similarity) but quite divergent from that of the type strain (91.9 and 91.7% similarity, respectively). Interestingly, 16S rRNA sequences determined in our laboratory confirmed these findings, with 99.9% similarity between M. triviale ATCC 23290 and 23291 but only 98.3% similarity of each of them with the type strain.
To evaluate the performance of our method for the identification of mycobacteria, 50 clinical isolates were obtained and sequenced from nine species of Mycobacterium (Table 3). These organisms were selected as representative of the most commonly isolated clinical species. In general, few nucleotide differences were observed between the clinical isolates of a given species and the corresponding type strain. The intraspecies percentage of similarity at the DNA level was very high (usually >99%; range, 96.0 to 100%). Comparison of the deduced amino acid sequences among isolates of the same species showed between 99.6 and 100% similarity for all species tested.
M. gordonae showed the highest degree of intraspecies variability, with percentages of similarity at the DNA level ranging from 96.0 to 99.0; however, all six clinical isolates of M. gordonae tested matched most closely with the type strain. Four clinical isolates of M. kansasii showed 100% similarity to the type strain, while two other isolates had identical sequences showing 97.3% similarity to the type strain. However, all six clinical isolates of M. kansasii were correctly identified.
DISCUSSION
Despite the close relationship between secretion and pathogenesis, the secA genes of Mycobacterium species have not been studied extensively. The secA1 sequence of only five species of mycobacteria was available in GenBank prior to this publication. For this work, the type strain was acquired for every species sequenced, with the exception of M. leprae (whose secA1 sequence was retrieved from GenBank). In addition, one or more additional strains were obtained for each of the species studied (Table 1). A 700-bp section of secA1 was amplified from 29 Mycobacterium spp. type strains by using primers Mtu.Forward1 and Mtu.Reverse3. In contrast to what was observed with other genes, such as recA (3), the size of the amplified fragment was identical among all isolates, making sequence alignment straightforward. Sequence analysis of the secA1 gene from 30 species of mycobacteria revealed the presence of a large number of nucleotide substitutions. DNA sequences allowed the differentiation at the species level for all species studied, with the exception of the members of the MTB complex, which showed identical sequences. Although the primers used in this study also amplified the secA1 gene of the two species of Nocardia tested, sequencing enabled the correct species assignment in both genera.
The overall nucleic acid similarities among the mycobacterial reference strains ranged from 83.3 to 100%. A particularly hypervariable region was observed at the beginning of the 700-bp region, between nucleotides 7 and 70 (corresponding to 447 to 510 of M. tuberculosis secA1). Many of the nucleotide substitutions generated actual differences in the amino acid sequence, in contrast to what was observed for recA (3) and rpoB (16) but similar to what was reported for gyrB (15). In fact, each of the species studied displayed unique amino acid sequences, with the exception of the members of the MTB complex.
High variability in protein sequence was observed in the first half of the 233-deduced-amino-acid sequence, with three particularly hypervariable regions between residues 5 to 23, 46 to 59, and 84 to 118 (corresponding to residues 152 to 170, 193 to 205, and 231 to 265 of MTB SecA1). Interestingly, the first two hypervariable regions are located inside nucleotide binding domain 1, between the two proposed ATP-binding Walker A and B motifs (40), which are characteristic of ATP-binding helicases (14) and essential for SecA function (20). The third hypervariable region is located in the N terminus of the substrate specificity domain that is essential for protein translocation and is unique to SecA (2, 28).
Comparison of the results of secA1 gene-based sequence analysis to those obtained with 16S rRNA gene-based sequence analysis (38) revealed a similarity in the relative position of each species within the tree (Fig. 2), with a general separation of rapid growers from slow growers. However, some differences were observed: M. triviale, a slow grower and member of the M. terrae complex, clustered with the rapid growers. The two rapid growers M. abscessus and M. chelonae, which form a separate branch among the rapid growers by 16S rRNA, showed enough differences in their secA1 sequences to form an early branch separated from all other species in this study. Excluding the members of the MTB complex, there was greater variability among the species by secA1 sequences that was manifested by longer branches in the tree. While 16S rRNA gene sequence analysis failed to distinguish between M. gastri and M. kansasii, these species could be differentiated by their secA1 sequences with high bootstrap values. Similarly, secA1 sequences allowed the differentiation between M. abscessus and M. chelonae and between M. marinum and M. ulcerans, while 16S sequences of the members of each pair are almost identical.
In general, the level of intraspecies divergence based on secA1 sequences was less than 1.0% (>99% similarity). Three particular cases showing a higher percentage of divergence are worth mentioning. The two reference strains of M. celatum [ATCC 51131 (T) and ATCC 51130] showed only 95.7% similarity (4.3% divergence); however, a low value of similarity (94.1%) has already been reported for these two strains based on partial sequences of rpoB (16). Analysis of secA1 sequences from three reference strains of M. triviale showed that two strains (ATCC 23290 and 23291) were almost identical to each other (99.9% similarity) but were quite divergent from that of the type strain (91.9 and 91.7% similarity, respectively). Interestingly, 16S rRNA sequences performed in our laboratory confirmed these findings, with 99.9% similarity between M. triviale ATCC 23290 and 23291 but only 98.3% similarity of each of them to the type strain.
Because of the high interspecies variability, which is evident by the long branches on the phylogenetic tree, even members of species that showed a higher degree of divergence could be assigned to the correct species, with the exception of the two reference strains of M. flavescens (ATCC 23008 and 23033), which appeared more closely related to the M. smegmatis type strain (Fig. 2). The percentages of similarity among the three reference strains of M. flavescens were quite low, with two strains (ATCC 23008 and 23033) appearing more related to one another (96.3% similarity) than to the type strain (91.0 and 90.4%). Interestingly, analysis of 16S rRNA gene sequences performed in our laboratory also showed low percentages of similarity among the three strains (range, 98.1 to 98.4%), which suggest the presence of genomic heterogeneity in the group. In agreement with that is the presence of two very distinct profiles for M. flavescens shown by restriction enzyme analysis of the hsp65 gene (32). Further study is needed to determine whether all three isolates would in fact be found to belong to the same species by DNA-DNA hybridization.
To evaluate the performance of our method in the clinical laboratory, we sequenced the 700-bp fragment of secA1 from 50 clinical isolates from nine frequently isolated species of mycobacteria (Table 3). DNA sequences were compared to one another and also to the sequences of the corresponding type strains. In addition, we also compared the deduced amino acid sequences. All clinical isolates were correctly identified at the species or complex level (for members of the MTB complex). The clinical isolates of the M. avium complex were divided into M. avium and M. intracellulare. Interestingly, five clinical isolates positive by the M. avium complex (MAC) Accuprobe but negative by either the M. avium or M. intracellulare probes (possibly members of the so-called X-cluster) grouped close to the M. intracellulare type strain, as previously reported (41) (data not shown).
In general, the intraspecies variation among the strains in each species was very low (intraspecies divergence, <1%). Higher intraspecies variability was observed with M. kansasii and M. gordonae, two species known for their high genetic heterogeneity. At least seven and eight different subtypes have been described for M. kansasii and M gordonae, respectively (12, 31, 32, 35). Nonetheless, all clinical isolates were correctly identified by our method. Given the data that we have obtained so far with the secA1 gene, well-characterized strains belonging to the same species show ≥97% similarity at the nucleotide level and ≥99.6% similarity at the amino acid level. Further work with additional species and with a larger number of isolates belonging to the species already studied will be necessary to determine with greater confidence the cutoff values for species assignment.
In this initial study, we have demonstrated that a 700-bp fragment of the secA1 gene of the genus Mycobacterium can be used for species-level or complex-level (for the MTB complex) identification. The degree of interspecies variation for secA1 gene sequences among mycobacteria was observed to be moderate, which makes this target suitable for diagnostic purposes. In addition to determining secA1 sequences of additional species and isolates as noted above, future work will also focus on the development and evaluation of a real-time PCR assay for direct testing of clinical samples.
Acknowledgments
We thank the technologists in the Mycology and Mycobacteriology section of the Microbiology Service, Department of Laboratory Medicine, Clinical Center, NIH, for technical assistance. We thank Gary A. Fahle and Charles Huber for their useful suggestions. We thank Patricia S. Conville for providing us with genomic DNA from the nonmycobacterial reference strains and also, along with Frank G. Witebsky and Patrick R. Murray, for critically reviewing the manuscript.
REFERENCES
- 1.Baele, M., V. Storms, F. Haesebrouck, L. A. Devriese, M. Gillis, G. Verschraegen, T. de Baere, and M. Vaneechoutte. 2001. Application and evaluation of the interlaboratory reproducibility of tRNA intergenic length polymorphism analysis (tDNA-PCR) for identification of Streptococcus species. J. Clin. Microbiol. 39:1436-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baud, C., S. Karamanou, G. Sianidis, E. Vrontou, A. S. Politou, and A. Economou. 2002. Allosteric communication between signal peptides and the SecA protein DEAD motor ATPase domain. J. Biol. Chem. 277:13724-13731. [DOI] [PubMed] [Google Scholar]
- 3.Blackwood, K. S., C. He, J. Gunton, C. Y. Turenne, J. Wolfe, and A. M. Kabani. 2000. Evaluation of recA sequences for identification of Mycobacterium species. J. Clin. Microbiol. 38:2846-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boddinghaus, B., T. Rogall, T. Flohr, H. Blocker, and E. C. Bottger. 1990. Detection and identification of mycobacteria by amplification of rRNA. J. Clin. Microbiol. 28:1751-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braunstein, M., A. M. Brown, S. Kurtz, and W. R. Jacobs, Jr. 2001. Two nonredundant SecA homologues function in mycobacteria. J. Bacteriol. 183:6979-6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braunstein, M., B. J. Espinosa, J. Chan, J. T. Belisle, and W. R. Jacobs, Jr. 2003. SecA2 functions in the secretion of superoxide dismutase A and in the virulence of Mycobacterium tuberculosis. Mol. Microbiol. 48:453-464. [DOI] [PubMed] [Google Scholar]
- 7.Cloud, J. L., H. Neal, R. Rosenberry, C. Y. Turenne, M. Jama, D. R. Hillyard, and K. C. Carroll. 2002. Identification of Mycobacterium spp. by using a commercial 16S ribosomal DNA sequencing kit and additional sequencing libraries. J. Clin. Microbiol. 40:400-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conville, P. S., S. H. Fischer, C. P. Cartwright, and F. G. Witebsky. 2000. Identification of Nocardia species by restriction endonuclease analysis of an amplified portion of the 16S rRNA gene. J. Clin. Microbiol. 38:158-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Economou, A. 1998. Bacterial preprotein translocase: mechanism and conformational dynamics of a processive enzyme. Mol. Microbiol. 27:511-518. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 1993. PHYLIP (phylogeny inference package), version 3.5c. Department of Genetics, University of Washington, Seattle.
- 11.Finlay, B. B., and S. Falkow. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61:136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hafner, B., H. Haag, H. K. Geiss, and O. Nolte. 2004. Different molecular methods for the identification of rarely isolated non-tuberculous mycobacteria and description of new hsp65 restriction fragment length polymorphism patterns. Mol. Cell Probes 18:59-65. [DOI] [PubMed] [Google Scholar]
- 13.Hall, L., K. A. Doerr, S. L. Wohlfiel, and G. D. Roberts. 2003. Evaluation of the MicroSeq system for identification of mycobacteria by 16S ribosomal DNA sequencing and its integration into a routine clinical mycobacteriology laboratory. J. Clin. Microbiol. 41:1447-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall, M. C., and S. W. Matson. 1999. Helicase motifs: the engine that powers DNA unwinding. Mol. Microbiol. 34:867-877. [DOI] [PubMed] [Google Scholar]
- 15.Kasai, H., T. Ezaki, and S. Harayama. 2000. Differentiation of phylogenetically related slowly growing mycobacteria by their gyrB sequences. J. Clin. Microbiol. 38:301-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, B. J., S. H. Lee, M. A. Lyu, S. J. Kim, G. H. Bai, G. T. Chae, E. C. Kim, C. Y. Cha, and Y. H. Kook. 1999. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB). J. Clin. Microbiol. 37:1714-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 18.McNabb, A., D. Eisler, K. Adie, M. Amos, M. Rodrigues, G. Stephens, W. A. Black, and J. Isaac-Renton. 2004. Assessment of partial sequencing of the 65-kilodalton heat shock protein gene (hsp65) for routine identification of Mycobacterium species isolated from clinical sources. J. Clin. Microbiol. 42:3000-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, J. F., and P. Cossart. 1999. Bacterial pathogenesis: before the post-genomic era. Curr. Opin. Microbiol. 2:15-17. [Google Scholar]
- 20.Mitchell, C., and D. Oliver. 1993. Two distinct ATP-binding domains are needed to promote protein export by Escherichia coli SecA ATPase. Mol. Microbiol. 10:483-497. [DOI] [PubMed] [Google Scholar]
- 21.Mori, H., and K. Ito. 2001. The Sec protein-translocation pathway. Trends Microbiol. 9:494-500. [DOI] [PubMed] [Google Scholar]
- 22.Owens, M. U., W. E. Swords, M. G. Schmidt, C. H. King, and F. D. Quinn. 2002. Cloning, expression, and functional characterization of the Mycobacterium tuberculosis secA gene. FEMS Microbiol. Lett. 211:133-141. [DOI] [PubMed] [Google Scholar]
- 23.Page, R. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 24.Pfyffer, G. E., B. A. Brown-Elliot, and J. Wallace. 2003. Mycobacterium: general characteristics, isolation, and staining procedures, p. 532-559. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. American Society for Microbiology, Washington, D.C.
- 25.Ringuet, H., C. Akoua-Koffi, S. Honore, A. Varnerot, V. Vincent, P. Berche, J. L. Gaillard, and C. Pierre-Audigier. 1999. hsp65 sequencing for identification of rapidly growing mycobacteria. J. Clin. Microbiol. 37:852-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogall, T., T. Flohr, and E. C. Bottger. 1990. Differentiation of Mycobacterium species by direct sequencing of amplified DNA. J. Gen. Microbiol. 136:1915-1920. [DOI] [PubMed] [Google Scholar]
- 27.Roth, A., M. Fischer, M. E. Hamid, S. Michalke, W. Ludwig, and H. Mauch. 1998. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S-23S rRNA gene internal transcribed spacer sequences. J. Clin. Microbiol. 36:139-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma, V., A. Arockiasamy, D. R. Ronning, C. G. Savva, A. Holzenburg, M. Braunstein, W. R. Jacobs, Jr., and J. C. Sacchettini. 2003. Crystal structure of Mycobacterium tuberculosis SecA, a preprotein translocating ATPase. Proc. Natl. Acad. Sci. USA 100:2243-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sloutsky, A., L. L. Han, and B. G. Werner. 2004. Practical strategies for performance optimization of the enhanced Gen-Probe amplified Mycobacterium tuberculosis direct test. J. Clin. Microbiol. 42:1547-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 31.Taillard, C., G. Greub, R. Weber, G. E. Pfyffer, T. Bodmer, S. Zimmerli, R. Frei, S. Bassetti, P. Rohner, J. C. Piffaretti, E. Bernasconi, J. Bille, A. Telenti, and G. Prod'hom. 2003. Clinical implications of Mycobacterium kansasii species heterogeneity: Swiss national survey. J. Clin. Microbiol. 41:1240-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Bottger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tortoli, E. 2003. Impact of genotypic studies on mycobacterial taxonomy: the new mycobacteria of the 1990s. Clin. Microbiol. Rev. 16:319-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tortoli, E. 2003. Mycobacterium kansasii, species or complex? Biomolecular and epidemiological insights. Kekkaku 78:705-709. [PubMed] [Google Scholar]
- 36.Tortoli, E., A. Bartoloni, E. C. Bottger, S. Emler, C. Garzelli, E. Magliano, A. Mantella, N. Rastogi, L. Rindi, C. Scarparo, and P. Urbano. 2001. Burden of unidentifiable mycobacteria in a reference laboratory. J. Clin. Microbiol. 39:4058-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tortoli, E., A. Mariottini, and G. Mazzarelli. 2003. Evaluation of INNO-LiPA MYCOBACTERIA v2: improved reverse hybridization multiple DNA probe assay for mycobacterial identification. J. Clin. Microbiol. 41:4418-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turenne, C. Y., L. Tschetter, J. Wolfe, and A. Kabani. 2001. Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J. Clin. Microbiol. 39:3637-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vincent, V., B. A. Brown-Elliot, J. Jost, and J. Wallace. 2003. Mycobacterium: phenotypic and genotypic identification, p. 560-584. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of Clinical Microbiology, 8th ed. American Society for Microbiology, Washington, D.C.
- 40.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wayne, L. G., R. C. Good, E. C. Bottger, R. Butler, M. Dorsch, T. Ezaki, W. Gross, V. Jonas, J. Kilburn, P. Kirschner, M. I. Krichevsky, M. Ridell, T. M. Shinnick, B. Springer, E. Stackebrandt, I. Tarnok, Z. Tarnok, H. Tasaka, V. Vincent, N. G. Warren, C. A. Knott, and R. Johnson. 1996. Semantide- and chemotaxonomy-based analyses of some problematic phenotypic clusters of slowly growing mycobacteria, a cooperative study of the International Working Group on Mycobacterial Taxonomy. Int. J. Syst. Bacteriol. 46:280-297. [DOI] [PubMed] [Google Scholar]