Abstract
The aroA gene of Aeromonas hydrophila SO2/2, encoding 5-enolpyruvylshikimate 3-phosphate synthase, was cloned by complementation of the aroA mutation in Escherichia coli K-12 strain AB2829, and the nucleotide sequence was determined. The nucleotide sequence of the A. hydrophila aroA gene encoded a protein of 440 amino acids which showed a high degree of homology to other bacterial AroA proteins. To obtain an effective attenuated live vaccine against A. hydrophila infections in fish, the aroA gene was inactivated by the insertion of a DNA fragment containing a kanamycin resistance determinant and reintroduced by allelic exchange into the chromosome of A. hydrophila AG2 by means of the suicide vector pSUP202. The A. hydrophila mutant AG2 aroA::Kar was highly attenuated when inoculated intraperitoneally into a rainbow trout, with a 50% lethal dose of >2 × 108 CFU. The mutants were not recoverable from the internal organs after 48 h postinoculation. Immunohistochemical studies demonstrated that immunopositive materials, but not whole cells, reacting with a polyclonal antiserum against A. hydrophila were present in the kidney and spleen 9 days postinjection. Vaccination of rainbow trout with the AroA mutant as a live vaccine conferred significant protection against the wild-type strain of A. hydrophila.
Aeromonas hydrophila is a gram-negative, facultatively anaerobic freshwater bacterium that causes disease in humans and terrestrial and aquatic animals. In humans it causes soft tissue wound infections and diarrheal disease (1, 15, 19), and in fish it affects several species, where it causes fatal hemorrhagic septicemia. However, the principal concern is in the intensive culture of salmonids, since the organism can inflict severe losses and can present a risk of infection not only for the fish but also for human handlers and consumers (3, 9, 13, 32).
Acute hemorrhagic septicemia is a systemic disease which may produce swelling of the body cavity and hemorrhage of organs. Mortality may occur with no external signs, but there may be localized infections at sites of injury. Disease symptoms can be induced in healthy rainbow trout by inoculating them with the microorganism or by injecting them with partially purified extracellular products (18). The pathogenicity of the organism may involve several extracellular products including proteases, hemolysins, enterotoxins, acetylcholinesterase, and a surface array protein layer (S layer). Some of the toxins have been isolated and biochemically characterized, but their roles in the pathogenesis of A. hydrophila have not been determined (8, 23, 30, 37, 38, 40).
Recently, there has been increasing interest in the use of live attenuated vaccines against fish bacterial pathogens, and some of them have been used with success in several fish farm trials (26, 31, 46, 47). The interest in constructing attenuated bacterial strains as vaccine candidates can be attributed to the superior protection afforded by live vaccines. In general, live vaccines elicit a stronger cell-mediated response than bacterins do (26), while the greater immunity provided by attenuated organisms compared with that provided by dead bacteria may be explained by the induced expression of stress proteins and, possibly, of certain abundant toxins in aeromonads within the host. The introduction of certain auxotrophic mutations in genes such as aroA, whose function is essential for bacteria to survive and grow in vivo and thus cause disease, produces attenuated organisms. Attenuated strains of the invasive bacteria Salmonella typhi (5), Salmonella typhimurium (14), Shigella flexneri (48), Yersinia enterocolitica (7), and Aeromonas salmonicida (47) were generated by introducing mutations in their respective aroA genes. Attenuation was also produced in this way in some noninvasive bacteria such as Bordetella pertussis (39), Pasteurella multocida (16), Pasteurella haemolytica (44), and Bacillus anthracis (17). The aroA mutant strains fail to grow in tissue because they are unable to synthesize chorismic acid, from which p-aminobenzoic acid, aromatic amino acids, and folate are produced. p-Aminobenzoic acid is required for folate biosynthesis, and exogenous folate cannot be taken up.
At present, no vaccines for protection of farmed fish against A. hydrophila infections are commercially available, although several studies have proved that injection or immersion vaccination with heat- or formalin-inactivated bacterins may provide protection (20, 29) and some researchers have described the responses of rainbow trout (Oncorhynchus mykiss) to immunization with live A. hydrophila organisms (24). A major problem for the development of commercial vaccines is the antigenic diversity of A. hydrophila strains (43), which may necessitate the development of vaccines containing the specific strains causing a disease outbreak in a particular geographical area. This problem may be overcome by using vaccines containing common antigens able to induce protection, but this has proved to be difficult, since procedures (heat or formalin inactivation, cell rupture) for the preparation of bacterins produce significant alterations of the antigens (43). The use of live attenuated vaccines may provide a rational solution to the problem, since they allow the transitory expression of a full range of protective antigens. In this paper, we report the molecular cloning and sequencing of the A. hydrophila aroA gene, the construction of an A. hydrophila aroA-deficient mutant by allelic replacement, and a demonstration of its applicability as an attenuated vaccine in fish.
MATERIALS AND METHODS
Bacteria, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. A. hydrophila strains were grown on Luria broth (LB) or Luria agar (LA) (27) or tryptic soy agar or broth (Biolife). Escherichia coli strains were grown in LB or LA. M9 minimal medium (41), used for E. coli, contained phosphate buffer, 1 mM MgSO4, 0.1 mM CaCl2, 0.1 mM MnSO4, 0.01 mM FeCl3, 0.2% (wt/vol) glucose, and 1.5% (wt/vol) Noble agar (Difco). Thiamine was added to a final concentration of 10 μg/ml. When required, an “aromix” consisting of tyrosine, tryptophan, and phenylalanine was added, each to a final concentration of 40 μg/ml, and p-aminobenzoic acid was added at 10 μg/ml. A. hydrophila strains were grown on minimal medium at the concentrations used for E. coli minimal medium when tested for the Aro− phenotype. For all strains used in this study, ampicillin (200 μg/ml), kanamycin (40 μg/ml), tetracycline (10 μg/ml), and chloramphenicol (20 μg/ml) were added as required. A. hydrophila and E. coli strains were routinely cultured at 28 and 37°C, respectively.
TABLE 1.
Characteristics of bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli AB2829 (aroA) | K-12 derivative with single mutation in aroA | 33 |
| E. coli C600-1 | Transformation recipient for plasmids | 28 |
| E. coli S17-1 | Mobilizing donor for conjugation | 42 |
| A. hydrophila AG2 | Virulent rainbow trout isolate | 2a |
| A. hydrophila aroA | aroA::Kar mutant | This study |
| Plasmids | ||
| pUC18 | Apr, cloning vector | 49 |
| pJRD215 | Kar Smr, broad-host-range mobilizable vector | 37 |
| pSUP202 | Tcr Apr Cmr, ColE1 ori, Mob+, broad-host-range mobilizable suicide vector | 42 |
| pARO39 | A. hydrophila aroA gene cloned into pUC18 | This study |
| pSARO39 | PvuII aroA-containing fragment from pARO39 cloned into pSUP202 | This study |
| pULMJ8 | Apr Kar Cmr, source of the 1.4-kb HincII Kar-containing fragment | 12 |
| pSKARO239 | 1.4-kb HincII Kar fragment from pULMJ8 cloned into the SmaI site of pSUP-ARO39 | This study |
The enzymes and biochemicals used in this work were obtained from either Boehringer GmbH, Promega, or Pharmacia and used as specified by the manufacturer.
Preparation and manipulation of DNA.
Chromosomal DNA from A. hydrophila AG2, the source of the aroA gene, was obtained from an overnight culture grown in LB at 28°C as reported previously (35), and E. coli-propagated plasmid DNAs were isolated by the alkali lysis method (6). Standard molecular cloning, transformation, and electrophoresis techniques were used (41). Southern blotting and hybridization were performed by random-primer DNA labeling with digoxigenin-dUTP, and hybrids were detected by an enzyme immunoassay as specified by the manufacturer (Boehringer).
Cloning of the A. hydrophila aroA gene in E. coli.
Chromosomal DNA prepared as described above was partially digested with Sau3A, and a library consisting of 4- to 9-kb fragments was prepared in BamHI-digested dephosphorylated pUC18 (Pharmacia). The ligation mixture was precipitated with ethanol, resuspended in 10 μl of distilled water, and used to transform electroporated E. coli aroA mutant AB2829. Electroporation was performed with a Gene Pulser apparatus (Bio-Rad Laboratories) set at 2.5 kV, 25 μF, and 1,000 Ω (field strength, 12.5 kV/cm), as described previously (10). Transformants were selected on LA plates supplemented with ampicillin. Recombinants which complemented the aroA lesion of AB2829 were selected by replica plating and growth of transformants on minimal medium supplemented with ampicillin and without aromatic supplements.
DNA sequencing.
Nucleotide sequences were determined by the dideoxynucleotide chain termination method with double-stranded templates by means of the fmol DNA sequencing system (Promega Corp.). A series of ordered deletions were generated in A. hydrophila aroA on pARO39 by using an Erase-a-Base kit (Promega Corp.). Gaps in the sequences were completed by using DNA primers synthesized by Promega Corp.
Bacterial conjugation.
Conjugation was performed as described previously (45). Briefly, the donor (E. coli S17-1 with the appropriate plasmid) and recipient (A. hydrophila AG2) strains were grown overnight in LB with shaking and incubated at 37 and 28°C, respectively. Then 10 μl each of overnight cultures of the donor and the recipient strains were mixed on the surface of a sterile 0.45-μm-pore-size filter (Millipore), placed on the surface of a dried LA plate with no antibiotics, and incubated for 4 h at 28°C. The growth was harvested in LB, and dilutions were spread on selective LA plates, which were then incubated at 28°C for 24 h.
LD50 determinations, vaccination trial, and measurement of humoral immune response.
Rainbow trout, 10 to 15 cm long, were obtained from a commercial fish farm. The fish were kept in 300-liter plastic tanks supplied with running well water at 18°C, maintained under constant photoperiod conditions (12 h of light/12 h of darkness), and fed with commercial trout pellets. Before manipulations, the fish were anesthetized with 1:15,000 tricaine methane sulfonate MS-222 (Sandoz) in water.
A. hydrophila AG2 was passaged in fish to increase virulence. Two colonies from a TSA plate were emulsified in 0.5 ml of phosphate-buffered saline (PBS), and groups of three trout were injected intramuscularly with 0.1 ml of the bacterial suspension. Bacteria were isolated from the muscle lesion of dead or moribund fish by streaking on TSA plates. The process was repeated six times until the fish died at 24 h.
For 50% lethal dose (LD50) determinations, seven groups of 10 fish were intraperitoneally injected with 0.1 ml each of a washed culture of A. hydrophila AG2 and A. hydrophila aroA emulsified in sterile PBS containing 103 to 109 CFU. The trout were observed for 7 days, and any dead specimens were removed for routine bacteriological examination. The experiment was done twice, and the LD50 were calculated by the statistical approach of Reed and Muench (36).
Two vaccination challenge trials were done. For each experiment, the vaccinated group consisted of 20 fish injected intraperitoneally with 0.1 ml of a bacterial suspension of 108 CFU of the A. hydrophila aroA mutant in PBS. Another 20 fish (the control group) were injected intraperitoneally with 0.1 ml of sterile PBS. The first vaccination challenge trial was performed at a water temperature of 12°C, and the second one was performed at a water temperature of 18°C. Control and vaccinated fish were challenged 5 weeks after vaccination, via intraperitoneal injection with 2 × 107 CFU (20 LD50) of A. hydrophila AG2 in 0.1 ml of PBS. The animals were observed daily up to 3 weeks after the challenge; dead specimens were removed, and hemorrhagic septicemia was diagnosed by routine bacteriological examination. The protective index was calculated as the relative percent survival (RPS) (2): RPS = [1 − (percent mortality in vaccinated fish/percent mortality in controls)] × 100.
For measurement of the humoral immune response, after immunization with the A. hydrophila aroA mutant, the titers of the serum antibodies against A. hydrophila AG2 were determined by microagglutination in fish vaccinated as described above. Four groups of six fish (two control groups and two vaccinated groups) were sacrificed at 24 and 42 days after vaccination and bled by puncture in the caudal vein, and the serum was serially diluted twice in sterile PBS. An equal volume of a washed suspension of formalin-killed cells of A. hydrophila AG2 was added to the serum dilutions in 96-well microtiter plates and incubated overnight at 22°C. The titer was recorded as the reciprocal of the last dilution which caused agglutination. Each serum sample was tested in duplicate assays, and the experiment was done twice.
Persistence of the A. hydrophila aroA mutant and antigen distribution in vivo.
The persistence of the A. hydrophila aroA mutant in groups of 20 trout injected intraperitoneally with 0.1 ml of PBS containing 108 CFU of bacteria was studied. Groups of three fishes were sacrificed at 24-h intervals, and viable bacteria were recovered from the head kidney by growth on medium described.
The distribution of A. hydrophila antigens on cryostat sections was studied by the indirect immunoperoxidase technique. Two fish from the above groups were sampled at 1, 2, 5, 7, and 9 days postinoculation, and tissue samples from the head kidney, spleen, liver, heart, muscle, skin, and gills were dissected under sterile conditions, frozen in liquid nitrogen, and stored at −80°C until needed. The primary antibody was a polyclonal antiserum against A. hydrophila AG2 obtained from a rabbit given weekly intravenous injections of 0.5 ml of sterile PBS containing increasing dilutions (106, 107, 108, and 108 per ml) of heat-killed A. hydrophila AG2 mixed with an equal volume of Freund’s incomplete adjuvant in the first three injections and of Freund’s complete adjuvant in the last immunization. The rabbit was bled, and the serum was stored at −20°C. The antibody titer (1:134,000) was determined by agglutination, and the dilution for immunohistochemistry was optimized (1:2,000) with A. hydrophila-coated glass slides. The secondary antibody was a goat anti-rabbit immunoglobulin G conjugated to peroxidase (Sigma), which was developed by incubation with 3,3-diaminobenzidine tetrahydrochloride (Sigma) in Tris-Cl buffer plus hydrogen peroxide. Control measurements were carried out by omitting the primary antibody, and endogenous peroxidase activity was inhibited by a 10-min incubation of the tissue sections in methanol plus 0.3% hydrogen peroxide before the incubation with the primary antibody.
RESULTS
Cloning of the aroA gene of A. hydrophila AG2.
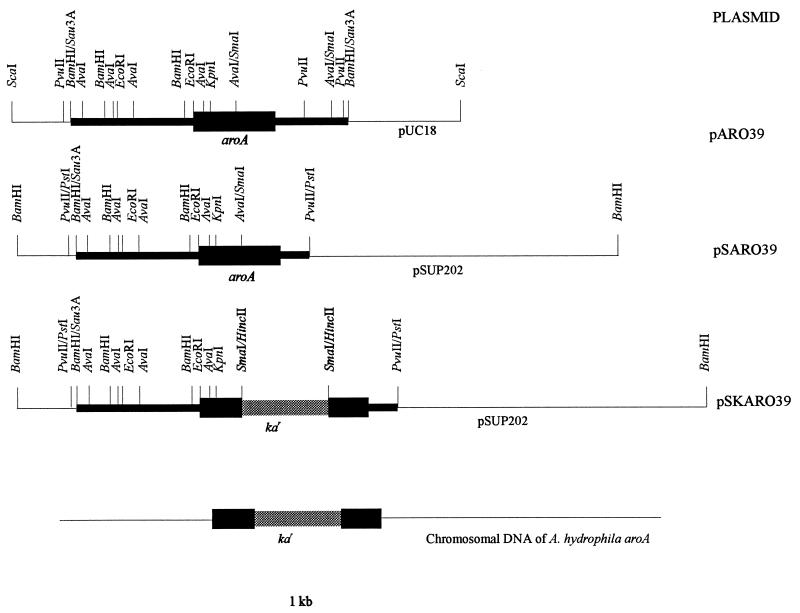
Restriction fragments of A. hydrophila AG2 genomic DNA, generated by partial digestion with Sau3A, were fractionated by agarose gel electrophoresis. Fragments of 4 to 9 kbp were used to construct a genomic library in the plasmid vector pUC18 as described in Materials and Methods, and the recombinant plasmids were used to transform the electroporated E. coli aroA mutant AB2829. A library consisting of 10,000 Apr colonies was obtained when these bacteria were plated on LA medium supplemented with ampicillin and incubated at 37°C for 24 h. Four recombinant clones which complemented the E. coli aroA defect were isolated by replica plating of transformants onto minimal medium supplemented with ampicillin and incubation at 37°C for 48 h. Recombinant plasmid DNA from each of four well-grown clones was isolated and used to retransform E. coli AB2829 to confirm the ability of the plasmids to complement the aroA defect in E. coli AB2829 when plated on defined minimal medium. All four recombinant plasmids (designated pARO39 to pARO42) were able to complement the growth of E. coli AB2829. Plasmid pARO39 was used to construct a restriction map (Fig. 1).
FIG. 1.
Restriction map of the aroA locus and construction of aroA::Kar, the base of allele exchange. Black boxes represent A. hydrophila cloned DNA. The thicker black box represents the A. hydrophila aroA gene, which is orientated from 5′ (left) to 3′ (right). The shaded box represents the Kar cassette. Horizontal lines represent different plasmid vectors or A. hydrophila aroA chromosomal DNA.
Nucleotide sequence of the A. hydrophila aroA gene.
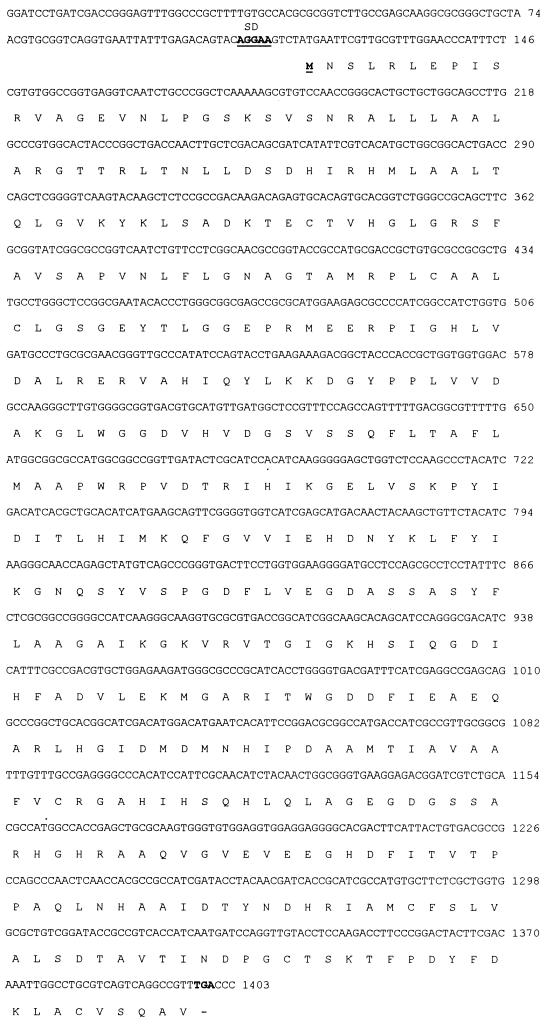
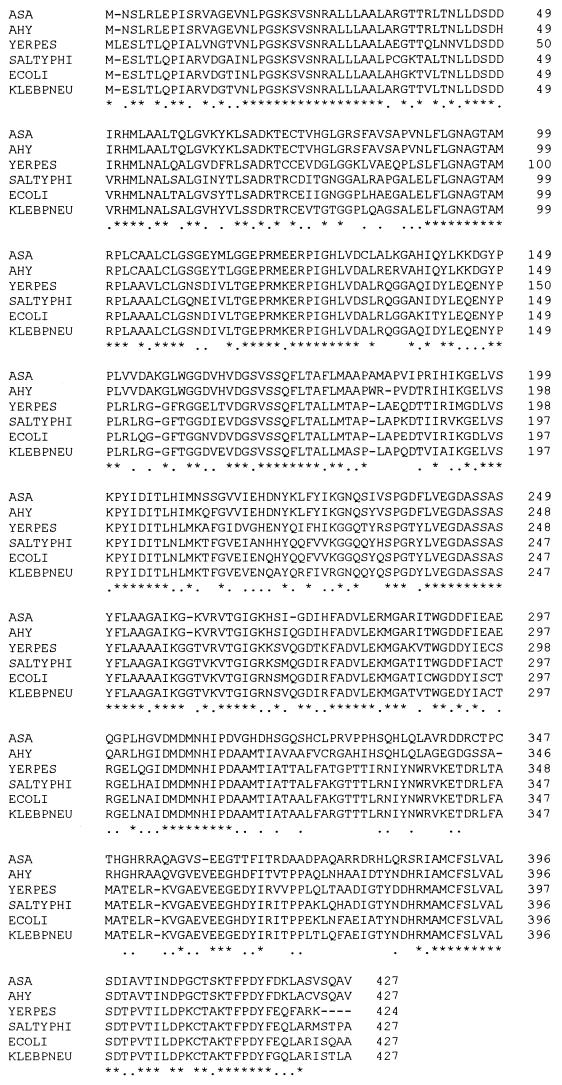
The nucleotide sequence of a 1.4-kb BamHI downstream fragment of pARO39 (Fig. 2) revealed an open reading frame of 1,281 nucleotides, which encodes a protein of 427 amino acids. The deduced molecular weight is 46,095, and the G+C content of the aroA coding region product is 61.29%. The predicted amino acid sequence of A. hydrophila AroA (5-enolpyruvylshikimate-3-phosphate synthase [EPSP synthase; EC 2.5.1.19]) showed a high degree of homology (83%) to that of the EPSP synthase of A. salmonicida (47). Also, a high degree of amino acid sequence conservation was revealed when the EPSP synthase of A. hydrophila was aligned with several bacterial EPSP synthases by means of the CLUSTAL multiple-alignment program (Fig. 3).
FIG. 2.
Nucleotide sequence of the 1.4-kb aroA-containing fragment of pARO39 and the amino acid sequence deduced from the open reading frame of the aroA gene. DNA bases (top line) and amino acids (one-letter code) (below) are listed and numbered to the right of the sequences. The ATG initiation codon (boldface and underlined) is preceded by a potential Shine-Dalgarno sequence (SD) (boldface and underlined). The symbol − indicates the TGA termination codon.
FIG. 3.
CLUSTAL computer alignment of the deduced amino acid sequences encoded by the aroA gene from A. salmonicida (ASA), A. hydrophila (AHY), Yersinia pestis (YERPES), Salmonella typhimurium (SALTYPHI), E. coli (ECOLI), and Klebsiella pneumoniae (KLEBPNEU). Amino acids identical in all species are indicated by an asterisk. Conservative substitutions are indicated by a dot.
Construction of an A. hydrophila aroA mutant.
This investigation was developed to isolate an aroA mutant strain of A. hydrophila by allelic exchange. To achieve this goal, the 3.7-kb PvuII aroA-containing fragment from pARO39 was subcloned into the Klenow-treated unique PstI site of pSUP202, which disrupted ampicillin resistance. The resultant plasmid (pSARO39 [Fig. 1]) was SmaI digested and ligated to the 1.4-kb HincII kanamycin-resistant (Kar) fragment from pULMJ8. The resultant Kar plasmid (pSKARO39 [Fig. 1]) was used to transform electroporated E. coli AB2829. None of the transformants were able to grow on minimal medium supplemented with kanamycin. These results demonstrated that the Kar cassette was inserted within the aroA gene.
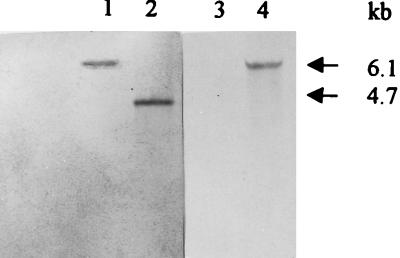
To reintroduce the inactivated aroA gene into the chromosome of A. hydrophila, the suicide plasmid pSKARO39 was mobilized from E. coli S17-1 into A. hydrophila AG2 (Apr), which was passaged in vivo previously to increase its virulence. Transconjugants were selected on LA plates containing ampicillin and kanamycin, as noted in Materials and Methods, and occurred at a frequency of 2 × 102 per recipient. pSKARO39, a pSUP202-based recombinant plasmid, is mobilizable by a sequence derived from RP4 on the chromosome of E. coli S17-1 and should not replicate in Aeromonas spp. owing to the absence of the pir gene product. Theoretically, all A. hydrophila colonies appearing on the selective medium should be AroA−, since the Kar cassette should be expressed only if the A. hydrophila AG2 aroA allele is replaced on the chromosome by the Kar cassette-mutated aroA gene. However, only 30% of transconjugants were Kar and AroA− and were susceptible to tetracycline and to chloramphenicol, the expected phenotype in bacteria in which allele replacement has occurred at the aroA locus. A total of 70% of transconjugants were Kar and AroA+ and were resistant to tetracycline and to chloramphenicol. Hybridization analysis of both AroA− and AroA+ transconjugants demonstrated that two types of genetic recombination events occurred. Chromosomal DNA was BamHI digested and hybridized to the 0.7-kb EcoRI-SmaI aroA gene moiety as a probe. The hybridization pattern is shown in Fig. 4. A 6.1-kb hybridization band was detected in all A. hydrophila aroA mutants (lane 1) so far analyzed, while a 4.7-kb hybridization band (1.4-kb sorter, corresponding to the length of the Kar cassette) (lane 2) was detected in A. hydrophila AG2 when probed with the EcoRI-SmaI 0.7-kb fragment from aroA gene. A 6.1-kb hybridization band was detected when BamHI-digested AroA− DNA was probed with the 1.4-kb Kar cassette (lane 4). However, no hybridization band was detected when BamHI-digested wild-type DNA was hybridized with the same probe (lane 3). Hybridization analysis of the AroA+ phenotype showed integration of the entire plasmid pSKARO39 into the chromosome at the aroA locus when plasmid pSUP202 was used as a probe (data not shown).
FIG. 4.
Southern hybridization analysis of chromosomal DNA from wild-type A. hydrophila AG2 (lanes 2 and 3) and an aroA mutant (lanes 1 and 4). Total-cell DNA was BamHI digested and probed with the EcoRI-SmaI 0.7-kb fragment from aroA (lanes 1 and 2) or with the HincII 1.4-kb Kar cassette (lanes 3 and 4).
To investigate the stability of A. hydrophila aroA mutants, several of them were passaged daily in LB without antibiotics, showing that Kar was stably maintained. Also, the reversion test performed with these mutants demonstrated no AroA+ revertants when 1011 bacteria were plated on minimal medium.
Virulence and survival of the A. hydrophila aroA strain in trout.
The LD50 of the A. hydrophila wild-type strain AG2 was 106 CFU, and infected fish died within 48 h. However, the aroA mutant of A. hydrophila had a LD50 of >2 × 108 CFU, and no signs of illness or death were detected in a period of 2 weeks. To demonstrate that attenuation was due exclusively to the mutated aroA gene, the wild-type aroA gene was cloned into a broad-host-range plasmid, pJRD215, and transferred from E. coli S17-1 into A. hydrophila aroA::Kar by conjugation. The LD50 of A. hydrophila aroA::Kar complemented with the wild-type aroA gene was very similar to that of A. hydrophila AG2 (Table 2).
TABLE 2.
Mortalities recorded for LD50 calculations of the AG2, aroA mutant, and AroA+ strains of A. hydrophila
| No. of bacteria/0.1 ml | No. of fish that died after inoculation witha:
|
|||||
|---|---|---|---|---|---|---|
| AG2
|
aroA mutant
|
AroA+b
|
||||
| Expt 1 | Expt 2 | Expt 1 | Expt 2 | Expt 1 | Expt 2 | |
| 109 | 10 | 10 | 6 | 5 | 10 | 10 |
| 108 | 9 | 10 | 4 | 4 | 8 | 9 |
| 107 | 8 | 8 | 2 | 2 | 8 | 8 |
| 106 | 6 | 6 | 0 | 0 | 6 | 5 |
| 105 | 3 | 3 | 0 | 0 | 3 | 3 |
| 104 | 0 | 0 | 0 | 0 | 0 | 0 |
| 103 | 0 | 0 | 0 | 0 | 0 | 0 |
| LD50 | 7 × 105 | 6 × 105 | 2 × 108 | 2 × 108 | 8 × 105 | 1 × 106 |
10 fish per group were inoculated.
Reverted strain (aroA::Kar complemented with the wild-type aroA gene cloned into pJRD215).
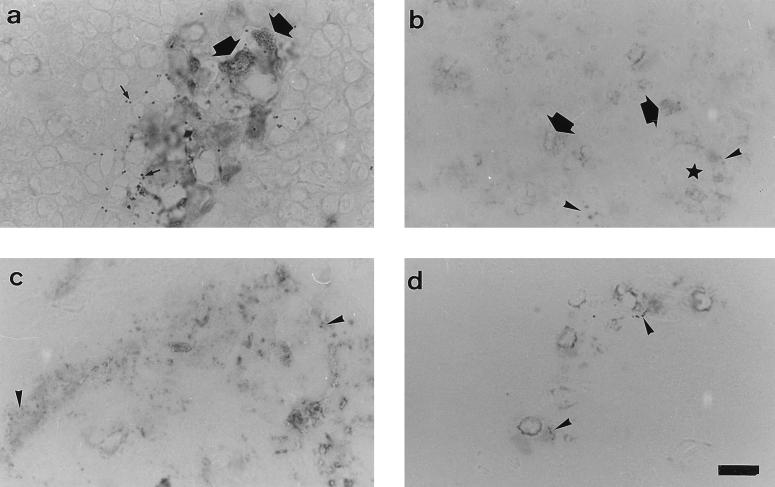
Viable aroA mutant bacteria were no longer recoverable after 48 h from intraperitoneally injected fishes. A. hydrophila antigens were observed mainly in the pronephros, spleen, liver, and heart. Up to day 5 after inoculation, the labeling was located as particulate spots in the vascular lumina and intracellularly in the endothelial cells of splenic ellipsoids, vascular sinusoids, large blood vessels, and hepatic sinusoids and in scattered macrophages (Fig. 5). From day 5 onward, A. hydrophila antigens could be detected as a diffuse staining, which was observable with decreasing intensity in the spleen, pronephros, and liver up to day 9.
FIG. 5.
Immunohistochemical demonstration of A. hydrophila antigens in different rainbow trout tissues after inoculation of A. hydrophila aroA. (a) Pronephros, 2 days postinoculation. Immunostaining occurs in macrophage-melanomacrophage clusters. The particulate labeling (large arrows) corresponds to intracellular bacteria; small arrows indicate melanin granules. (b) Spleen, 5 days postinoculation. Immunolabeling occurs as diffuse and particulate staining in macrophages (large arrows) and endothelial cells (arrowheads). The asterisk indicates the lumen of a blood vessel. (c) Heart, 5 days postinoculation. Abundant diffuse and particulate labeling occurs in the endocardium. (d) Liver, 1 day postinoculation; immunostaining occurs in the sinusoidal cells. Bar, 10 μm.
Vaccination trials with the A. hydrophila aroA strain.
In two different trials, more than 75% of fish seroconverted by 42 days after vaccination with the aroA mutant. Immunized fish showed a notable increase in the level of the agglutinating antibody titers cross-reacting with the wild-type A. hydrophila AG2 strain (Table 3).
TABLE 3.
Antibody titers against A. hydrophila AG2 after intraperitoneal immunization with the aroA mutant
| Time (days) postimmunization | Antibody titera in:
|
|||
|---|---|---|---|---|
| Expt 1
|
Expt 2
|
|||
| Controls | Immunized | Controls | Immunized | |
| 24 | 1.1 ± 0.4 (n = 6) | 22.3 ± 16.7 (n = 6) | 1.7 ± 1.1 (n = 6) | 19 ± 8.8 (n = 4) |
| 42 | 2 ± 0 (n = 4) | 24.2 ± 24.2 (n = 6) | 1.5 ± 0.5 (n = 5) | 22 ± 12 (n = 4) |
The antibody titer is shown as the reciprocal of the last dilution which caused agglutination. Each value represents the mean ± standard deviation of titers from fish sampled on the same day. n, number of fish.
The results of the two vaccination challenge experiments are shown in Table 4. In the first trial, which was performed at 12°C, 35% of the unvaccinated fish and 65% of the vaccinated fish survived more than 3 weeks after intraperitoneal challenge with the wild-type A. hydrophila AG2 strain. In the second trial, which was performed at 18°C, all unvaccinated fish had died by 10 days and the accumulated RPS was 75%. Most dying fishes showed typical clinical signs of hemorrhagic septicemia, mainly external lesions (abdominal distension and skin ulceration at the injection site), and internal hemorrhages. Colonies of A. hydrophila AG2 were recovered from all dead fish. No evident external lesions were observed in the surviving fish.
TABLE 4.
Mortalities observed in fish vaccinated with the A. hydrophila aroA mutant after challenge with the AG2 wild-type strain
| Time (days) postchallenge | No. of fish that died ina:
|
|||
|---|---|---|---|---|
| Expt 1
|
Expt 2
|
|||
| Controls | Vaccinated | Controls | Vaccinated | |
| 1 | NDb | ND | 5 | 0 |
| 2 | 6 | 3 | 6 | 3 |
| 3 | 2 | 0 | 1 | 0 |
| 4 | 2 | 3 | 1 | 2 |
| 5 | 2 | 0 | 1 | 0 |
| 6 | 1 | 1 | 1 | 0 |
| 7 | 0 | 0 | 0 | 0 |
| 8 | 0 | 0 | 2 | 0 |
| 9 | 0 | 0 | 1 | 0 |
| 10 | 0 | 0 | 2 | 0 |
| No. of survivors on day 21 (RPSc) | 7 | 13 (47%) | 0 | 15 (75%) |
20 fish per challenge group.
ND, not determined.
Calculated from the percentage of dead fish in each group 21 days after challenge.
DISCUSSION
In this report, we have described the molecular cloning and sequencing of the aroA gene as well as the construction of a mutant strain of A. hydrophila AG2, by insertional inactivation of the aroA gene with a kanamycin resistance cassette, which may be used as an effective live attenuated vaccine. The aroA gene is highly conserved in all species so far studied, and in some of them it forms part of an operon with serC, which is proximal to the promoter of the operon and upstream from aroA (11). No consensus sequences have been described for Aeromonas promoters; however, a potential ribosome binding site is located 6 bp upstream from the initiation ATG codon in the aroA gene (Fig. 2). The results also showed considerable homology between the deduced amino acid sequence from the A. hydrophila aroA gene and those from other species (Fig. 3). Nucleotide sequence analysis of the aroA gene flanking sequences revealed no significant homology to any other database sequences. However, an open reading frame of 1,050 bp was found 228 bp downstream from the end of the aroA gene (data not shown). The deduced amino acid sequence (350 amino acids) was 80% homologous to the phenylalanine-repressible phospho-2-dehydro-3-deoxyheptonate aldolase (DAHP synthetase) of E. coli, the product of the aroG gene. Neither nucleotide nor amino acid sequence homology was found up to 1 kb upstream of the aroA gene.
LD50 determinations and persistence in fish demonstrated that the A. hydrophila aroA mutant is highly attenuated for colonization and infection of internal organs relative to the wild-type strain. The persistence of the mutant in fish tissues, as determined by the recovery of viable bacteria and immunohistochemical studies, was shorter than that described by other authors for genetically attenuated A. salmonicida aroA mutants (25, 47). Rapid clearance of the A. hydrophila aroA mutant may be related to bacterial species differences and to the temperature of the water at which experiments were conducted, which was higher in the present study (25). Moreover, the immunohistochemical studies of A. hydrophila antigens indicated that bacterial cells, detected as particulate immunopositive materials, were present up to 5 days postinoculation and that after this period only degraded products were present (detected as a diffuse staining). The tissue distribution of the antigens is comparable to that found in carp after intramuscular injection with an A. hydrophila bacterin, confirming the importance of the lymphoid organs in bacterial antigen trapping but also of the liver and heart. In our study, a decrease in the staining intensity was apparent up to day 9 postinoculation, but Lamers and De Hass (21) described the persistence of A. hydrophila antigens in the spleen and kidney up to 1 year after intramuscular immunization with a bacterin consisting of heat-inactivated and disrupted cells. This is an important point that remains to be resolved, since the length of persistence of the antigens in the lymphoid tissues may determine the magnitude of the immune response and the duration of the protection induced by the vaccine.
The A. hydrophila aroA mutant can act as a live vaccine to prevent hemorrhagic septicemia of fish. A single immunization conferred a significant protection (RPS of 75% at 18°C) when fish were challenged with a 20-fold LD50 of the wild-type strain. The difference observed between the RPS values from the two trials can be explained by the decreased virulence of A. hydrophila at lower water temperatures, as confirmed by the low mortality found in the control group of the first trial. Although different authors (4, 43) have been unable to establish a clear correlation between the humoral response and protection against A. hydrophila, there was a moderate increase in the titers of the agglutinating antibodies against A. hydrophila in vaccinated fish, which are in the range of those reported in rainbow trout (34) and carp (22) immunized with A. hydrophila bacterins but somewhat lower than those reported in similar experiments with an A. salmonicida aroA mutant (47). This may reflect a predominance of the cellular immune reactions over the humoral response, as reported for A. salmonicida (25, 26). Although further studies are necessary, including the testing of immersion and oral vaccination methods, the use of aro bacterial mutants for vaccine production may offer a number of advantages over classical vaccine preparations, because the mutation renders the organism avirulent without affecting its ability to produce virulent determinants (47). In the case of A. hydrophila, this may be particularly important, because its pathogenicity appears to be closely related to the production of extracellular products (3), which are lost in conventional bacterin preparations.
ACKNOWLEDGMENTS
This work was supported by DGICYT grant PB94-0136 from the Spanish Ministerio de Educación y Cultura and by grant LE-24/94 from the Junta de Castilla y León.
REFERENCES
- 1.Altwegg M, Geiss H K. Aeromonas as a human pathogen. Crit Rev Microbiol. 1989;16:253–286. doi: 10.3109/10408418909105478. [DOI] [PubMed] [Google Scholar]
- 2.Amend D F. Potency testing of fish vaccines. International Symposium on Fish Biologics: serodiagnostics and vaccines. Dev Biol Stand. 1981;49:447–454. [Google Scholar]
- 2a.Austin, B. Unpublished data.
- 3.Austin B, Austin D A. Bacterial fish pathogens. 2nd ed. Chichester, United Kingdom: Ellis Horwood; 1993. [Google Scholar]
- 4.Baba T, Imamura J, Izawa K, Ikeda K. Immune protection in the carp, Cyprinus carpio L., after immunization with Aeromonas hydrophila crude lypopolysaccharide. J Fish Dis. 1988;11:237–244. [Google Scholar]
- 5.Bacon G A, Burrows T W, Yates M. The effects of the biochemical mutations on the virulence of Bacterium typhosa: the virulence of mutants. Br J Exp Pathol. 1950;31:703–711. [PMC free article] [PubMed] [Google Scholar]
- 6.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowe F, O’Gaora P, Maskell D, Cafferkey M, Dougan G. Virulence, persistence, and immunogenicity of Yersinia enterocolitica O:8 aroA mutants. Infect Immun. 1989;57:3234–3236. doi: 10.1128/iai.57.10.3234-3236.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakraborty T, Huhle B, Hof H, Bergbauer H, Goebel W. Marker exchange mutagenesis of the aerolysin determinant in Aeromonas hydrophila demonstrates the role of aerolysin in A. hydrophila-associated systemic infections. Infect Immun. 1987;55:2274–2280. doi: 10.1128/iai.55.9.2274-2280.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Del Corral F, Shotts E B, Jr, Brown J. Adherence, haemagglutination and cell surface characteristics of motile aeromonads virulent for fish. J Fish Dis. 1990;13:255–268. [Google Scholar]
- 10.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;7:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duncan K, Coggins R. The serC-aroA operon of Escherichia coli. Biochem J. 1986;234:49–57. doi: 10.1042/bj2340049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernández C, Cadenas R F, Noirot-Grost M F, Martin J F, Gil J A. Characterization of a region of plasmid pBL1 of Brevibacterium lactofermentum involved in replication via the rolling-circle model. J Bacteriol. 1994;176:3154–3161. doi: 10.1128/jb.176.11.3154-3161.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Handfield M, Simard P, Couillard M, Letarte R. Aeromonas hydrophila isolated from food and drinking water: hemagglutination, hemolysis, and cytotoxicity for a human intestinal cell line (HT-29) Appl Environ Microbiol. 1996;62:3459–3461. doi: 10.1128/aem.62.9.3459-3461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoiseth S K, Stocker B A D. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature (London) 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 15.Holmberg S, Schell W L, Ganning G R. Aeromonas intestinal infections in the United States. Ann Intern Med. 1986;150:683–689. doi: 10.7326/0003-4819-105-5-683. [DOI] [PubMed] [Google Scholar]
- 16.Homchampa P, Strugnell R A, Adler B. Molecular analysis of the aroA gene of Pasteurella multocida and vaccine potential of a constructed aroA mutant. Mol Microbiol. 1992;6:3585–3593. doi: 10.1111/j.1365-2958.1992.tb01794.x. [DOI] [PubMed] [Google Scholar]
- 17.Ivins B E, Welkos S L, Knudson G B, Little S F. Immunization against anthrax with aromatic compound-dependent (Aro−) mutants of Bacillus anthracis and with recombinant strains of Bacillus subtilis that produce anthrax protective antigen. Infect Immun. 1990;58:303–308. doi: 10.1128/iai.58.2.303-308.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janda J M, Guthertz L S, Kokka R P, Shimada T. Aeromonas species in septicemia: laboratory characteristics and clinical observations. Clin Infect Dis. 1994;19:77–83. doi: 10.1093/clinids/19.1.77. [DOI] [PubMed] [Google Scholar]
- 19.Janda J M, Duffey P S. Mesophilic aeromonads in human disease: current taxonomy, laboratory identification, and infectious disease spectrum. Rev Infect Dis. 1988;10:980–997. doi: 10.1093/clinids/10.5.980. [DOI] [PubMed] [Google Scholar]
- 20.Lamers C H J. Natural and acquired agglutinins to Aeromonas hydrophila in carp (Ciprinus carpio) Can J Fish Aquat Sci. 1986;43:619–624. [Google Scholar]
- 21.Lamers C H J, De Hass M J M. Antigen localization in the lymphoid organs of carp (Cyprinus carpio) Cell Tissue Res. 1985;242:491–498. doi: 10.1007/BF00225413. [DOI] [PubMed] [Google Scholar]
- 22.Lamers C H J, De Hass M J M, Van Muiswinkel W B. Humoral response and memory formation in carp Cyprinus carpio after injection of Aeromonas hydrophila bacterin. Dev Comp Immunol. 1985;9:65–76. doi: 10.1016/0145-305x(85)90060-6. [DOI] [PubMed] [Google Scholar]
- 23.Leung K Y, Stevenson R M W. Tn5-induced protease-deficient strains of Aeromonas hydrophila with reduced virulence for fish. Infect Immun. 1988;56:2639–2644. doi: 10.1128/iai.56.10.2639-2644.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loghothetis P N, Austin B. Immune response of rainbow trout (Onchorhynchus mykiss, Wallaby) to Aeromonas hydrophila. Fish Shellfish Immunol. 1994;4:239–254. [Google Scholar]
- 25.Marsden M J, Devoy A, Vaughan L M, Foster T J, Secombes C J. Use of genetically attenuated strain of Aeromonas salmonicida to vaccinate salmonid fish. Aquacult Int. 1996;4:55–66. [Google Scholar]
- 26.Marsden M J, Vaughan L M, Foster T J, Secombes C J. A live (ΔaroA) Aeromonas salmonicida vaccine for furunculosis preferentially stimulates T-cell responses relative to B-cell responses in rainbow trout (Oncorhynchus mykiss) Infect Immun. 1996;64:3863–3869. doi: 10.1128/iai.64.9.3863-3869.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 28.Nagahase K, Tanaka T, Hishinuma F, Kuroda M, Sakaguchi K. Control of tryptophan synthetase amplified by varying the number of composite plasmids in E. coli cells. Gene. 1977;1:141–152. doi: 10.1016/0378-1119(77)90025-7. [DOI] [PubMed] [Google Scholar]
- 29.Newman S G. Bacterial vaccines for fish. Annu Rev Fish Dis. 1993;3:145–185. [Google Scholar]
- 30.Nieto T P, Santos Y, Rodríguez L A, Ellis A E. An extracellular acetylcholinesterase produced by Aeromonas hydrophila is a major lethal toxin for fish. Microb Pathog. 1991;11:101–110. doi: 10.1016/0882-4010(91)90003-s. [DOI] [PubMed] [Google Scholar]
- 31.Norquist A, Bergman A, Skogman G, Wolf-Watz H. A field trial with the live attenuated fish vaccine strain Vibrio anguillarum VAN1000. Bull Eur Assoc Fish Pathol. 1994;14:156–158. [Google Scholar]
- 32.Paniagua C, Rivero O, Anguita J, Naharro G. Pathogenicity factors and virulence for rainbow trout (Salmo gairdneri) of motile Aeromonas spp. isolated from a river. J Clin Microbiol. 1990;28:350–355. doi: 10.1128/jcm.28.2.350-355.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pittard J, Wallace B J. Distribution and function of genes concerned with aromatic biosynthesis in Escherichia coli. J Bacteriol. 1966;91:1494–1508. doi: 10.1128/jb.91.4.1494-1508.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Post G. Response of rainbow trout (Salmo gairdneri) to antigens of Aeromonas hydrophila. J Fish Res Board Can. 1966;23:1487–1494. [Google Scholar]
- 35.Priefer U, Simon R, Puyhler A. Cloning with cosmid. In: Puhler A, Timmis K N, editors. Advanced molecular genetics. Berlin, Germany: Springer-Verlag KG; 1984. pp. 190–201. [Google Scholar]
- 36.Reed L J, Muench H. A simple method of estimating fifty percent end points. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 37.Rivero O, Anguita J, Mateos D, Paniagua C, Naharro G. Cloning and characterization of an extracellular temperature-labile serine protease gene from Aeromonas hydrophila. FEMS Microbiol Lett. 1991;81:1–8. doi: 10.1016/0378-1097(91)90461-i. [DOI] [PubMed] [Google Scholar]
- 38.Rivero O, Anguita J, Paniagua C, Naharro G. Molecular cloning and characterization of an extracellular protease gene from Aeromonas hydrophila. J Bacteriol. 1990;172:3905–3908. doi: 10.1128/jb.172.7.3905-3908.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts R, Maskell D, Novotny P, Dougan G. Construction and characterization in vivo of Bordetella pertussis aroA mutants. Infect Immun. 1990;58:732–739. doi: 10.1128/iai.58.3.732-739.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rose J M, Houston C W, Coppenhaver D H, Dixon J D, Kurosky A. Purification and chemical characterization of a cholera toxin-cross-reactive cytolytic enterotoxin produced by a human isolate of Aeromonas hydrophila. Infect Immun. 1989;57:1165–1169. doi: 10.1128/iai.57.4.1165-1169.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 42.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 43.Stevenson R M W. Vaccination against Aeromonas hydrophila. In: Ellis A E, editor. Fish vaccination. London, United Kingdom: Academic Press, Ltd.; 1988. pp. 112–123. [Google Scholar]
- 44.Tatum F M, Briggs R E, Halling S M. Molecular gene cloning and nucleotide sequencing and construction of an aroA mutant of Pasteurella haemolytica serotype A1. Appl Environ Microbiol. 1994;60:2011–2016. doi: 10.1128/aem.60.6.2011-2016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor R K, Manoil C, Mekalanos J J. Broad-host-range vectors for delivery of TnphoA: use in genetic analysis of secreted virulence determinants of Vibrio cholerae. J Bacteriol. 1989;171:1870–1878. doi: 10.1128/jb.171.4.1870-1878.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thornton J C, Garduno R A, Newman S G, Kay W W. Surface disorganised, attenuated mutants of Aeromonas salmonicida as furunculosis vaccines. Microb Pathog. 1991;11:85–89. doi: 10.1016/0882-4010(91)90002-r. [DOI] [PubMed] [Google Scholar]
- 47.Vaughan L M, Smith P R, Foster T J. An aromatic-dependent mutant of the fish pathogen Aeromonas salmonicida is attenuated in fish and is effective as a live vaccine against the salmonid disease furunculosis. Infect Immun. 1993;61:2172–2181. doi: 10.1128/iai.61.5.2172-2181.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verma N K, Lindberg A A. Construction of aromatic dependent Shigella flexneri 2a live vaccine candidate strains: deletion mutations in the aroA and the aroD genes. Vaccine. 1991;9:6–9. doi: 10.1016/0264-410x(91)90308-s. [DOI] [PubMed] [Google Scholar]
- 49.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]