Abstract
Of the nasopharyngeal cultures recovered from 942 day care center (DCC) attendees in Lisbon, Portugal, 591 (62%) yielded Streptococcus pneumoniae during a surveillance performed in February and March of 1999. Forty percent of the isolates were resistant to one or more antimicrobial agents. In particular, 2% were penicillin resistant and 20% had intermediate penicillin resistance. Multidrug resistance to macrolides, lincosamides, and tetracycline was the most frequent antibiotype (17% of all isolates). Serotyping and molecular typing by pulsed-field gel electrophoresis were performed for 202 out of 237 drug-resistant pneumococci (DRPn). The most frequent serotypes were 6B (26%), 14 (22%), 19F (16%), 23F (10%), and nontypeable (12%). The majority (67%) of the DRPn strains were representatives of nine international clones included in the Pneumococcal Molecular Epidemiology Network; eight of them had been detected in previous studies. Fourteen novel clones were identified, corresponding to 26% of the DRPn strains. The remaining 7% of the strains were local clones detected in our previous studies. Comparison with studies conducted since 1996 in Portuguese DCCs identified several trends: (i) the rate of DRPn frequency has fluctuated between 40 and 50%; (ii) the serotypes most frequently recovered have remained the same; (iii) nontypeable strains appear to be increasing in frequency; and (iv) a clone of serotype 33F emerged in 1999. Together, our observations highlight that the nasopharynxes of children in DCCs are a melting pot of successful DRPn clones that are important to study and monitor if we aim to gain a better understanding on the epidemiology of this pathogen.
The ecological niche of Streptococcus pneumoniae is the nasopharynxes of humans. Although pneumococci are widely spread in the community, carriage rates are particularly high in preschool children attending day care centers (DCCs) (24, 45). S. pneumoniae can cause a spectrum of diseases including invasive disease, such as meningitis, bloodstream infections, and pneumonia, as well as infections of the upper respiratory tract. S. pneumoniae is a major cause of morbidity and mortality in young children (6). In the last decade an increasing proportion of pneumococci that are not susceptible to penicillin has been observed, reaching up to 50% in some areas of the world (1, 10, 16, 37, 39, 43).
In 1996 our group began yearly surveillance of the nasopharyngeal carriage of pneumococci and other bacterial respiratory tract pathogens in DCCs located in the Lisbon area of Portugal. Since 1996, a total of 23 DCCs and over 1,800 children participated in this study. Serotypes, antibiotypes, and molecular types of the majority of drug-resistant S. pneumoniae isolates were determined during these annual surveillances (8, 9, 34, 35), allowing one to obtain a view of temporal changes of drug-resistant pneumococci (DRPn) inhabiting the nasopharyngeal flora, an important ecological reservoir which may be a major source of drug-resistant strains causing pediatric and adult disease. The importance of such surveillances for both the national and international communities has been highlighted in recent reports (15, 32, 38).
In the study described here we provide new data on the nasopharyngeal carriage of drug-resistant pneumococci during the 1999 surveillance. We also summarize temporal changes noted in the same surveillance system between 1996 and 1999.
MATERIALS AND METHODS
Study population.
Between February and March of 1999, nasopharyngeal samples were recovered from children attending DCCs in the Lisbon area. Lisbon is the capital of Portugal and, according to the last census (of 2001), approximately 2 million people live in the city and its suburbs. The 14 DCCs were selected to be of diverse geographic locations and sizes and to include children of different social strata. The target population was 1,454 children, of which a total of 942 children (480 females and 462 males), with ages ranging from 6 months to 6 years (average, 3.3 years old), participated in the study.
Questionnaire.
Questionnaires on antibiotic consumption habits of the children were addressed to their parents and/or guardians at the time of sampling. The questions were as follows. (i) Is the child taking any antibiotics now? (ii) Has the child taken antibiotics within the last month? (iii) Has the child received repeated courses (more than three) of antibiotics in the last 6 months? (iv) If the child took antibiotics recently, which antibiotic did the child take and what was the reason?
Nasopharyngeal swab and isolation of S. pneumoniae.
Nasopharyngeal secretions were taken by a pediatric nurse using swabs and modified Stuart's medium (Mini-Tip Culturette; Becton Dickinson Microbiology Systems, Cockeysville, Md.). Swabs were transferred to the central research laboratory at the Instituto de Tecnologia Química e Biológica and were inoculated into appropriate culture media within 4 h after the sample collection, to select and identify S. pneumoniae. Blood agar with gentamicin (5 mg/ml) and anaerobic incubation were used for selective culture of pneumococci. The pneumococci were identified by optochin susceptibility and colony morphology. One colony was picked and a pure culture was frozen. All isolates were frozen in Mueller-Hinton broth (Difco, Detroit, Mich.) and 15% (vol/vol) glycerol (Merck, Darmstadt, Germany) and kept at −70°C (8).
Antimicrobial susceptibility testing.
Susceptibility testing was performed using the Kirby-Bauer technique, according to the NCCLS recommendations and definitions (28). The antimicrobial agents tested were chloramphenicol, erythromycin, clindamycin, tetracycline, and sulfamethoxazole-trimethoprim (SXT). Antibiotic disks were purchased from Oxoid (Hampshire, England). Isolates were also screened for MICs of penicillin and ceftriaxone with the E-test (AB Biodisk, Solna, Sweden) according to the manufacturer's recommendations. In the interpretation of decreased penicillin susceptibility, isolates were divided into intermediately resistant (0.1 μg/ml ≤ MIC < 1.5 μg/ml) and resistant (MIC ≥ 1.5 μg/ml) strains, according to NCCLS guidelines (28).
Serotyping.
Strains resistant to at least one of the antimicrobial agents tested (except those resistant only to SXT) were serotyped by the Quellung reaction using commercially available antisera (Statens Seruminstitut, Copenhagen, Denmark) (40).
PFGE.
The strains that were serotyped were also typed by pulsed-field gel electrophoresis (PFGE). Preparation of chromosomal DNA, restriction with SmaI endonuclease, and PFGE were done as previously described (35). PFGE patterns were assigned by visual inspection of the macrorestriction profiles, by using currently accepted criteria (42). The particular PFGE patterns, which were detected in previous studies (8, 35), received the same uppercase letter; the new PFGE patterns were assigned two distinct uppercase letters.
MLST.
Multilocus sequence typing (MLST) was performed on selected isolates and interpreted as described previously (14, 33). DNA sequencing was done either at the Rockefeller University Protein/DNA Technology Center (New York, N.Y.) or at Macrogene, Inc. (Seoul, Korea).
RESULTS
Study population.
Between February and March of 1999, 942 children were screened, corresponding to 65% of the children attending the 14 DCCs studied. The number of children enrolled at each day care center varied from 44 to 178, with an average of 104 children. Participation rates in this study in each DCC ranged from 40 to 75%, and the number of children sampled in each center ranged from 20 to 132.
Pneumococcal carriage and resistance to antimicrobial agents.
A total of 591 children (63%) of the 942 sampled were pneumococcal carriers. Of the 591 pneumococcal isolates, 60% were susceptible to all antimicrobial agents tested, 18% were susceptible to penicillin but resistant to at least one of the other antimicrobials, and 22% had decreased susceptibility to penicillin: the penicillin MIC for 2% of the isolates was equal to or higher than 1.5 μg/ml and 20% had intermediate resistance. Rates of resistance to antimicrobial agents other than penicillin were 26% for tetracycline, 25% for SXT, 8.5% for chloramphenicol, and 2% for ceftriaxone. Concerning macrolide-lincosamide resistance, 22% of the isolates were resistant to erythromycin and 20.5% were resistant to clindamycin. Multidrug resistance (defined as resistance to three or more classes of antimicrobial agents) was common (26% of all pneumococcal isolates) and frequently (17%) included resistance to erythromycin, clindamycin, and tetracycline (Table 1).
TABLE 1.
Antibiotypes of pneumococcal isolates (n = 591)
| Antibiotype resistant toa: | No. of isolates |
|---|---|
| PG R, SXT | 6 |
| PG R, CHL, TET, SXT | 4 |
| PG I, CHL, ERY, CLI, TET, SXT | 5 |
| PG I, CHL, ERY, CLI, TET | 3 |
| PG I, CHL, ERY, CLI, SXT | 2 |
| PG I, ERY, CLI, TET, SXT | 18 |
| PG I, CHL, TET, SXT | 29 |
| PG I, ERY, CLI, TET | 26 |
| PG I, ERY, CLI, SXT | 2 |
| PG I, ERY, TET, SXT | 4 |
| PG I, CHL, SXT | 1 |
| PG I, CHL, TET | 1 |
| PG I, ERY, CHL | 1 |
| PG I, ERY, TET | 1 |
| PG I, TET, SXT | 5 |
| PG I, ERY | 1 |
| PG I, TET | 1 |
| PG I, SXT | 12 |
| PG I | 8 |
| CHL, ERY, CLI, TET, SXT | 4 |
| CHL, ERY, CLI, TET | 1 |
| ERY, CLI, TET, SXT | 12 |
| ERY, CLI, TET | 29 |
| ERY, CLI, SXT | 3 |
| ERY, TET, SXT | 1 |
| ERY, CLI | 13 |
| ERY, SXT | 2 |
| TET, SXT | 4 |
| ERY | 3 |
| SXT | 35 |
| Susceptible to all antimicrobial agents | 354 |
PG R, penicillin MIC ≥ 1.5μg/ml; PG I, penicillin 0.1 μg/ml ≤ MIC < 1.5 μg/ml; TET, tetracycline; ERY, erythromycin; CLI, clindamycin; CHL, chloramphenicol. Multidrug resistance to erythromycin, clindamycin, and tetracycline is highlighted in bold.
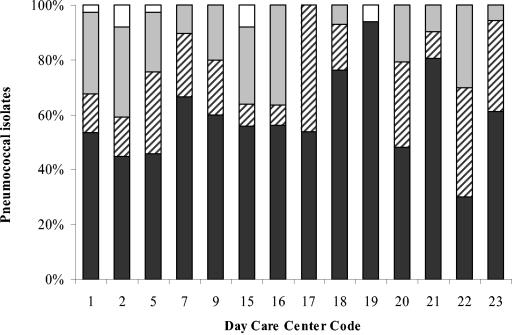
Carriage of antibiotic-resistant S. pneumoniae varied widely from one DCC to another (Fig. 1), reinforcing the idea that DCCs are autonomous epidemiological units (35). In DCC17, for example, all strains recovered (n = 13) were susceptible to penicillin, but half of them were resistant to at least another antimicrobial agent. By contrast, in DCC19, all strains (n = 17) but one were susceptible to all antimicrobial agents tested; in DCC22, similar proportions of isolates susceptible to all drugs (n = 13), susceptible to penicillin but resistant to some other drugs (n = 17), and intermediately resistant to penicillin (n = 13) were found. We used chi-square and Fisher exact tests to determine whether this diversity was associated with antibiotic intake in the month prior to the sampling and found no significant associations at a 95% confidence level (data not shown).
FIG. 1.
Distribution of antimicrobial resistance among S. pneumoniae isolates recovered in different day care centers. White bars, isolates resistant to penicillin; gray bars, isolates intermediately resistant to penicillin; dashed bars, isolates susceptible to penicillin but resistant to other antimicrobial agents; black bars, isolates susceptible to all antimicrobial agents tested.
Questionnaire.
Seven percent of the 942 children were taking antimicrobials at the time of sampling; close to 23% of the children had taken antimicrobial agents in the previous month, and 21% had taken three or more courses of antimicrobial agents in the previous 6 months. The antibiotics most frequently reported by the parents were amoxicillin (39%) and amoxicillin-clavulanic acid (41%); antibiotic use was mainly due to otitis media (50%) and tonsillitis (24%).
Serotyping and molecular typing of DRPn isolates.
A total of 202 DRPn out of 237 were characterized by serotyping and PFGE. The 35 out of 237 DRPn that were resistant to SXT only were not further characterized, because previous studies demonstrated a high degree of diversity among SXT-resistant strains in terms of serotypes and molecular types, suggesting frequent independent emergence of resistance to this drug (8). Among the 12 serotypes found the most frequent were 6B (26%), 14 (22%), 19F (16%), and 23F (10%). Other capsular types detected were 3 (<1%), 6A (1%), 9V (4%), 15A (4%), 15B (<1%), 19A (<1%), 23A (<1%), and 33F (2%). Twelve percent of the DRPn were nontypeable.
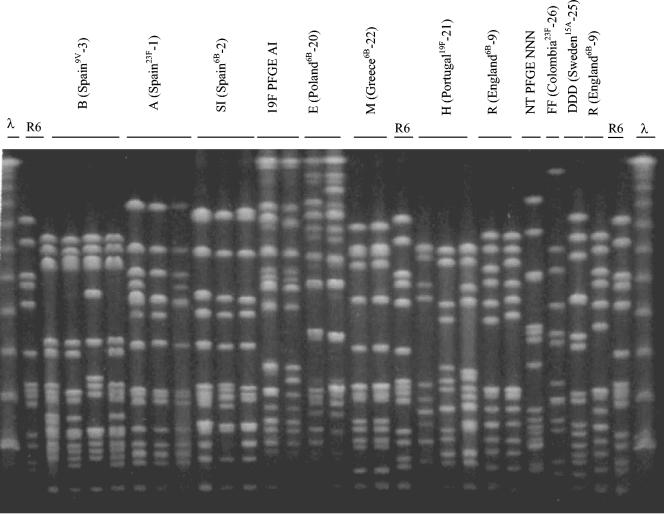
PFGE analysis grouped the isolates in 29 patterns, some of which are shown in Fig. 2. Table 2 summarizes relevant properties of the 202 DRPn strains, grouping them by level of resistance to penicillin and PFGE pattern. Serotypes, antibiotic resistance profiles, and distribution of isolates by DCC are also shown. PFGE patterns were associated with particular serotypes; the association with antibiotypes was less obvious. In all DCCs but one (DCC19, in which only one drug-resistant strain was detected), four or more PFGE patterns were identified among the DRPn isolates.
FIG. 2.
PFGE profiles of some drug-resistant S. pneumoniae clones isolated from children attending day care centers in Lisbon, Portugal. The λ ladder and R6 are indicated and were used as molecular weight markers. Clonal types of strains identical to PMEN representatives are designated by the PFGE code letter and the PMEN code between the brackets; other clonal types are designated by the serotype and PFGE code letter. NT, nontypeable.
TABLE 2.
Summary of clonal analysis of 202 drug-resistant pneumococcal isolates
| PFGE type | Capsular type | Antibiotype resistant toe: | No. of isolates | DCC code (no. of strains) |
|---|---|---|---|---|
| Resistant to penicillina | ||||
| B (Spain9V-3) | 9V/14 | PG, SXT | 6 | 2 (4), 5 (1), 19 (1) |
| A (Spain23F-1) | 23F | PG, CHL, TET, SXT | 4 | 1 (2), 15 (2) |
| Intermediate resistance to penicillinb | ||||
| A (Spain23F-1) | 23F | PG, CHL, TET (12), SXT | 13 | 1 (8), 2 (1), 5 (1), 7 (2), 15 (1) |
| B (Spain9V-3) | 9V/14 | PG, ERY (11), CHL (9), TET (10), SXT (13) | 26 | 1 (8), 2 (1), 5 (3), 7 (2), 9 (3), 15 (2), 16 (3), 18 (2), 21 (1), 22 (1) |
| SI (Spain6B-2) | 6B | PG, CHL, ERY (4), CLI (4), TET (8), SXT | 10 | 16 (10) |
| R (England14-9) | 14 | PG, ERY, CLI, SXT | 2 | 22 (2) |
| FF (Colombia23F-26) | 23F | PG | 3 | 2 (2), 16 (1) |
| E (Poland6B-20) | 6B | PG, CHL (2), ERY, CLI, TET, SXT (3) | 14 | 2 (3), 5 (2), 7 (1), 16 (2), 22 (6) |
| DDD (Sweden15A-25) | 15A | PG, ERY, CLI, TET | 6 | 1 (1), 20 (3), 21 (2) |
| NNN | NTd | PG, ERY, CLI, TET, SXT | 3 | 1 (1), 2 (1), 9 (1) |
| NF | NT | PG, ERY, TET, SXT | 2 | 2 (2) |
| BZ | 15A | PG, ERY, CLI, TET | 2 | 9 (1), 22 (1) |
| D | 19F | PG, ERY, CLI, TET, SXT | 4 | 15 (4) |
| AI | 19F | PG, CHL, ERY (3), CLI (3), TET, SXT | 14 | 16 (9), 20 (3), 22 (2) |
| NK | NT | PG, TET, SXT | 3 | 2 (3) |
| AQ | 19A | PG, ERY, CLI, TET, SXT (1) | 2 | 21 (1), 22 (1) |
| BE | NT | PG, ERY, CLI, TET, SXT | 6 | 21 (5), 5 (1) |
| Others with only one isolate | 10 | |||
| Penicillin susceptible but resistant to other drugsc | ||||
| B (Spain9V-3) | 9V/14 | ERY, CLI, TET (8), SXT (6) | 9 | 1 (4), 17 (2), 21 (3) |
| H (Portugal19F-21) | 19F | ERY, CLI, TET (4) | 9 | 1 (1), 2 (1), 17 (2), 18 (3), 20 (2) |
| R (England14-9) | 14 | ERY, CLI (5), SXT (2) | 8 | 9 (6), 21 (2) |
| M (Greece6B-22) | 6B | CHL (2), ERY, CLI, TET, SXT (5) | 11 | 5 (1), 7 (6), 16 (1), 21 (1), 17 (2) |
| E (Poland6B-20) | 6B | ERY (12), CLI (12), TET, SXT (1) | 13 | 2 (3), 5 (2), 22 (7), 23 (1) |
| AG | 33F | ERY, CLI | 5 | 7 (3), 18 (1), 23 (1) |
| AQ | 19F | ERY, CLI, TET | 2 | 16 (2) |
| X | 19F | ERY, CLI, TET | 2 | 1 (2) |
| BE | NT | ERY, CLI, TET, SXT | 2 | 21 (2) |
| NNN | NT | ERY, CLI (2), TET, SXT | 3 | 2 (1), 15 (2) |
| CA | 6A | ERY (1), CLI (1), TET, SXT | 3 | 22 (1), 23 (2) |
| Others with only one isolate | 5 |
MIC ≥ 1.5 μg/ml. Total isolates, 10 (5%).
0.1 μg/ml ≤ MIC < 1.5 μg/ml. Total isolates, 120 (59%).
Total isolates, 72 (36%).
NT, nontypeable.
PG, penicillin; TET, tetracycline; ERY, erythromycin; CLI, clindamycin; CHL, chloramphenicol. Numbers in parentheses indicate the number of isolates resistant to the antimicrobial agent (no number, all isolates were resistant).
By comparing the 29 PFGE patterns (clonal types) with patterns from previous studies (8, 34), the 29 clones were grouped as follows: representatives of Pneumococcal Molecular Epidemiology Network (PMEN) clones; Lisbon DCC clones, which included strains that do not belong to PMEN clones and have PFGE patterns identified in our previous studies (8, 34); and novel clones, which included strains with PFGE profiles that we could not identify in our previous studies (1996 to 1998). The majority (67%) of drug-resistant pneumococci were PMEN clones, 26% were novel clones, and 7% were Lisbon DCC clones. PMEN clones were present in all day care centers and, together, they accounted from 40 to 100% of the DRPn strains in various DCCs, thus representing the majority of the isolates. Strains belonging to the Lisbon DCC clones were detected in 5 DCCs, and the novel clones were identified in 11 of the 14 DCCs (Table 3). Some relevant properties of the clones of each group are described below.
TABLE 3.
Prevalence of PMEN clones, Lisbon DCC clones, and novel clones in each DCCa
| DCC code | Strain classification
|
||||
|---|---|---|---|---|---|
| No. of DRPn fully characterized | No. of PFGE types | % Found of PMEN clones | % Found of Lisbon DCC clones | % Found of novel clones | |
| 1 | 31 | 9 | 77 | 13 | 10 |
| 2 | 25 | 9 | 60 | 12 | 28 |
| 5 | 13 | 7 | 77 | 8 | 15 |
| 7 | 14 | 5 | 79 | 0 | 21 |
| 9 | 11 | 4 | 82 | 9 | 9 |
| 15 | 11 | 4 | 46 | 54 | 0 |
| 16 | 28 | 7 | 57 | 0 | 43 |
| 17 | 6 | 3 | 100 | 0 | 0 |
| 18 | 8 | 5 | 63 | 0 | 37 |
| 19 | 1 | 1 | 100 | 0 | 0 |
| 20 | 9 | 3 | 56 | 0 | 44 |
| 21 | 19 | 8 | 47 | 0 | 53 |
| 22 | 21 | 7 | 76 | 0 | 24 |
| 23 | 5 | 5 | 40 | 0 | 60 |
| Total | 202 | 29 | 135 (67%) | 15 (7%) | 52 (26%) |
PMEN clones found in our study: A, clone Spain23F-1; SI, clone Spain6B-2; B, clone Spain9V-3; R, clone England14-9; E, clone Poland6B-20; H, clone Portugal19F-21; M, clone Greece6B-22; DDD, clone Sweden15A-25; FF, clone Colombia23F-26. Lisbon DCC clones, clones detected in our previous day care center studies; Novel clones, clones found in this study.
PMEN clones.
Nine PMEN clones were detected: clone Spain23F-1 present in five DCCs; Spain6B-2 in one DCC; Spain9V-3 in 12 DCCs; England14-9 in three DCCs; Poland6B-20 in six DCCs; clone Portugal19F-21 in five DCCs; clone Greece6B-22 in five DCCs; clone Sweden15A-25 in three DCCs, and clone Colombia23F-26 in two DCCs.
Clone B (Spain9V-3) was the most prevalent (20% of the DRPn strains) and also the most widely disseminated—detected in 12 out of 14 DCCs. Strains were either of serotype 9V or 14. Although the majority of the strains had intermediate resistance to penicillin and were also resistant to erythromycin and clindamycin, strains with other biotypes were also detected. Clone SI (Spain6B-2) was detected in DCC16. In our previous studies (8, 34) no strains of this clone were identified. MLST was performed for two strains and the allelic profile ST90 characteristic of the Spain6B-2 clone reference strain was identified (Table 4) (27). The remaining PMEN clones (Spain23F-1, England6B-9, Poland6B-20, Portugal19F-21, Greece6B-22, Sweden15A-25, and Colombia23F-26) had all been recovered in 1996 to 1998 and had the same properties described previously (8, 34).
TABLE 4.
Multilocus sequence typing of selected isolatesa
| Strain | Antibiotype | Serotype | PFGE | ST | Observationsb |
|---|---|---|---|---|---|
| 2613 | PG, CHL, TET, ERY, CLI, SXT | 6B | SI | 90 | Spain6B-2 |
| 2623 | PG, CHL, TET, SXT | 6B | SI | 90 | Spain6B-2 |
| 2119 | PG, ERY, CLI, TET, SXT | NT | NNN | 344 | Norway, Australia |
| 2430 | ERY, CLI, TET, SXT | NT | NNN | 344 | Norway, Australia |
| 2859 | PG, ERY, CLI, TET, SXT | NT | BE | 897 | dlv of ST344 |
| 2870 | ERY, CLI, TET, SXT | NT | BE | 897 | dlv of ST344 |
| 2659 | PG, ERY, TET, SXT | 19F | AI | 87 | Denmark |
| 2798 | PG, CHL, ERY, CLI, TET, SXT | 19F | AI | 1198 | slv of ST88 (Spain, Italy) |
| 3012 | ERY, CLI | 33F | AG | 1199 | slv of ST717 (United Kingdom) |
| 2253 | ERY, CLI | 33F | AG | 717 | United Kingdom |
PG, penicillin; TET, tetracycline; ERY, erythromycin; CLI, clindamycin; CHL, chloramphenicol; ST, sequence type; dlv, double locus variant; slv, single locus variant; NT, nontypeable.
Country where ST or related ST was detected. Further details on epidemiological data are provided in the text.
Lisbon DCC clones.
Six clonal types were identified: NNN, D, X, XXX, EEE, and WWW. The six strains of clone NNN were all nontypeable, were resistant to erythromycin, clindamycin, tetracycline, and SXT, and were either susceptible to penicillin or intermediately resistant to penicillin. This clone was detected in four DCCs. In our earlier studies clone NNN was recovered in a single DCC only (DCC2) in 1997 and 1998 (35). By MLST this clone had ST344, i.e., had the same allelic profile of two nontypeable strains recovered from patients with invasive disease in Norway in 1996 and Australia in 1998 (Table 4). Four strains of clone D and capsular type 19F were recovered in DCC15. The strains had intermediate resistance to penicillin and were also resistant to erythromycin, clindamycin, tetracycline, and SXT. This clone was also detected in our earlier studies since 1996 but only in DCC7, and the previous isolates were resistant to penicillin and SXT only (8).
Novel clones.
Three of the 14 novel clonal types (NF, NK, and BE) were nontypeable. The other clones were of serotypes 6A, 15A, 19A, 19F, and 33F. Three clones (BE, AI, and AG) had a high number of isolates accounting for 52% of these isolates. Representatives of these three clones were selected for MLST.
Clone BE was found in two DCCs and all the strains were nontypeable and resistant to erythromycin, clindamycin, tetracycline, and SXT; six out of eight strains also had intermediate resistance to penicillin. This clone, although distinct by PFGE, according to MLST is a double-locus variant of the clone of PFGE type NNN (also nontypeable), described above (Table 4).
All strains of clone AI had capsular type 19F and were resistant to chloramphenicol, tetracycline, and SXT. These strains were recovered in three DCCs (DCC16, -20, and -22). Strains recovered from children in DCC20 were also resistant to erythromycin and clindamycin. MLST of two representative isolates yielded two sequence types (ST87 and ST1198), differing in the gki and ddl alleles. Comparison with data from the MLST database identified one strain also with ST87, which was found to cause invasive disease in Denmark in 1997, and three isolates with double-locus variants of ST87 that were detected in Spain in 1989 and 1997 and in 1999 in Italy (Table 4).
All strains of clone AG had capsular type 33F and were resistant to erythromycin and clindamycin and were found in three DCCs (DCC7, -18, and -23). By MLST, two ST types were identified, ST1199 and ST717 (differing only in the ddl allele). ST717 was previously identified in one invasive strain from the United Kingdom in 2003, which was also of type 33F (Table 4).
DISCUSSION
This study was a follow-up of the Lisbon Day Care Center Initiative which started in 1996 and was conducted on a yearly basis (8, 9, 34, 35). The main purpose of the initiative was to monitor the prevalence of drug-resistant pneumococcal strains among children attending DCCs and to characterize the strains by serotyping and molecular typing techniques. No such studies had ever been conducted in Portugal and, to our best knowledge, this remains the only surveillance program of this type in the country. In addition, although other studies on the pneumococci carried by DCC attendees have been conducted in other countries (3, 4, 19, 29), we are not aware of efforts that have lasted for several years and are of such large magnitude, where all DRPn (including strains susceptible to penicillin) have been extensively characterized by serotyping and genotyping (and antibiotyping), with the possible exception of a study described by Nilsson and Laurell (29) in Malmo, Sweden, which included close to 3,000 children attending 63 DCCs during a 3-year period (1995 to 1997). In that study, molecular typing was done by AP-PCR and BOX-A PCR. Other DCC studies carried out in Greece (4), The Netherlands (3), and Israel (19) have been described. These studies had smaller numbers of children and of pneumococcal isolates. In addition, these studies were point-prevalence ones and did not include yearly surveillances like ours, and they also had different designs. In particular, the Israeli study was a 5-month longitudinal study of 252 children; the authors used ribotyping to characterize the 594 pneumococci (19). Two Greek studies were performed since December of 1995. The first one was performed with 338 children attending seven day care centers. The study was conducted from December of 1995 to February of 1996. The second study, was conducted from February of 1997 to February of 1999, but only 95 of the 2,448 children sampled attended day care centers; the authors used RFEL and MLST to characterize 128 isolates susceptible to penicillin but resistant to other antimicrobials (4). A study from The Netherlands compared 259 children attending 16 DCCs with 276 children who did not attend DCCs. Children were sampled twice from January to March of 1999, and a total of 305 pneumococci were recovered (3).
In fact, in the 4 years of the study, 23 DCCs and 1,800 children were enrolled, generating 3,053 nasopharyngeal samples of which 1,687 were positive for pneumococci. All pneumococcal isolates were characterized by antibiotype. Of the 740 DRPn recovered, 615 pneumococci strains were completely characterized by serotyping and PFGE typing (8, 35).
These data have been a rich source of information on the pneumococcal population and have enabled us to observe trends of the DRPn in its ecological niche. In addition, these studies have provided a good baseline for two intervention studies that began in 2001 aimed to decrease carriage of DRPn: one based on education on antibiotic use and hygiene control measures (the EURIS project, European Resistance Intervention Study) and the other based on the licensed 7-valent conjugate vaccine Prevenar (in preparation). The EURIS project is now being continued in a sequel named PREVIS (Pneumococcal Resistance, Epidemicity, and Virulence—an International Study) and, by 2006, we will have completed a decade of annual-biannual surveillance of pneumococci carried by DCC attendees.
The data from the 1999 surveillance described here indicate that the carriage rate of S. pneumoniae in Portuguese DCCs has increased continuously since 1996, from 47% to 48, 60, and 63% in subsequent years (35). The reasons for this increase are not clear. We can only speculate that the nurses who collected the samples have been mastering their sampling technique, yielding higher pneumococcal recovery rates. Similar high rates of pneumococcal carriage in DCC children have been reported in other countries such as Iceland (50%) (23), Sweden (52%) (22), Lithuania (51%) (23), France (54.7%) (12), Israel (63%) (45), and Greece (48%) (44).
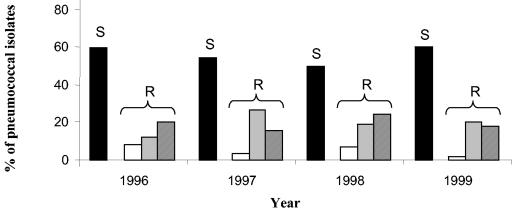
Comparison of the 1999 results with results of the previous years of surveillance indicated that the proportion of DRPn ranged between 40 to 50% of all isolates. The carriage rate of penicillin-resistant and penicillin-intermediate pneumococci also fluctuated—ranging from 2 to 8% and 12 to 27%, respectively—over time. Interestingly, 1999 was the year with the lowest rate of penicillin resistance (2%) (Fig. 3).
FIG. 3.
Comparison of 4 years of the study. White bars, isolates resistant to penicillin; light gray bars, isolates intermediately resistant to penicillin; dark gray bars, isolates susceptible to penicillin but resistant to other antimicrobial agents; black bars, isolates susceptible to all antimicrobial agents tested. The total number of pneumococci was 277, 354, 465, and 591 in the four consecutive years, respectively.
Analysis of Portuguese invasive strains, recovered between 1999 and 2001 from pediatric patients of the same age group, identified higher values of DRPn (63.8%) and of resistance to penicillin (4.7%) (38). However, drug resistance to antimicrobials other than penicillin was 15.9% among invasive pediatric isolates, a value comparable to the 18% carriage rate of such strains identified in our study. In 1999, 64% of the penicillin-susceptible strains were coresistant to macrolides, clindamycin, and tetracycline, a trend previously observed in 1997 and 1998 studies (17, 33-35). DCC studies in Greece (44) found antibiotypes and levels of erythromycin and tetracycline resistance comparable to those of our study. The Portuguese and Greek data contrast with data from pneumococcal isolates from DCC attendees in Sweden which had similar levels of penicillin nonsusceptibility (20%) (36) but only rarely were resistant to erythromycin, tetracycline, and/or SXT.
The antimicrobial consumption was similar in all years and the antibiotics most frequently used were always amoxicillin-clavulanic acid and amoxicillin. In our studies otitis has always been the major cause for antibiotic consumption reported by the parents. Otitis is highly recognized as the most frequent affliction responsible for visits to pediatricians and for antibiotic prescription, and thus is a major cause of morbidity and has high costs associated with it (6). In addition, a study on the similarity of strains recovered from otitis and carriage (20) suggests that intervention aimed to decrease carriage will also target otitis media.
The most frequent serotypes recovered in 1999 (6B, 14, 19F, 23F, and nontypeable) were all (except for nontypeable isolates) included in the 7-valent conjugate vaccine. The 7-valent conjugate vaccine covered 78% of the DRPn isolates. These capsular types had already been included among the most frequent serotypes recovered in the past years (8, 34). These were also the serotypes most commonly found in healthy children in Sweden (36), Israel (31), the United Kingdom (5), and Greece (41), although in a different rank order. Serotypes 14, 23F, and 6B were also among the five most common capsular types in invasive strains recovered in 1999 to 2001 in Portugal from children younger than 6 years old (38). In our study, nonvaccine types (3, 6A, 15A, 15B, 19A, and 33F) together with nontypeable strains accounted for 22% of the DRPn. All these nonvaccine types had been detected in previous years apart from serotype 33F (8, 35).
Molecular typing identified several PMEN clones among the isolates. The international clone Spain9V-3, first detected in Spain (7) and France (18, 25), was the clone most frequently isolated in our study. Interestingly, strains belonging to this clonal type also included isolates that had intermediate levels of resistance or were fully susceptible to penicillin. Strains of Spain9V-3 have most often been described as fully resistant to penicillin (27) and isolates of invasive origin have been recovered in several countries (11, 13, 26). In our study, the majority of strains belonging to this clonal type expressed serotype 14, which represents one of the most frequent serotypes among invasive isolates in Europe, the United States, and Canada (21).
Interestingly, the relative proportion of strains of serotype 23F (10%) decreased when compared to those from 1998 (P = 0.0038), reflecting the lower prevalence of international clone Spain23F-1 in the present study (Table 5). On the other hand, the multiresistant Spain6B-2 clone was now detected, for the first time, in Portuguese DCCs. The clone was detected in a single DCC. Another six PMEN clones were identified (England14-9, Poland6B-20, Portugal19F-21, Greece6B-22, Sweden15A-25, and Colombia23F-26); all of them had also been recovered in earlier Portuguese DCC studies (8, 34). Clones D and NNN, found since 1996 in a single DCC (35), continued to be detected in other DCCs and thus are persisting and appear to be spreading through time (Table 5).
TABLE 5.
Prevalence over time of major clones found in 1999
| Clone | Percent found in:
|
|||
|---|---|---|---|---|
| 1996 (n = 91) | 1997 (n = 131) | 1998 (n = 191) | 1999 (n = 202) | |
| Spain23F-1 | 22.0 | 16.0 | 14.7 | 8.4 |
| Spain9V-3 | 11.0 | 1.5 | 13.6 | 20.3 |
| Poland6B-20 | 5.5 | 9.9 | 2.1 | 13.4 |
| Portugal19F-21 | 4.4 | 8.4 | 5.2 | 4.5 |
| Colombia23F-26 | 0.0 | 10.7 | 4.7 | 1.5 |
| Greece6B-22 | 19.8 | 7.6 | 18.8 | 5.4 |
| England14-9 | 1.1 | 0.0 | 14.1 | 5.0 |
| Sweden15A-25 | 0.0 | 6.1 | 3.7 | 3.0 |
| Spain6B-2 | 0.0 | 0.0 | 0.0 | 1.0 |
| D | 12.1 | 9.9 | 0.0 | 2.0 |
| NNN | 0.0 | 0.8 | 1.6 | 3.0 |
| New | 12.2 | 15.2 | 24.8 | |
| Unique | 13.2 | 10.7 | 5.2 | 7.4 |
Novel clones were detected and accounted for 26% of the DRPn strains isolated. Three of them, BE, AI, and AG, had MLST sequence types that were identical or differed in only one or two loci from strains of invasive origin recovered in other countries. Interestingly, clone AG was associated with serotype 33F, an unusual capsular type in carriage (2); strains of clone BE were nontypeable. Both clones have “capsules” not included in 7-valent conjugate vaccine and are multiresistant to antimicrobial agents. An increased prevalence of these strains among carried isolates could be, as postulated by others (36), a preview of what will soon appear in the pneumococcal disease population.
In summary, the majority of drug-resistant strains recovered from DCC children in Portugal are members of multidrug-resistant international clones that can cause illness (invasive and noninvasive). The emergence of a serotype 33F clone and the increasing number of nontypeable clones, both with capacity to cause acute otitis media (5, 30), highlight that the nasopharynxes of DCC children are a melting pot that is important to monitor (2) if we aim to gain a better understanding of the epidemiology of this pathogen.
Acknowledgments
Partial support for this work was provided by contracts EURIS (QLK2-CT-2000-01020) from the European Community, PRAXIS/P/SAU/14051/1998 from Fundação para a Ciência e Tecnologia, Portugal, and a grant from Fundação Calouste Gulbenkian, Portugal, awarded to H. de Lencastre. R. Sá-Leão and J. Carriço were supported by grants SFRH/BPD/14596/2003 and SFRH/BD/3123/2000, respectively, both from Fundação para a Ciência e Tecnologia, Portugal. S. Nunes was supported by grant 011/BIC/01 from contract QLK2-CT-2000-01020 and C. R. Alves was supported by grant 001/99/BIC/P from contract PRAXIS/P/SAU/14051/1998.
We thank Natacha Sousa for assistance in MLST typing, Idalina Bonfim for helping with isolation and characterization of the strains, and the experienced state-registered pediatric nurse Anabela Gonçalves. We thank Alexander Tomasz of The Rockefeller University for helpful suggestions and critical reading of the manuscript.
REFERENCES
- 1.Baquero, F., J. A. Garcia-Rodriguez, J. Garcia de Lomas, and L. Aguilar, and The Spanish Surveillance Group for Respiratory Pathogens. 1999. Antimicrobial resistance of 1,113 Streptococcus pneumoniae isolates from patients with respiratory tract infections in Spain: results of a 1-year (1996-1997) multicenter surveillance study. Antimicrob. Agents Chemother 43:357-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogaert, D., R. De Groot, and P. W. Hermans. 2004. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect. Dis. 4:144-154. [DOI] [PubMed] [Google Scholar]
- 3.Bogaert, D., M. N. Engelen, A. J. Timmers-Reker, K. P. Elzenaar, P. G. Peerbooms, R. A. Coutinho, R. de Groot, and P. W. Hermans. 2001. Pneumococcal carriage in children in The Netherlands: a molecular epidemiological study. J. Clin. Microbiol. 39:3316-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogaert, D., P. W. Hermans, I. N. Grivea, G. S. Katopodis, T. J. Mitchell, M. Sluijter, R. De Groot, N. G. Beratis, and G. A. Syrogiannopoulos. 2003. Molecular epidemiology of penicillin-susceptible non-beta-lactam-resistant Streptococcus pneumoniae isolates from Greek children. J. Clin. Microbiol. 41:5633-5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brueggemann, A. B., D. T. Griffiths, E. Meats, T. Peto, D. W. Crook, and B. G. Spratt. 2003. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J. Infect. Dis. 187:1424-1432. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 1997. Prevention of pneumococcal disease: recommendations of the Advisory Committee on Immunization Practices (ACIP). Morb. Mortal. Wkly. Rep. 46:1-24. [Google Scholar]
- 7.Coffey, T. J., C. G. Dowson, M. Daniels, J. Zhou, C. Martin, B. G. Spratt, and J. M. Musser. 1991. Horizontal transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol. Microbiol. 5:2255-2260. [DOI] [PubMed] [Google Scholar]
- 8.de Lencastre, H., K. G. Kristinsson, A. Brito-Avô, I. S. Sanches, R. Sá-Leão, J. Saldanha, E. Sigvaldadottir, S. Karlsson, D. Oliveira, R. Mato, M. Aires de Sousa, and A. Tomasz. 1999. Carriage of respiratory tract pathogens and molecular epidemiology of Streptococcus pneumoniae colonization in healthy children attending day care centers in Lisbon, Portugal. Microb. Drug Resist. 5:19-29. [DOI] [PubMed] [Google Scholar]
- 9.de Lencastre, H., I. Santos Sanches, A. Brito-Avô, R. Sá-Leão, J. Saldanha, K. G. Kristinsson, and A. Tomasz. 1999. Carriage and antibiotic resistance of respiratory pathogens and molecular epidemiology of antibiotic-resistant Streptococcus pneumoniae colonizing children in day-care centers in Lisbon: the Portuguese day-care center initiative. Clin. Microbiol. Infect. 5:S55-S63. [DOI] [PubMed] [Google Scholar]
- 10.Di Fabio, J. L., E. Castaneda, C. I. Agudelo, F. De La Hoz, M. Hortal, T. Camou, G. Echaniz-Aviles, M. Noemi, C. Barajas, I. Heitmann, J. C. Hormazabal, M. C. Brandileone, V. S. Dias Vieira, M. Regueira, R. Ruvinski, A. Corso, M. Lovgren, J. A. Talbot, C. De Quadros, et al. 2001. Evolution of Streptococcus pneumoniae serotypes and penicillin susceptibility in Latin America, Sireva-Vigia Group, 1993 to 1999. Pediatr. Infect. Dis. J. 20:959-967. [DOI] [PubMed] [Google Scholar]
- 11.Doit, C., B. Picard, C. Loukil, P. Geslin, and E. Bingen. 2000. Molecular epidemiology survey of penicillin-susceptible and -resistant Streptococcus pneumoniae recovered from patients with meningitis in France. J. Infect. Dis. 181:1971-1978. [DOI] [PubMed] [Google Scholar]
- 12.Dunais, B., C. Pradier, H. Carsenti, M. Sabah, G. Mancini, E. Fontas, and P. Dellamonica. 2003. Influence of child care on nasopharyngeal carriage of Streptococcus pneumoniae and Haemophilus influenzae. Pediatr. Infect. Dis. J. 22:589-592. [DOI] [PubMed] [Google Scholar]
- 13.Enright, M. C., A. Fenoll, D. Griffiths, and B. G. Spratt. 1999. The three major Spanish clones of penicillin-resistant Streptococcus pneumoniae are the most common clones recovered in recent cases of meningitis in Spain. J. Clin. Microbiol. 37:3210-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 15.Felmingham, D., C. Feldman, W. Hryniewicz, K. Klugman, S. Kohno, D. E. Low, C. Mendes, and A. C. Rodloff. 2002. Surveillance of resistance in bacteria causing community-acquired respiratory tract infections. Clin. Microbiol. Infect. 8:12-42. [DOI] [PubMed] [Google Scholar]
- 16.Felmingham, D., R. R. Reinert, Y. Hirakata, and A. Rodloff. 2002. Increasing prevalence of antimicrobial resistance among isolates of Streptococcus pneumoniae from the PROTEKT surveillance study, and comparative in vitro activity of the ketolide, telithromycin. J. Antimicrob. Chemother. 50:25-37. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Rey, C., E. Bouza, L. Aguilar, J. Garcia-de-Lomas, and F. Baquero. 2003. Evolution of penicillin and erythromycin co-resistance in Streptococcus pneumoniae in Spain. Int. J. Antimicrob. Agents 22:541-544. [DOI] [PubMed] [Google Scholar]
- 18.Gasc, A. M., P. Geslin, and A. M. Sicard. 1995. Relatedness of penicillin-resistant Streptococcus pneumoniae serogroup 9 strains from France and Spain. Microbiology 141:623-627. [DOI] [PubMed] [Google Scholar]
- 19.Givon-Lavi, N., R. Dagan, D. Fraser, P. Yagupsky, and N. Porat. 1999. Marked differences in pneumococcal carriage and resistance patterns between day care centers located within a small area. Clin. Infect. Dis. 29:1274-1280. [DOI] [PubMed] [Google Scholar]
- 20.Hanage, W. P., K. Auranen, R. Syrjanen, E. Herva, P. H. Makela, T. Kilpi, and B. G. Spratt. 2004. Ability of pneumococcal serotypes and clones to cause acute otitis media: implications for the prevention of otitis media by conjugate vaccines. Infect. Immun. 72:76-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hausdorff, W. P., J. Bryant, C. Kloek, P. R. Paradiso, and G. R. Siber. 2000. The contribution of specific pneumococcal serogroups to different disease manifestations: implications for conjugate vaccine formulation and use, part II. Clin. Infect. Dis. 30:122-140. [DOI] [PubMed] [Google Scholar]
- 22.Henriques Normark, B., B. Christensson, A. Sandgren, B. Noreen, S. Sylvan, L. G. Burman, and B. Olsson-Liljequist. 2003. Clonal analysis of Streptococcus pneumoniae nonsusceptible to penicillin at day-care centers with index cases, in a region with low incidence of resistance: emergence of an invasive type 35B clone among carriers. Microb. Drug Resist. 9:337-344. [DOI] [PubMed] [Google Scholar]
- 23.Hjaltested, E. K., J. Bernatoniene, H. Erlendsdottir, P. Kaltenis, G. Bernatoniene, T. Gudnason, A. Haraldsson, and K. G. Kristinsson. 2003. Resistance in respiratory tract pathogens and antimicrobial use in Icelandic and Lithuanian children. Scand. J. Infect. Dis. 35:21-26. [DOI] [PubMed] [Google Scholar]
- 24.Kellner, J. D., E. L. Ford-Jones, et al. 1999. Streptococcus pneumoniae carriage in children attending 59 Canadian child care centers. Arch. Pediatr. Adolesc. Med. 153:495-502. [DOI] [PubMed] [Google Scholar]
- 25.Lefevre, J. C., M. A. Bertrand, and G. Faucon. 1995. Molecular analysis by pulsed-field gel electrophoresis of penicillin-resistant Streptococcus pneumoniae from Toulouse, France. Eur. J. Clin. Microbiol. Infect. Dis. 14:491-497. [DOI] [PubMed] [Google Scholar]
- 26.Marchese, A., M. Ramirez, G. C. Schito, and A. Tomasz. 1998. Molecular epidemiology of penicillin-resistant Streptococcus pneumoniae isolates recovered in Italy from 1993 to 1996. J. Clin. Microbiol. 36:2944-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefevre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.National Committee for Clinical Laboratory Standards. 1997. Performance standards for antimicrobial disk susceptibility tests, 6th ed., vol. 17, no. 1. Approved standard M2-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 29.Nilsson, P., and M. H. Laurell. 2001. Carriage of penicillin-resistant Streptococcus pneumoniae by children in day-care centers during an intervention program in Malmo, Sweden. Pediatr. Infect. Dis. J. 20:1144-1149. [DOI] [PubMed] [Google Scholar]
- 30.Porat, N., G. Barkai, M. R. Jacobs, R. Trefler, and R. Dagan. 2004. Four antibiotic-resistant Streptococcus pneumoniae clones unrelated to the pneumococcal conjugate vaccine serotypes, including 2 new serotypes, causing acute otitis media in southern Israel. J. Infect. Dis. 189:385-392. [DOI] [PubMed] [Google Scholar]
- 31.Regev-Yochay, G., M. Raz, R. Dagan, N. Porat, B. Shainberg, E. Pinco, N. Keller, and E. Rubinstein. 2004. Nasopharyngeal carriage of Streptococcus pneumoniae by adults and children in community and family settings. Clin. Infect. Dis. 38:632-639. [DOI] [PubMed] [Google Scholar]
- 32.Sahm, D. F., J. A. Karlowsky, L. J. Kelly, I. A. Critchley, M. E. Jones, C. Thornsberry, Y. Mauriz, and J. Kahn. 2001. Need for annual surveillance of antimicrobial resistance in Streptococcus pneumoniae in the United States: 2-year longitudinal analysis. Antimicrob. Agents Chemother. 45:1037-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sá-Leão, R., A. Tomasz, and H. de Lencastre. 2001. Multilocus sequence typing of Streptococcus pneumoniae clones with unusual drug resistance patterns: genetic backgrounds and relatedness to other epidemic clones. J Infect. Dis. 184:1206-1210. [DOI] [PubMed] [Google Scholar]
- 34.Sá-Leão, R., A. Tomasz, I. S. Sanches, A. Brito-Avô, S. E. Vilhelmsson, K. G. Kristinsson, and H. de Lencastre. 2000. Carriage of internationally spread clones of Streptococcus pneumoniae with unusual drug resistance patterns in children attending day care centers in Lisbon, Portugal. J. Infect. Dis. 182:1153-1160. [DOI] [PubMed] [Google Scholar]
- 35.Sá-Leão, R., A. Tomasz, I. S. Sanches, S. Nunes, C. R. Alves, A. B. Avô, J. Saldanha, K. G. Kristinsson, and H. de Lencastre. 2000. Genetic diversity and clonal patterns among antibiotic-susceptible and -resistant Streptococcus pneumoniae colonizing children: day care centers as autonomous epidemiological units. J. Clin. Microbiol. 38:4137-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandgren, A., K. Sjostrom, B. Olsson-Liljequist, B. Christensson, A. Samuelsson, G. Kronvall, and B. Henriques Normark. 2004. Effect of clonal and serotype-specific properties on the invasive capacity of Streptococcus pneumoniae. J. Infect. Dis. 189:785-796. [DOI] [PubMed] [Google Scholar]
- 37.Schito, G. C., E. A. Debbia, and A. Marchese. 2000. The evolving threat of antibiotic resistance in Europe: new data from the Alexander Project. J. Antimicrob Chemother. 46:3-9. [DOI] [PubMed] [Google Scholar]
- 38.Serrano, I., M. Ramirez, and J. Melo-Cristino. 2004. Invasive Streptococcus pneumoniae from Portugal: implications for vaccination and antimicrobial therapy. Clin. Microbiol. Infect. 10:652-656. [DOI] [PubMed] [Google Scholar]
- 39.Song, J. H., N. Y. Lee, S. Ichiyama, R. Yoshida, Y. Hirakata, W. Fu, A. Chongthaleong, N. Aswapokee, C. H. Chiu, M. K. Lalitha, K. Thomas, J. Perera, T. T. Yee, F. Jamal, U. C. Warsa, B. X. Vinh, M. R. Jacobs, P. C. Appelbaum, and C. H. Pai. 1999. Spread of drug-resistant Streptococcus pneumoniae in Asian countries: Asian Network for Surveillance of Resistant Pathogens (ANSORP) Study. Clin. Infect. Dis. 28:1206-1211. [DOI] [PubMed] [Google Scholar]
- 40.Sorensen, U. B. 1993. Typing of pneumococci by using 12 pooled antisera. J. Clin. Microbiol. 31:2097-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Syrogiannopoulos, G. A., G. D. Katopodis, I. N. Grivea, and N. G. Beratis. 2002. Antimicrobial use and serotype distribution of nasopharyngeal Streptococcus pneumoniae isolates recovered from Greek children younger than 2 years old. Clin. Infect. Dis. 35:1174-1182. [DOI] [PubMed] [Google Scholar]
- 42.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thornsberry, C., D. F. Sahm, L. J. Kelly, I. A. Critchley, M. E. Jones, A. T. Evangelista, and J. A. Karlowsky. 2002. Regional trends in antimicrobial resistance among clinical isolates of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis in the United States: results from the TRUST Surveillance Program, 1999-2000. Clin. Infect. Dis. 34:S4-S16. [DOI] [PubMed] [Google Scholar]
- 44.Tsolia, M., G. Kouppari, A. Zaphiropoulou, S. Gavrili, M. Tsirepa, D. Kafetzis, and T. Karpathios. 1999. Prevalence and patterns of resistance of Streptococcus pneumoniae strains isolated from carriers attending day care centers in the area of Athens. Microb. Drug Resist. 5:271-278. [DOI] [PubMed] [Google Scholar]
- 45.Yagupsky, P., N. Porat, D. Fraser, F. Prajgrod, M. Merires, L. McGee, K. P. Klugman, and R. Dagan. 1998. Acquisition, carriage, and transmission of pneumococci with decreased antibiotic susceptibility in young children attending a day care facility in southern Israel. J. Infect. Dis. 177:1003-1012. [DOI] [PubMed] [Google Scholar]