Abstract
We studied the pheno- and genotypes of an oral Granulicatella elegans strain in comparison with those of a blood-derived isolate which caused infective endocarditis. The two isolates exhibited identical biochemical characteristics and had the same drug MICs. Their genotypes were indistinguishable, indicating that these were from the same clone. The transmission of G. elegans from the oral cavity thus should be noted as a possible cause of infective endocarditis.
Infective endocarditis (IE) is a relatively uncommon but life-threatening disease of the endocardium of the heart. Strong risk factors include cardiac valvular abnormalities, where certain microorganisms entering into the bloodstream can potentially attach and grow and then cause the disease (8, 20, 26). The most predominant pathogens of IE are bacterial species in the oral cavity, such as viridans streptococci (1, 26, 27). The tooth-tissue interface is a typical portal for bacteria to enter the body, and nearly all physical entries of organisms into the bloodstream, i.e., of those that can cause transient bacteremia, occur during routine tooth brushing and food chewing in healthy individuals (6), though bacteremia is more often the result of dental treatment (14, 15). Therefore, resident oral bacteria which inevitably and frequently enter the body possibly become major pathogens of IE (9, 10). However, direct proof of this theory has not been shown and the pathogenicity of the bacteria in resident floras remains to be elucidated. In the present study, we investigated a case of IE caused by one of the most fastidious bacteria, Granulicatella elegans. This bacterium was first described as a member of a family of nutritionally variant streptococci (NVS) (22) but was later reclassified into the new genus Granulicatella (7).
A 53-year-old woman showing clinical signs of acute endocarditis was transferred from an outside facility and admitted to the Iwate Medical University Memorial Heart Center in Morioka, Japan. Medical historical data revealed a dental procedure 3 months before without any other anamnesis. Four consecutive arterial and venous blood cultures were successively performed at the outside facility as well as at our department with BACTEC PLUS Anaerobic/F culture bottles (Becton Dickinson and Company, Sparks, Md.) and Brucella HK agar plates (Kyokuto Pharma. Indust. Co., Tokyo, Japan) at 35°C under anaerobic conditions. Two sets of cultures tested positive and yielded gram-positive coccoids in short chains. The patient was empirically treated intravenously with penicillin (30 × 104 U/kg of body weight once a day) and gentamicin sulfate (3 mg/kg of body weight once a day). Cardiac surgery was performed 7 days after admission. She was released from the hospital without a fever after 27 days.
PCR amplification of the 16S rRNA gene was performed with 50 ng of purified genomic DNA as described previously (18). DNA sequencing of the amplicons was carried out using the primer EB1.5-1 (30). Obtained sequences of about 600 nucleotides were compared with known sequences by use of the BLAST algorithm (National Center for Biotechnology Information, Bethesda, Md.). MICs were determined by a microdilution method of the National Committee for Clinical Laboratory Standards (17) as described previously (29) with the minor modification of adding l-cysteine HCl (0.01%) to the medium. Pulsed-field gel electrophoresis (PFGE) was performed following restriction digestion with SmaI and ApaI as described elsewhere (24). For an arbitrarily primed (AP)-PCR analysis, purified genomic DNA (5 ng) was amplified randomly with primer AP3 (2).
Species identification of the isolate, designated as strain IMU02b01, from blood cultures was confirmed by determination of the 16S rRNA gene sequence, which demonstrated 99.4% identity with the sequence of G. elegans (GenBank accession number Y1543.1). To investigate whether the pathogen in this case of IE was of oral origin, we attempted to isolate G. elegans from the oral cavity of the patient. We collected specimens of dental plaque and cultured them as described previously (18) and chose 11 isolates of gram-positive coccoids in chains which demonstrated bacteriolytic activity and satellitism. Their 16S rRNA gene sequences were verified by PCR direct sequencing. Among the candidates, the sequence of one isolate, designated IMU02p18, demonstrated 99.1% similarity to that of G. elegans. Further, eight isolates showed sequence similarities to Granulicatella adiacens ranging from 92 to 99%; we did not obtain sequence data for the other two strains.
Strain IMU02p18 exhibited the same biochemical characteristics as strain IMU02b01 and G. elegans CCUG 26024 (Table 1). Among the characteristics examined, urease production (25), acidification of raffinose and sucrose (7, 22), and hydrolysis of hippurate (7) have been reported to be strain dependent. Strain IMU02p18, as well as strain IMU02b01, were negative for urease, and the two isolates fermented sucrose but not raffinose. In addition, hippurate hydrolysis was positive with them. The antimicrobial susceptibility tendencies of these two isolates were also identical (Table 2). Both were highly susceptible to penicillin and other β-lactams; they were, however, intermediate to amikacin and resistant to arbekacin. In accordance with the results, combination treatment with penicillin and gentamicin was effective in this case. Notably, the present susceptibility profile is in agreement with those of other NVS isolates (23, 29), as many NVS strains are susceptible to penicillin whereas they exhibit various susceptibility characteristics with respect to aminoglycosides.
TABLE 1.
Biochemical characterization of blood- and oral cavity-derived G. elegans isolatesa
| Characteristic | Result for:
|
||
|---|---|---|---|
| Strain IMU02b01 | Strain IMU02p18 | G. elegans CCUG 26024 | |
| Enzyme production | |||
| Pyrrolidonyl aminopeptidase | + | + | + |
| Alkaline phosphatase | − | − | − |
| Urease | − | − | − |
| Arginine dihydrolase | + | + | + |
| α-Galactosidase | − | − | − |
| β-Galactosidase | − | − | − |
| β-Glucuronidase | − | − | − |
| α-Glucosidase | + | + | + |
| β-Glucosidase | − | − | − |
| Hippurate hydrolysis | + | + | + |
| Acetoin production | − | − | − |
| Acidification of: | |||
| Trehalose | − | − | − |
| Lactose | − | − | − |
| Raffinose | − | − | − |
| Sucrose | + | + | + |
| Melibiose | − | − | − |
| Arabinose | − | − | − |
| Sorbitol | − | − | − |
| Mannitol | − | − | − |
| Growth in THB supplemented with: | |||
| l-Cysteine HCl (0.01%) | + | + | + |
| Pyridoxal HCl (0.001%) | − | − | − |
+, positive result; −, negative result.
TABLE 2.
Antimicrobial susceptibility of blood- and oral cavity-derived G. elegans isolates
| Agent | MIC (μg/ml)a
|
||
|---|---|---|---|
| Strain IMU02b01 | Strain IMU02p18 | G. elegans CCUG 26024 | |
| Penicillin | ≤0.06 | ≤0.06 | ≤0.06 |
| Ampicillin | ≤0.25 | ≤0.25 | ≤0.25 |
| Cefazolin | ≤0.5 | ≤0.5 | ≤0.5 |
| Ceftazidime | ≤0.5 | ≤0.5 | ≤0.5 |
| Cefozopran | ≤0.5 | ≤0.5 | ≤0.5 |
| Cefdinir | ≤0.12 | ≤0.12 | ≤0.12 |
| Cefepime | ≤0.12 | ≤0.12 | ≤0.12 |
| Imipenem | ≤0.12 | ≤0.12 | ≤0.12 |
| Gentamicin | ≤4 | ≤4 | ≤4 |
| Amikacin | 32 | 32 | ≤16 |
| Arbekacin | >16 | >16 | ≤4 |
| Erythromycin | ≤0.25 | ≤0.25 | ≤0.25 |
| Clarithromycin | ≤0.25 | ≤0.25 | ≤0.25 |
| Clindamycin | ≤0.25 | ≤0.25 | ≤0.25 |
| Minocycline | ≤2 | ≤2 | ≤2 |
| Vancomycin | 1 | 1 | ≤0.5 |
| Teicoplanin | ≤1 | ≤1 | ≤1 |
| Fosfomycin | 16 | ≤8 | ≤8 |
| Levofloxacin | ≤2 | ≤2 | ≤2 |
| Sulfamethoxazole-Trimethoprim | >40 | >40 | >40 |
MICs were determined by a microdilution method of the National Committee for Clinical Laboratory Standards (21).
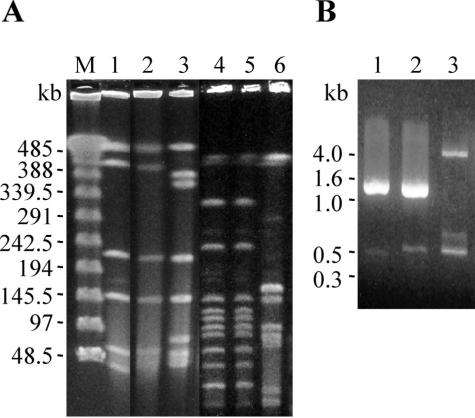
The PFGE patterns of the two isolates were indistinguishable after digestion by SmaI (Fig. 1A, lanes 1 and 2) and ApaI (Fig. 1A, lanes 4 and 5), while the patterns of G. elegans CCUG 26024 with SmaI and ApaI (Fig. 1A, lanes 3 and 6, respectively) were obviously different from those of the two isolates. These results, according to the criteria of Tenover et al. (28), indicated that G. elegans IMU02b01 and IMU02p18 were from the same clone.
FIG. 1.
Patterns obtained by PFGE and AP-PCR for the blood- and oral cavity-derived G. elegans isolates. (A) Chromosomal DNAs were digested with SmaI (lanes 1 to 3) and ApaI (lanes 4 to 6) and then separated by PFGE. Lanes: M, λDNA-PFGE makers (Pharmacia Biotech); 1 and 4, IMU02b01; 2 and 5, IMU02p18; 3 and 6, G. elegans CCUG 26024. (B) AP-PCR was performed with primer AP3 (2). Lanes: 1, IMU02b01; 2, IMU02p18; 3, G. elegans CCUG 26024.
The finding of genotypes that were indistinguishable between the two isolates was further supported by the AP-PCR results (Fig. 1B).
Colonization frequencies of G. elegans in the human oral cavity and other habitats have not been well documented. It was reported that NVS strains were isolated from the human mouth with an incidence rate ranging from 1 to 10% of the total floras (21). We observed that the ratio of healthy adults carrying NVS was 98% (91 of 93), indicating a significantly high frequency of these bacteria in dental plaque (18). On the basis of studies that found that 8% of previously noticed NVS were G. elegans (25) and that G. elegans strains tended to be biochemically identified as G. adiacens (16), the frequency of G. elegans in adult dental plaque is estimated to be approximately 10%. This figure is comparable with results seen with Abiotrophia defectiva (16, 18, 25) and each species of viridans streptococci (12). In accordance with the estimation, the ratio of G. elegans to G. adiacens in the NVS candidates from the plaque specimens was 1:8 in the present study.
When an animal endocarditis model was used, oral G. elegans isolates were reported to be less infective than those of oral G. adiacens and A. defectiva (19). Nevertheless, this organism has been continuously isolated and identified from IE patient samples (references 4, 5, and 22 and the present study). Therefore, it is of interest to clarify whether the critical characteristics of this organism, such as the production of bacteriolytic activity (the present study) and exopolysaccharide (3), are involved in its pathogenicity. In addition, we recently observed that S. aureus and S. epidermidis colonized in the human oral cavity with notable rates of occurrence (13), which agrees with other reports that showed a relatively high prevalence of staphylococcal IE (11, 26). Therefore, given that a proportion of IE cases are caused by staphylococcal species, most of the causative agents of this disease are likely to be oral in origin.
Acknowledgments
We thank Masakazu Inoue, Kagoshima University School of Dentistry, Kagoshima, Japan, for providing G. elegans CCUG 26024.
This study was supported in part by a grant-in-aid for the Open Research Project from the Ministry of Education, Culture, Sports, Science and Technology of Japan and from the Promotion and Mutual Aid Corporation for Private Schools of Japan.
REFERENCES
- 1.Bayliss, R., C. Clarke, C. Oakley, W. Somerville, and A. G. W. Whitfield. 1983. The teeth and infective endocarditis. Br. Heart J. 50:506-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bidet, P., V. Lalande, B. Salauze, B. Burghoffer, V. Avesani, M. Delmée, A. Rossier, F. Barbut, and J.-C. Petit. 2000. Comparison of PCR-ribotyping, arbitrarily primed PCR, and pulsed-field gel electrophoresis for typing Clostridium difficile. J. Clin. Microbiol. 38:2484-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouvet, A. 1995. Human endocarditis due to nutritionally variant streptococci: Streptococcus adjacens and Streptococcus defectivus. Eur. Heart J. 16(Suppl. B):24-27. [DOI] [PubMed] [Google Scholar]
- 4.Casalta, J. P., G. Habib, B. La Scola, M. Drancourt, T. Caus, and D. Raoult. 2002. Molecular diagnosis of Granulicatella elegans on the cardiac valve of a patient with culture-negative endocarditis. J. Clin. Microbiol. 40:1845-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christensen, J. J., and R. R. Facklam. 2001. Granulicatella and Abiotrophia species from human clinical specimens. J. Clin. Microbiol. 39:3520-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cobe, H. M. 1954. Transitory bacteremia. J. Oral Surg. 7:609-615. [DOI] [PubMed] [Google Scholar]
- 7.Collins, M. D., and P. A. Lawson. 2000. The genus Abiotrophia (Kawamura et al.) is not monophyletic: proposal of Granulicatella gen. nov., Granulicatella adiacens comb. nov., Granulicatella elegans comb. nov. and Granulicatella balaenopterae comb. nov. Int. J. Sys. Evol. Microbiol. 50:365-369. [DOI] [PubMed] [Google Scholar]
- 8.Coykendall, A. L. 1989. Classification and identification of the viridans streptococci. Clin. Microbiol. Rev. 2:315-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dajani, A. S., K. A. Taubert, W. Wilson, A. F. Bolger, A. Bayer, P. Ferrieri, M. H. Gewitz, S. T. Shulman, S. Nouri, J. W. Newburger, C. Hutto, T. J. Pallasch, T. W. Gage, M. E. Levison, G. Peter, and G. Zuccaro. 1997. Prevention of bacterial endocarditis. Recommendations by the American Heart Association. JAMA 277:1794-1801. [PubMed] [Google Scholar]
- 10.Durack, D. T. 1995. Prevention of infective endocarditis. N. Engl. J. Med. 332:38-44. [DOI] [PubMed] [Google Scholar]
- 11.Etienne, J., J. Fleurette, J. F. Ninet, P. Favet, and L. D. Gruer. 1986. Staphylococcal endocarditis after dental extraction. Lancet ii:511-512. [DOI] [PubMed] [Google Scholar]
- 12.Frandsen, E. V., V. Pedrazzoli, and M. Kilian. 1991. Ecology of viridans streptococci in the oral cavity and pharynx. Oral Microbiol. Immunol. 6:129-133. [DOI] [PubMed] [Google Scholar]
- 13.Ikeda, Y., Y. Ohara-Nemoto, S. Kimura, K. Ishibashi, and K. Kikuchi. 2004. PCR-based identification of Staphylococcus epidermidis targeting gseA encoding the glutamic acid-specific protease. Can. J. Microbiol. 50:493-498. [DOI] [PubMed] [Google Scholar]
- 14.Kaplan, E. L., and R. C. Anderson. 1977. Infective endocarditis after use of dental irrigation device. Lancet ii:610. [DOI] [PubMed] [Google Scholar]
- 15.Lineberger, L. T., and T. J. De Marco. 1973. Evaluation of transient bacteremia following routine periodontal procedures. J. Periodontol. 44:757-762. [DOI] [PubMed] [Google Scholar]
- 16.Mikkelsen, L., E. Theilade, and K. Poulsen. 2000. Abiotrophia species in early dental plaque. Oral Microbiol. Immunol. 15:263-268. [DOI] [PubMed] [Google Scholar]
- 17.NCCLS. 2000. Performance standard for antimicrobial susceptibility testing; ninth informational supplements. NCCLS document M100-S10. NCCLS, Wayne, Pa.
- 18.Ohara-Nemoto, Y., S. Tajika, M. Sasaki, and M. Kaneko. 1997. Identification of Abiotrophia adiacens and Abiotrophia defectiva by 16S rRNA gene PCR and restriction fragment length polymorphism analysis. J. Clin. Microbiol. 35:2458-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okada, Y., K. Kitada, M. Takagaki, H. Ito, and M. Inoue. 2000. Endocardiac infectivity and binding to extracellular matrix proteins of oral Abiotrophia species. FEMS Immunol. Med. Microbiol. 27:257-261. [DOI] [PubMed] [Google Scholar]
- 20.Okell, C. C., M. B. Camb, and S. D. Elliott. 1935. Bacteriaemia and oral sepsis with special reference to the etiology of subacute endocarditis. Lancet ii:869-872. [Google Scholar]
- 21.Pompei, R., E. Caredda, V. Piras, C. Serra, and L. Pintus. 1990. Production of bacteriolytic activity in the oral cavity by nutritionally variant streptococci. J. Clin. Microbiol. 28:1623-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roggenkamp, A., M. Abele-Horn, K.-H. Trebesius, U. Tretter, I. B. Autenrieth, and J. Heesemann. 1998. Abiotrophia elegans sp. nov., a possible pathogen in patients with culture-negative endocarditis. J. Clin. Microbiol. 36:100-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruoff, K. L. 1991. Nutritionally variant streptococci. Clin. Microbiol. Rev. 4:184-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sasaki, M., C. Yamaura, Y. Ohara-Nemoto, S. Tajika, Y. Kodama, Y. Ohya, and S. Kimura. Streptococcus anginosus infection in oral cancer and its infection route. Oral Dis., in press. [DOI] [PubMed]
- 25.Sato, S., T. Kanamoto, and M. Inoue. 1999. Abiotrophia elegans strains comprise 8% of the nutritionally variant streptococci isolated from the human mouth. J. Clin. Microbiol. 37:2553-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strom, B. L., E. Abrutyn, J. A. Berlin, J. L. Kinman, R. S. Feldman, P. D. Stolley, M. E. Levison, O. M. Korzeniowski, and D. Kaye. 1998. Dental and cardiac risk factors for infective endocarditis. A population-based, case-control study. Ann. Intern. Med. 129:761-769. [DOI] [PubMed] [Google Scholar]
- 27.Taubert, K. A., and A. S. Dajani. 1998. Preventing bacterial endocarditis: American Heart Association guidelines. Am. Fam. Physician 57:457-468. [PubMed] [Google Scholar]
- 28.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tuohy, M. J., G. W. Procop, and J. A. Washington. 2000. Antimicrobial susceptibility of Abiotrophia adiacens and Abiotrophia defectiva. Diagn. Microbiol. Infect. Dis. 38:189-191. [DOI] [PubMed] [Google Scholar]
- 30.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S Ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]