Abstract
Of 162 multidrug-resistant Mycobacterium tuberculosis isolates from Taiwan, 60.5% were found to belong to the Beijing family on the basis of spoligotyping results. IS6110 restriction fragment length polymorphism fingerprinting showed genetic diversity among the multidrug-resistant isolates. Furthermore, 90.1% of the multidrug-resistant isolates had mutations in the rpoB gene, and 11 novel alleles were recognized.
The emergence of multidrug resistance in Mycobacterium tuberculosis has become a global problem (5). The prevalence rates of multidrug-resistant (MDR) M. tuberculosis isolates have been reported to range from 0 to 26.8% (21). An estimated 90% of rifampin (RMP)-resistant isolates are also isoniazid (INH) resistant; therefore, RMP resistance in M. tuberculosis can be referred to as a surrogate marker for MDR. The resistance of M. tuberculosis to RMP is caused by mutations confined in a short 81-bp-long DNA region in the gene rpoB encoding the β-subunit of RNA polymerase (14, 16, 17, 20). Beijing family genotypes strains have been reported to be associated with transmissions of drug-resistant tuberculosis in Germany, Azerbaijan, Cuba, Estonia, Russia, New York, and South Africa (7).
The prevalence of antituberculosis drug resistance in M. tuberculosis has been on the increase in Taiwan. The rates of primary anti-tuberculosis drug resistance from 1990 to 2002 were 9.2 to 19% resistance to INH, 5.7 to 10% resistance to streptomycin (SM), 1.5 to 6.1% resistance to RMP, 0.7 to 15.7% resistance to ethambutol (EMB), and 1.2 to 5.1% resistance to MDR (11, 27). Molecular epidemiology of MDR M. tuberculosis in Taiwan has not been well known and was thus investigated by using spoligotyping and standard IS6110 restriction fragment length polymorphism (RFLP) analysis in this study. The prevalence of rpoB mutations associated with RMP resistance among MDR isolates was also investigated.
MDR isolates.
A total of 162 MDR isolates of M. tuberculosis were collected during 1998 to 2003 at the Center for Chest Diseases, Taipei, Taiwan, and 40 susceptible isolates were collected from Taipei Veteran General Hospital, Taipei, Taiwan, in 2003. Among the 162 MDR isolates, 37 (22.8%) isolates were resistant to INH and RMP, 62 (38.3%) isolates were resistant to INH, RMP, and EMB, 15 (9.3%) isolates were resistant to INH, RMP, and SM, and 48 (29.6%) isolates were resistant to all four drugs.
Spoligotyping and RFLP fingerprinting.
Spoligotyping was performed with a commercial kit (10) according to the manufacturer's instructions. Standard IS6110 RFLP fingerprinting was performed as described previously (25).
Computer analysis.
The spoligotypes were scanned and analyzed using Bionumerics software, version 2.0 (Applied Maths, Kortijk, Belgium). A statistical analysis was performed using EpiInfo 6.04 (Centers for Disease Control and Prevention, Atlanta, Ga.).
rpoB genotyping.
The rpoB gene was amplified with primers rpoB-F (5′-TCGGCGAGCCCATCACGTCG-3′) and rpoB-R (5′-GCGTACACCGACAGCGAGCC-3′), which yielded a 541-bp fragment containing the hot-spot region. PCR products were purified with a commercial kit, and both strands of each product were sequenced.
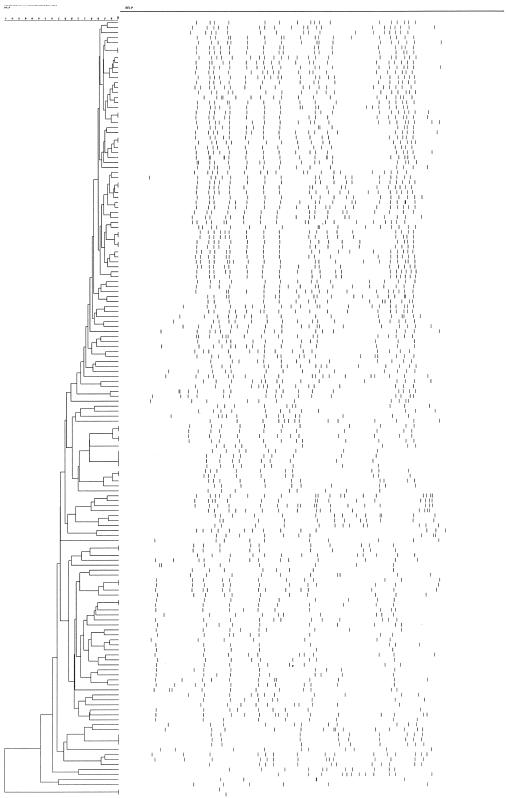
In the spoligotyping analysis, 39 spoligotypes were resolved, including 17 clusters (Fig. 1). Overall, 98 (60.5%) of the 162 MDR isolates showed the Beijing family genotypes. The proportion of isolates resistant to at least three drugs was higher among the Beijing family genotypes (80%) than among non-Beijing genotypes (73.4%). A sufficient quality of genomic DNA for IS6110 RFLP fingerprinting was obtained from 155 MDR isolates, and genetic diversity of the MDR isolates was found. Overall, a RFLP dendrogram revealed 139 patterns that included 14 clusters (18.7% of isolates) at a 98% similarity level: 12 clusters had two strains, one cluster had three strains, and one cluster had four strains (Fig. 2). One cluster with a single IS6110 band was excluded because of distinct spoligotypes.
FIG. 1.
Dendrogram of 162 MDR-TB isolates analyzed by spoligotyping.
FIG. 2.
Dendrogram of 155 MDR-TB isolates analyzed by IS6110 RFLP genotyping.
Of the 162 MDR isolates, 146 (90.1%) had mutations in the 81-bp core region whereas no mutation was found in the 40 susceptible strains. A total of 80 (49.4%), 33 (20.4%), and 14 (8.6%) MDR isolates carried the mutated codons at positions 531, 526, and 516, respectively (Table 1). Overall, 91.8% of the mutated isolates exhibited single site changes. There was no statistical association between Beijing family genotypes and the mutation frequencies of each mutated codon.
TABLE 1.
Mutations in an 81-bp region of the rpoB gene in 162 M. tuberculosis MDR isolates
| Mutated codon | Specific mutation | Amino acid change | No. of strains | Frequency 4 (%) |
|---|---|---|---|---|
| 508 | ACC→ATC | Thr→Ile | 1 | 0.6 |
| 513 | CAA→CCA | Gln→Pro | 5 | 3.1 |
| CAA→AAA | Gln→Lys | 2 | 1.2 | |
| CAA→CCG | Gln→Pro | 1 | 0.6 | |
| 515 | ATG→GTG | Met→Val | 1 | 0.6 |
| 516 | GAC→TAC | Asp→Tyr | 5 | 3.1 |
| GAC→GTC | Asp→Val | 4 | 2.5 | |
| GAC→GGC | Asp→Gly | 2 | 1.2 | |
| GAC→GCC | Asp→Ala | 1 | 0.6 | |
| GAC→TTC | Asp→Phe | 1 | 0.6 | |
| GAC→GAG | Asp→Glu | 1 | 0.6 | |
| 517 | CAG→CCG | Gln→Pro | 1 | 0.6 |
| 522 | TCG→TTG | Ser→Leu | 2 | 1.2 |
| TCG→TGG | Ser→Trp | 1 | 0.6 | |
| TCG→TTC | Ser→Phe | 1 | 0.6 | |
| 526 | CAC→TAC | His→Tyr | 14 | 8.6 |
| CAC→GAC | His→Asp | 5 | 3.1 | |
| CAC→CTC | His→Leu | 5 | 3.1 | |
| CAC→CGC | His→Arg | 3 | 1.9 | |
| CAC→TGC | His→Cys | 2 | 1.2 | |
| CAC→AAC | His→Asn | 1 | 0.6 | |
| CAC→ACC | His→Thr | 1 | 0.6 | |
| CAC→CGA | His→Arg | 1 | 0.6 | |
| CAC→GGC | His→Gly | 1 | 0.6 | |
| 531 | TCG→TTG | Ser→Leu | 75 | 46.3 |
| TCG→TGG | Ser→Trp | 2 | 1.2 | |
| TCG→CAG | Ser→Gln | 1 | 0.6 | |
| TCG→GTG | Ser→Val | 1 | 0.6 | |
| TCG→GGG | Ser→Gly | 1 | 0.6 | |
| 533 | CTG→CCG | Ser→Pro | 6 | 3.7 |
| 509-511 | DELETION | 1 | 0.6 | |
| 513-514 | INSERTION | TTC (Phe) | 4 | 2.5 |
The mutated codons with corresponding amino acids are indicated. New alleles were indicated in boldface characters.
Genetic diversities of drug resistance isolates might be attributable to some host factors beside strain evolution in different geographic regions. The frequency of occurrence of MDR isolates without mutation is comparable to the results seen in studies conducted in some Asia countries (Table 2). It is lower than the results seen in another two studies of RMP-resistant isolates reported from Taiwan but higher than results obtained from other countries. One of the studies analyzing 20 strains revealed four substitutions and one insertion (19), while the other study analyzing 53 RMP-resistant isolates revealed 16 types of mutations and five novel alleles within the 69-bp core region (19). In contrast, our study revealed a total of 32 mutations, 11 new alleles, and the highest frequencies of mutations at codons 513, 526, and 531 (Table 2). Together with results obtained from worldwide studies, these results might be helpful in developing a thorough rapid single-nucleotide-polymorphism detection method, such as microarray, for high-throughput testing. For isolates without mutations, N-terminal codon 146 for low-level resistance and codon 562 might be also involved in RMP drug resistance (30). An additional gene, the arr gene (1, 23), found to be associated with RMP resistance in Pseudomonas aeruginosa and other mycobacteria, may be also involved in the development of MDR tuberculosis.
TABLE 2.
Frequency of codon mutations in RMP-resistant M. tuberculosis isolates from different geographic regionsa
| Country (reference; no. of isolates) | Frequency (%) of mutated codon:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 533 | 531 | 526 | 522 | 516 | 513 | 511 | 508 | No mutation within hot-spot region | |
| United States (26; n = 61) | 8.2 | ||||||||
| Greece (13; n = 17) | 52.9 | 17.6 | 12.0 | 5.9 | 5.9 | ||||
| Asia (8; n = 90) | 53.3 | 16.7 | 1.1 | 14.0 | 5.6 | 1.1 | 6.5 | ||
| Australia (29; n = 33) | 30.3 | 6.1 | 9.1 | 3.0 | |||||
| Italy (18; n = 37) | 2.7 | 59.4 | 35.1 | 8.1 | 2.7 | 0.0 | |||
| Brazil (24; n = 82) | 1.2 | 55.7 | 23.2 | 2.4 | 7.4 | 1.2 | 1.2 | 3.6 | |
| Hungary (2; n = 29) | 31.0 | 6.9 | 38.0 | 6.8 | 10.3 | ||||
| India (12; n = 44) | 2.2 | 63.6 | 22.7 | 4.5 | 2.2 | 6.8 | 2.2 | 2.3 | |
| Spain (6; n = 50) | 2.0 | 48.0 | 22.0 | 14.0 | 2.0 | 6.0 | 0.0 | ||
| East Asia (19; n = 66) (China, 20; Japan, 3; Korea, 18) | 3.0 | 51.5 | 10.6 | 17.0 | 6.0 | 10.6 | |||
| East Asia (19; n = 66) (Taiwan, 20) | 5.0 | 40.0 | 10.0 | 15.0 | 20.0 | ||||
| Latvia (22; n = 34) | 41.2 | 20.6 | 32.0 | ||||||
| Turkey (4; n = 41) | 4.8 | 56.1 | 19.5 | 4.9 | 7.2 | 2.4 | 2.4 | ||
| China (28; n = 86) | 2.0 | 41.0 | 40.0 | 3.0 | 4.0 | 2.0 | 2.0 | 10.0 | |
| Kaohsiung, Taiwan (9; n = 63) | 7.5 | 41.5 | 18.9 | 1.9 | 15.1 | 6.3 | 15.9 | ||
| Taiwan (this research) (n = 162) | 3.7 | 49.4 | 20.4 | 2.4 | 8.6 | 4.9 | 0.0 | 0.6 | 9.9 |
Studies on RMP drug-resistant strains isolated in Taiwan as to the multimutation frequency of the rpoB gene are indicated with boldface characters.
In the RFLP analysis, a cluster with three strains belonged to Beijing genotypes; the three isolates had the same drug resistance profiles (they were resistant to INH, RMP, and EMB and susceptible to SM) and had a single mutation at codon 531 (TCG→TTG, Ser→Leu). In the cluster with four strains, which were resistant to all four drugs (except for one isolate, which was susceptible to EMB), all had a universal TTC (Phe) insertion mutation between codons 513 and 514. The epidemiological relatedness of these four isolates was investigated on the basis of the exhibited RFLP patterns, and possible household contact transmission was linked to two of four strains, while the other clusters had no apparent epidemiological links. Besides, no RFLP pattern identical to that of the MDR W strain (3, 15) was observed in this study population. However, 54.9% of the 162 MDR isolates analyzed had the same mutation site as the W strain in rpoB of either codon 526(His→Tyr) or 531(Ser→Leu) (3). MDR M. tuberculosis could be developed through an acquired resistance or come from an exogenous new infection. These data suggest that the prevalence of RMP resistance among M. tuberculosis isolates in Taiwan might be due to the development of mutations in the rpoB gene in various M. tuberculosis strains rather than due to the transmission of MDR clones.
Nucleotide sequence accession numbers.
The sequences with mutations in new alleles found in this study were deposited in GenBank under accession numbers AY823310, AY823311, AY823312, AY823313, AY823314, AY823315, AY823316, AY823317, and AY823318.
Acknowledgments
This work was supported by grant DOH92-DC-2035 from Center for Disease Control Taiwan, Department of Health.
We thank Jing-Jou Yan for helpful discussion; Sharon Liu, Meng-Hsiun Chen, Su-Yin Chang, Pei-Ju Chin, and Wei-Lun Huang for their excellent technical support; and Wei-Jun Su for providing drug-susceptible M. tuberculosis isolates.
REFERENCES
- 1.Alexander, D. C., J. R. Jones, and J. Liu. 2003. A rifampin-hypersensitive mutant reveals differences between strains of Mycobacterium smegmatis and presence of a novel transposon, IS1623. Antimicrob. Agents Chemother. 47:3208-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartfai, Z., A. Somoskovi, C. Kodmon, N. Szabo, E. Puskas, L. Kosztolanyi, E. Farago, J. Mester, L. M. Parsons, and M. Salfinger. 2001. Molecular characterization of rifampin-resistant isolates of Mycobacterium tuberculosis from Hungary by DNA sequencing and the line probe assay. J. Clin. Microbiol. 39:3736-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bifani, P. J., B. B. Plikaytis, V. Kapur, K. Stockbauer, X. Pan, M. L. Lutfey, S. L. Moghazeh, W. Eisner, T. M. Daniel, M. H. Kaplan, J. T. Crawford, J. M. Musser, and B. N. Kreiswirth. 1996. Origin and interstate spread of a New York City multidrug-resistant Mycobacterium tuberculosis clone family. JAMA 275:452-457. [PubMed] [Google Scholar]
- 4.Cavusoglu, C., S. Hilmioglu, S. Guneri, and A. Bilgic. 2002. Characterization of rpoB mutations in rifampin-resistant clinical isolates of Mycobacterium tuberculosis from Turkey by DNA sequencing and line probe assay. J. Clin. Microbiol. 40:4435-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Espinal, M., A. Laszlo, L. Simonsen, F. Boulahbal, S. J. Kim, A. Reniero, S. Hoffner, H. L. Rieder, N. Binkin, C. Dye, R. Williams, and M. C. Raviglione. 2001. Global trends in resistance to antituberculosis drugs. World Health organization—International Union Against Tuberculosis and lung Disease Working Group on Anti-Tuberculosis Drug Resistance Surveillance. N. Engl. J. Med. 344:1294-1303. [DOI] [PubMed] [Google Scholar]
- 6.Garcia, L., M. Alonso-Sanz, M. J. Rebollo, J. C. Tercero, and F. Chaves. 2001. Mutations in the rpoB gene of rifampin-resistant Mycobacterium tuberculosis isolates in Spain and their rapid detection by PCR-enzyme-linked immunosorbent assay. J. Clin. Microbiol. 39:1813-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glynn, J. R., J. Whiteley, P. J. Bifani, K. Kremer, and D. van Soolingen. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirano, K., C. Abe, and M. Takahashi. 1999. Mutations in the rpoB gene of rifampin-resistant Mycobacterium tuberculosis strains isolated mostly in Asian countries and their rapid detection by line probe assay. J. Clin. Microbiol. 37:2663-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hwang, H. Y., C. Y. Chang, L. L. Chang, S. F. Chang, Y. H. Chang, and Y. J. Chen. 2003. Characterization of rifampicin-resistant Mycobacterium tuberculosis in Taiwan. J. Med. Microbiol. 52:239-245. [DOI] [PubMed] [Google Scholar]
- 10.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liaw, Y. S., P. R. Hsueh, C. J. Yu, S. K. Wang, P. C. Yang, and K. T. Luh. 2004. Drug resistance pattern of Mycobacterium tuberculosis in a university hospital in Taiwan, 1998-2002. J. Formosa Med. Assoc. 103:671-677. [PubMed] [Google Scholar]
- 12.Mani, C., N. Selvakumar, S. Narayanan, and P. R. Narayanan. 2001. Mutations in the rpoB gene of multidrug-resistant Mycobacterium tuberculosis clinical isolates from India. J. Clin. Microbiol. 39:2987-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsiota-Bernard, P., G. Vrioni, and E. Marinis. 1998. Characterization of rpoB mutations in rifampin-resistant clinical Mycobacterium tuberculosis isolates from Greece. J. Clin. Microbiol. 36:20-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller, L. P., J. T., Crawford, and T. Shinnick. 1994. The rpoB gene of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 38:805-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moss, A. R., D. Alland, E. Telzak, D. Hewlett, Jr., V. Sharp, P. Chiliade, V. LaBombardi, D. Kabus, B. Hanna, L. Palumbo, K. Brudney, A. Weltman, K. Stoeckle, K. Chirgwin, M. Simberkoff, S. Moghazeh, W. Eisner, M. Lutfey, and B. Kreiswirth. 1997. A city-wide outbreak of a multiple-drug-resistant strain of Mycobacterium tuberculosis in New York. Int. J. Tuberc. Lung Dis. 1:115-121. [PubMed] [Google Scholar]
- 16.Musser, J. M. 1995. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin. Microbiol. Rev. 8:496-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ovchinnikov, Y. A., G. S. Monastyrskaya, V. V. Gubanov, V. M. Lipkin, E. D. Sverdlov, I. F. Kiver, I. A. Bass, S. Z. Mindlin, O. N. Danilevskaya, and R. B. Khesin. 1981. Primary structure of Escherichia coli RNA polymerase nucleotide substitution in the beta subunit gene of the rifampicin resistant rpoB255 mutant. Mol. Gen. Genet. 184:536-538. [DOI] [PubMed] [Google Scholar]
- 18.Pozzi, G., M. Meloni, E. Iona, G. Orru, O. F. Thoresen, M. L. Ricci, M. R. Oggioni, L. Fattorini, and G. Orefici. 1999. rpoB mutations in multidrug-resistant strains of Mycobacterium tuberculosis isolated in Italy. J. Clin. Microbiol. 37:1197-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qian, L., C. Abe, T. P. Lin, M. C. Yu, S. N. Cho, S. Wang, and J. T. Douglas. 2002. rpoB genotypes of Mycobacterium tuberculosis Beijing family isolates from east Asian countries. J. Clin. Microbiol. 40:1091-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Telenti, A., P. Imboden, F. Marchesi, D. Lowrie, S. Cole, M. J. Colston, L. Matter, K. Schopfer, and T. Bodmer. 1993. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 341:647-650. [DOI] [PubMed] [Google Scholar]
- 21.The Who/IUATLD Global Project on Anti-tuberculosis Drug Resistance Surveillance. 2004.. Anti-tuberculosis drug resistance in the world, Third Global Report. World Health Organization, Geneva, Switzerland.
- 22.Tracevska, T., I. Jansone, L. Broka, O. Marga, and V. Baumanis. 2002. Mutations in the rpoB and katG genes leading to drug resistance in Mycobacterium tuberculosis in Latvia. J. Clin. Microbiol. 40:3789-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tribuddharat, C., M. Fennewald. 1999. Integron-mediated rifampin resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:960-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valim, A. R., M. L. Rossetti, M. O. Ribeiro, and A. Zaha. 2000. Mutations in the rpoB gene of multidrug-resistant Mycobacterium tuberculosis isolates from Brazil. J. Clin. Microbiol. 38:3119-3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Embden, J. D. A., Crawford, J. T., Dale, J. W., Gicquel, B., Hermans, P. W. M., McAdam, R., Shinnick, T., and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams, D. L., C. Waguespack, K. Eisenach, J. T. Crawford, F. Portaels, M. Salfinger, C. M. Nolan, C. Abe, V. Sticht-Groh, and T. P. Gillis. 1994. Characterization of rifampin-resistance in pathogenic mycobacteria. Antimicrob. Agents Chemother. 38:2380-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu, M. C., J. Suo, C. Y. Chiang, K. J. Bai, T. P. Lin, and K. T. Luh. 1997. Initial drug resistance of Mycobacterium tuberculosis in Taiwan. J. Formos. Med. Assoc. 96:890-894. [PubMed] [Google Scholar]
- 28.Yue, J., W. Shi, J. Xie, Y. Li, E. Zeng, and H. Wang. 2003. Mutations in the rpoB of multidrug-resistant Mycobacterium tuberculosis isolates from China. J. Clin. Microbiol. 41:2209-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuen, L. K., D. Leslie, and P. J. Coloe. 1999. Bacteriological and molecular analysis of rifampin-resistant Mycobacterium tuberculosis strains isolated in Australia. J. Clin. Microbiol. 37:3844-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, Y., A. Telenti. 2000. Genetics of drug resistance in Mycobacterium tuberculosis, p. 235-254. In G. F. Hatfull and W. R. Jacobs, Jr. (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, D.C.