Abstract
We have evaluated a diagnostic system based on the loop-mediated isothermal amplification (LAMP) assay for the rapid, simple, and sensitive detection of Newcastle disease virus (NDV) directly from culture isolates as well as clinical samples. By using one set of specific primers targeting the fusion protein gene, the LAMP assay rapidly amplified the target gene within 2 h, requiring only a regular laboratory water bath or heat block for reaction. The results obtained from testing the genomes of 38 NDV strains, other different viruses, and clinical samples of experimentally infected chickens showed that LAMP was as sensitive and specific as nested PCR. All LAMP-positive samples were positive by nested PCR. The LAMP assay is faster than nested PCR, cost-effective, and easy to perform. Our results clearly demonstrate that the LAMP-based assay is a useful tool for the rapid and sensitive diagnosis of NDV infection.
Newcastle disease is a highly contagious viral infection of poultry caused by a paramyxovirus called avian paramyxovirus type 1 (APMV-1), one of the nine serotypes of the virus identified (4). Presently, this disease is still widespread in many countries of Asia, Africa, the Americas, and also Europe (3, 25). Newcastle disease virus (NDV) is present in a variety of strains which differ widely in virulence from causing symptomatic to lethal infection (6). NDV strains can be classified into highly virulent (velogenic), intermediate (mesogenic), and nonvirulent (lentogenic) on the basis of their pathogenicity in chickens (8).
Although vaccination programs and importation quarantine requirements have provided significant protection against any outbreak, the virus remains a potential threat to commercial poultry producers as well as the backyard type operation owners. This was proven by the most recent outbreak of NDV reported in many countries, in which eradication measures resulted in the depopulation of birds (16). Thus, rapid, sensitive, and easy diagnostic methods for detecting NDV surely would pave the way for more effective control of the disease.
Recently, a novel nucleic acid amplification method termed loop-mediated isothermal amplification (LAMP) was developed by Notomi et al. (21). This method relies on autocycling strand displacement DNA synthesis performed by using the Bst DNA polymerase large fragment (18, 19, 21). The characteristics of LAMP are rapidity under isothermal conditions, low reaction temperature (63 to 65°C), and high specificity for the target sequence. This is attributable to recognition of the target sequence at six independent sites in the initial stage and at four independent sites during the last stages. It requires only four primers, a DNA polymerase, and a regular laboratory water bath or heat block for reaction, making it simple to perform (18, 21). Hence, LAMP can be a simple and valuable tool for the rapid diagnosis of infectious diseases by using very basic facilities and should be easily applicable in the clinical laboratories of developing countries.
In this study, we endeavored to develop a diagnostic method based on the LAMP reaction for the detection of NDV and compared the sensitivity and specificity of LAMP and nested PCR in the detection of an RNA target in total RNA. Further, we evaluated the sensitivity and specificity of NDV detection by LAMP, comparing it with nested PCR using an in vivo model system.
MATERIALS AND METHODS
Viral strains.
Thirty-eight NDV strains, three other serotypes of APMV (10, 17), and three other clinically related viruses which could be isolated from chickens maintained as master stocks were used in this study (Table 1). The stocks were passaged once in specific-pathogen-free eggs at low titer for experimental purposes.
TABLE 1.
Comparison of the specificity of LAMP and nested PCR
| Strain no. | Name of NDV strain | Virulencea | Result byb:
|
|
|---|---|---|---|---|
| LAMP | Nested PCR | |||
| 1 | NDV/chicken/Japan/Sato/30 | V | + | + |
| 2 | NDV/chicken/Komarov/40 | M | + | + |
| 3 | NDV/chicken/Hitchner B1/48 | L | + | + |
| 4 | NDV/chicken/Japan/Miyadera/51 | V | + | + |
| 5 | NDV/chicken/Japan/Ishii/62 | L | + | + |
| 6 | NDV/chicken/Japan/Narashino/67 | V | + | + |
| 7 | NDV/chicken/Japan/Chiba/69 | V | + | + |
| 8 | NDV/sparrow-hawks/Japan/Taka/73 | V | + | + |
| 9 | NDV/sparrow-hawks/Japan/Taka small plaque | V | + | + |
| 10 | NDV/chicken/Japan/Chiba/81 | V | + | + |
| 11 | NDV/chicken/Japan/Chiba/84 | V | + | + |
| 12 | NDV/pigeon/Japan/Ibaraki/84 | M | + | + |
| 13 | NDV/chicken/Japan/Ibaraki/85 | V | + | + |
| 14 | NDV/chiken/Japan/Chiba/85 | V | + | + |
| 15 | NDV/pheasant/Japan/Gunma/85 | V | + | + |
| 16 | NDV/chicken/Japan/Niigata/85 | V | + | + |
| 17 | NDV/chicken/Japan/Miyadera/85 | ND | + | + |
| 18 | NDV/chicken/Japan/Chiba/86 | V | + | + |
| 19 | NDV/pigeon/Japan/Tochigi/86 | M | + | + |
| 20 | NDV/chicken/Japan/Chiba/87 | V | + | + |
| 21 | NDV/pigeon/Japan/Niigata/88 | M | + | + |
| 22 | NDV/chicken/Japan/Niigata/89 | V | + | + |
| 23 | NDV/chicken/Japan/Kagoshima/91 | V | + | + |
| 24 | NDV/pigeon/Japan/Tokachi/91 | M | + | + |
| 25 | NDV/pigeon/Japan/Kumamoto/95 | M | + | + |
| 26 | NDV/pigeon/Japan/Tochigi/95 | M | + | + |
| 27 | NDV/chicken/Japan/Tokyo/96 | V | + | + |
| 28 | NDV/parakeet/Japan/Chiba/97 | V | + | + |
| 29 | NDV/pheasant/Japan/Ibaraki/97 | V | + | + |
| 30 | NDV/pigeon/Japan/Saitama/97 | M | + | + |
| 31 | NDV/chicken/Japan/Ibaraki-2/99 | V | + | + |
| 32 | NDV/chicken/Japan/Chiba-222/99 | V | + | + |
| 33 | NDV/chicken/Japan/Ibaraki/2000 | V | + | + |
| 34 | NDV/pigeon/Japan/Gunma/2000 | M | + | + |
| 35 | NDV/chicken/Japan/Ibaraki/2000 | V | + | + |
| 36 | NDV/pigeon/Japan/Kumamoto/2000 | M | + | + |
| 37 | NDV/quail/Japan/Chiba/2001 | V | + | + |
| 38 | NDV/chicken/Japan/Nagano/2001 | V | + | + |
| 39 | APMV2/chicken/California/Yucaipa/56 | NA | − | − |
| 40 | APMV3/turkey/Wisconsin/68 | NA | − | − |
| 41 | APMV7/dove/Tennessee/75 | NA | − | − |
| 42 | Fowlpoxvirus | NA | − | − |
| 43 | Avian reoviruses | NA | − | − |
| 44 | Marek's disease virus | NA | − | − |
L, lentogenic; M, mesogenic; V, velogenic; ND, not determined; NA, not applicable.
+, LAMP or nested PCR amplification; −, no LAMP or nested PCR amplification.
RNA extraction and cDNA synthesis.
RNA was extracted directly from 200 μl of allantoic fluid or 100 mg of organ aliquot of the homogenate by using Trizol reagent (Invitrogen, Carlsbad, Calif.). Complementary cDNA was synthesized using 12.5 μl of eluted RNA with oligo(dT)18 primer and the Rous-associated virus-2 reverse transcriptase kit (Takara, Kyoto, Japan) according to the manufacturer's instructions.
Oligonucleotide primers.
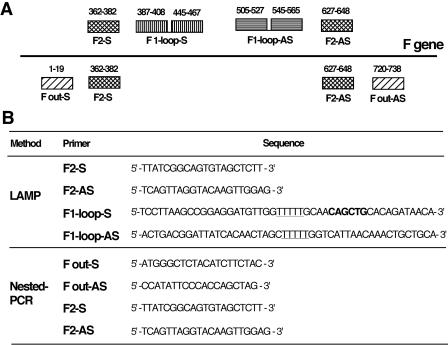
Primers used for LAMP and nested PCR were designed from the target gene (fusion [F] gene). The name, location, and sequence of each primer are shown in Fig. 1. All primers for LAMP were designed as described by Notomi et al. (21). Of the two inner primers of LAMP, each primer has two binding regions connected by a TTTT spacer.
FIG. 1.
(A) Schematic representation of the relationship between LAMP and nested PCR primers. The outer primers for LAMP were the same as the inner primers for nested PCR. (B) Name and sequence of each primer used in this study. The restriction site for PvuII is indicated in bold type, and the two TTTT spacers are underlined.
Standard plasmid preparation.
A fragment (1,700 bp) of the F gene of the APMV-1/chicken/Japan/Miyadera/51 strain was cloned into pGEM-T Easy vectors (Promega, Madison, Wis.). The recombinant Escherichia coli strain carrying the recombinant plasmids was inoculated into Luria-Bertani broth (12 ml) and incubated at 37°C overnight with horizontal shaking. Plasmid DNAs were extracted from the culture with the QIAprep Spin Miniprep kit (QIAGEN, Valencia, Calif.) and checked by sequencing. Correct plasmid was diluted to serve as standard for determining the sensitivities of the assays.
LAMP reaction.
LAMP was carried out in a 25-μl reaction mixture containing 40 pmol each of F1 loop-S (F1-S) and F1-AS, 20 pmol each of F2-S and F2-AS, 0.4 mM concentrations of each deoxynucleoside triphosphate, 1 M betaine (Sigma), 20 mM Tris-HCl (pH 7.5), 10 mM KCl, 10 mM (NH4)2SO4, 8 mM MgSO4, 0.1% Triton X-100, 8 U of Bst DNA polymerase, and 2 μl of the plasmid standard or of undiluted cDNA. The mixture was incubated at 65°C for 120 min and heated at 80°C for 10 min to terminate the reaction. The LAMP reactions were carried out in a Petty Bath Shaker (Wakenyaku Co., Ltd.) or Chill Heat CHT-101 (IWAKI Asahi Techno glass). To confirm the sequence, the amplified products were digested with the restriction enzyme PvuII (for 1 h at 37°C), which cuts F1-loop-S, and their sizes were analyzed by electrophoresis on a 2% agarose gel. If the amplified products had the sequence as expected, the products would produce 248-, 262-, or 300-bp fragments by this digestion.
Nested PCR.
We tested the sensitivity and specificity of the LAMP method and compared it with nested PCR by using the same templates at identical concentrations. In the first round of PCR, 748-bp fragment of the F gene was amplified in 20 μl of reaction mixture containing 2 μl of 10× PCR buffer, 0.19 mM concentrations of each deoxynucleoside triphosphate, 20 pmol of F out-S primer and 20 pmol of F out-AS primer, 2 μl of undiluted cDNA, and 0.5 U of TaKaRa Taq. In the second round of PCR, 2 μl of undiluted first-round PCR product or 2 μl of the plasmid standard was added to 18 μl of a PCR mixture similar to the one described for the first round with F2-S primer and F2-AS primer to amplify a 285-bp fragment. The cycle conditions were identical for both rounds and were 1 cycle at 95°C for 1 min; 35 cycles at 95°C for 45 s, 50°C for 45 s, and 72°C for 4 min; and a final extension at 72°C for 7 min. The PCRs were carried out by using the Gene Amp PCR system 9700 (Applied Biosystems).
Experimentally infected chicken and tissue samples.
Eight 4-week-old White Leghorn chickens (Hokuren Co.ltd., Sapporo, Japan) were each inoculated orally with 0.2 ml of lentogenic B1 strain containing 108 50% egg infective doses grown in allantoic fluid and diluted 1:100 in sterile water. Three days after inoculation, four inoculated chickens were sacrificed and the remaining chickens were sacrificed at day 6 postinoculation. The necropsy was performed on each chicken. Ten different organs of each chicken were dissected using different scissors and forceps to avoid contamination and carefully homogenized in 1.5-ml microtubes in phosphate-buffered saline. Individual organ aliquots of the homogenates were stored at −20°C until their use for RNA extraction (Table 2). As negative controls, 10 different organs of two uninoculated chickens and the tracheas and lungs of 20 uninoculated chickens were collected during the experiment.
TABLE 2.
Comparative sensitivity for virus detection in organs of experimentally infected chickens by LAMP and nested PCR
| Specimen | 3 days after inoculation
|
6 days after inoculation
|
||
|---|---|---|---|---|
| LAMP | Nested PCR | LAMP | Nested PCR | |
| Brain | − | − | − | − |
| Trachea | + (3/4) | + (3/4) | + (4/4) | + (4/4) |
| Lung | + (3/4) | + (3/4) | + (4/4) | + (4/4) |
| Kidney | − | − | − | − |
| Spleen | − | − | − | − |
| Liver | − | − | − | − |
| Pancreas | − | − | − | − |
| Intestine | − | − | − | − |
| Cecal tonsil | − | − | − | − |
| Bursa | − | − | − | − |
−, negative; +, positive. The number positive out of the number tested is given in parentheses.
DNA sequencing.
The nucleotide sequences of plasmids or nested PCR products were determined by using the Big Dye Terminator kit (Applied Biosystems; Japan Ltd.) with an automated DNA sequencer (ABI PRISM 310 genetic analyzer). The Genetyx-Mac 10 package (Software Development Co., Ltd, Tokyo, Japan) was used to align the sequences and combined with the BLAST program (National Center for Biotechnology Information) search of GenBank for a homology check with known NDV gene sequences.
RESULTS
Analytical sensitivity of the LAMP method compared to nested PCR.
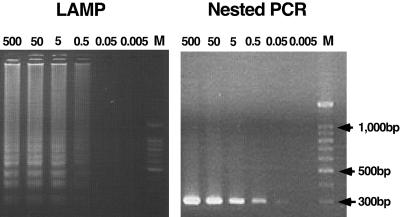
The F plasmid of the Miyadera/51 strain was constructed to facilitate initial evaluation and optimization of the LAMP and nested PCR methods. One set of primers including four primers with six recognition sites of LAMP and one set of primers including four primers with four recognition sites of nested PCR were examined by using a standard plasmid as template. The detection limit of both methods was 0.5 pg or 9 × 104 copies/reaction determined by using a serial 10-fold dilution of the plasmids (Fig. 2). This was also confirmed by the fact that, when 0.5 pg to 0.5 ng of DNA plasmid/reaction was detected 100% of the time by nested PCR or LAMP, 0.05 pg or approximately 9 × 103 copies of DNA plasmid/reaction was detected infrequently (three out of five times) and 0.005 pg or 9 × 102 copies/reaction was not detected by both systems. As outlined in Fig. 2, there was no difference in the sensitivity when LAMP was compared to the nested PCR system.
FIG. 2.
Sensitivities of LAMP and nested PCR methods. The LAMP and nested PCR methods were carried out using different concentrations of plasmid at picograms of plasmid per reaction. Each concentration of plasmid was tested five times. (A) LAMP reaction; (B) nested PCR reaction. Marker, 100-bp ladder size markers (Promega).
Analytical specificity of the LAMP method compared to nested PCR.
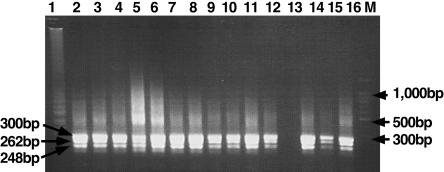
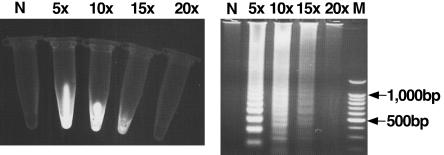
Specificity of primers was tested by LAMP and nested PCR with RNAs extracted from 38 NDV strains and other viruses (Table 1). All the NDV strains gave a positive reaction, while no DNA band of three other serotypes of APMV and three other clinically related viruses including avian reovirus, fowlpoxvirus, and Marek's disease virus was observed by both methods (Table 1). In order to confirm that the products amplified by LAMP primers and nested PCR primers had correct sequences, the LAMP products were digested with the restriction enzyme PvuII and nested PCR products were directly sequenced. The sizes of the LAMP-digested fragments were analyzed by electrophoresis and compared with the sizes predicted from the expected sequence. The nucleotide sequences of nested PCR products were compared to other NDV sequences obtained from GenBank. The sizes of LAMP fragments generated after digestion were in agreement with the predicted sizes: 248, 262, and 300 bp (Fig. 3). The determined nucleotide sequences were highly homologous to other NDV strains. Nonspecific reactions were not observed by LAMP or nested PCR methods. These results indicate that the two methods were highly specific among the strains we tested. Since LAMP had high specificity for the target sequence, a detection system of LAMP products was conducted by ethidium bromide inspection with the naked eye and was compared to the detection by electrophoresis using serial fivefold dilution of LAMP products. As shown in Fig. 4, a LAMP-positive reaction can be directly detected with the naked eye by observing the color of the solution whereas a negative reaction has no color and the detection limits of both systems were 15-fold dilutions.
FIG. 3.
Restriction enzyme analysis of the LAMP products. Electrophoretic analysis of LAMP-amplified cDNA NDV strains and digestion with PvuII. Lane 1, undigested LAMP product; lanes 2 through 12 and 14 through 16, LAMP products after digestion with PvuII; lane 13, negative control; lane M, 100-bp ladder size markers (Promega).
FIG. 4.
Sensitivities of visual inspection and electrophoresis detection of LAMP-amplified products. Lanes N, no template; lane M, 100-bp ladder size. The number above each tube and each lane represents the dilution of LAMP product: 5×, 5-fold dilution; 10×, 10-fold dilution; 15×, 15-fold dilution; 20×, 20-fold dilution.
Detection of NDV from clinical specimens.
In order to evaluate the optimal tissues for NDV diagnosis by LAMP and nested PCR methods, RNAs of 80 specimens from infected chickens and 60 specimens from uninfected chickens were extracted and subjected to LAMP and nested PCR. Only trachea and lung specimens were positive. At 3 days postinfection, samples were positive from three of four chickens by both methods (Table 2). Other tissue samples were negative. At day 6 postinoculation, all tracheas and lungs were positive by both methods. All specimens from the 22 uninfected control chickens were negative. There was 100% concordance between nested PCR and LAMP performed on tissue extracts.
DISCUSSION
Newcastle disease is complicated in that different isolates and strains of the virus may induce enormous variation in the severity of disease, even in a given host such as chicken (4). Rapid detection and identification of NDV isolates is always essential to control this disease so that quick identification of the affected birds can be done and appropriate control measures are effectively applied.
Our results demonstrated that LAMP has a high sensitivity and specificity equal to those for the nested PCR method. The results obtained from diluted plasmid and RNAs showed that both systems have a similar detection limit (0.5 pg of plasmid/reaction or 9 × 104 copies) with all nested-PCR-positive clinical samples being positive by LAMP. All specimens from uninfected chickens were negative by both methods. NDV was detected at an early stage (3 days postinfection) and in various tissue types (trachea and lung).
The development of both LAMP and nested PCR assays requires knowledge of the gene sequences, careful primer design, and assay optimization. Designing LAMP primers is crucial in optimizing the LAMP reaction because LAMP uses four primers that recognize six distinct regions on the target gene. Nevertheless, LAMP operation is quite simple, requiring only a regular laboratory water bath or heat block for incubation under isothermal conditions. The time for LAMP reaction was quicker, taking only 2 h, compared to 6 h for nested PCR. Another useful feature of LAMP was that LAMP products stained with ethidium bromide were simply detected by the naked eye (Fig. 4). This could be achieved due to the high specificity and high amplification efficiency of LAMP. However, the product detection system with ethidium bromide had several limitations, such as generation of hazardous waste and sensitivity 25 to 100 times less than that of SYBR Green I. Mori et al. (18) have reported that LAMP products can be observed by the naked eye when a white precipitate of magnesium pyrophosphate is present in the reaction mixture. However, this detection limit is limited when the turbidity of reaction is low. To increase the sensitivity and the rate of recognition by the naked eye and to avoid the more hazardous aspects of postamplification analysis, we could add SYBR Green I to the LAMP reaction mixture (11).
In comparison with the traditional technique based on virus isolation in embryonated chicken eggs, LAMP can directly detect NDV from samples and would be relatively inexpensive and efficient. Virus isolation in embryonated chicken eggs requires 4 to 7 days for detection of positive results (4), while LAMP requires only several hours. New technologies using monoclonal antibodies have also been developed to improve the detection and identification of NDV (2, 5, 13, 23). However, the results provide little information on the infecting strain of NDV and therefore have limited diagnostic value (1, 4). Molecular techniques based on reverse transcription-PCR have already been established to detect and identify NDV in allantoic fluid or clinical samples (7, 9, 12, 14, 15, 16, 20, 22, 24, 26, 27). However, PCR methods have several disadvantages such as the requirement of thermal cycling, variable specificity, and low-amplification efficiency, as well as the need for special equipment.
In summary, we describe a LAMP-based method for the detection of NDV. This method is rapid, sensitive, and specific. Experimentally infected chickens produced positive results as early as 3 days postinfection. Although limited numbers of clinical samples were used in the present study, it has been shown that LAMP could be a convenient tool for detecting NDV infection because it requires minimal laboratory facilities and is relatively simple and inexpensive to perform.
REFERENCES
- 1.Aldous, E. W., and D. J. Alexander. 2001. Detection and differentiation of Newcastle disease virus (avian paramyxovirus type 1). Avian Pathol. 30:117-128. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, D. J. 1989. Newcastle disease, p. 114-120. In H. G. Purchase, L. H. Arp, C. H. Domermuth, and J. E. Pearson (ed.), A laboratory manual for the isolation and identification of avian pathogens, 3rd ed. Kendall/Hunt Publishing Company, Dubuque, Iowa.
- 3.Alexander, D. J. 1995. Newcastle disease in countries of the European Union. Avian Pathol. 24:3-10. [DOI] [PubMed] [Google Scholar]
- 4.Alexander, D. J. 1997. Newcastle disease and other avian Paramyxoviridae infections, p. 541-570. In B. W. Calnek, H. J. Barnes, C. W. Beard, L. R. McDougald, and Y. M. Saif (ed.), Diseases of poultry, 10th ed. Iowa State University Press, Ames.
- 5.Alexander, D. J., and R. J. Manvell. 1997. Antigenic diversity and similarities detected in avian paramyxovirus type 1 (Newcastle disease virus) isolates using monoclonal antibodies. Avian Pathol. 26:399-418. [DOI] [PubMed] [Google Scholar]
- 6.Alexander, D. J., and R. C. Jones. 2001. Paramyxoviridae, p. 257-272. In F. Jordan, M. Pattison, D. Alexander, and T. Faragher (ed.), Poultry diseases, 5th ed. W. B. Saunders, Harcourt Publishers Ltd., London, United Kingdom.
- 7.Barbezange, C., and V. Jestin. 2002. Development of a RT-nested PCR test detecting pigeon Paramyxovirus-1 directly from organs of infected animals. J. Virol. Methods 106:197-207. [DOI] [PubMed] [Google Scholar]
- 8.Beard, C. W., and R. P. Hanson. 1984. Newcastle disease, p. 452-470. In M. S. Hofstad, B. W. Calnek, C. F. Helmboldt, W. M. Reid, H. W. Yoder (ed.), Disease of poultry, 8th ed. Iowa State University Press, Ames.
- 9.Gohm, D. S., B. Thur, and M. A. Hofmann. 2000. Detection of Newcastle disease virus in organs and faeces of experimentally infected chickens using RT-PCR. Avian Pathol. 29:143-152. [DOI] [PubMed] [Google Scholar]
- 10.Hoshi, S., T. Mikami, K. Nagata, M. Onuma, and H. Izawa. 1983. Monoclonal antibodies against a paramyxovirus isolated from Japanese sparrow-hawks. Arch. Virol. 76:145-151. [DOI] [PubMed] [Google Scholar]
- 11.Iwamoto, T., T. Sonobe, and K. Hayashi. 2003. Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J. Clin. Microbiol. 41:2616-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarecki-Black, J. C., J. D. Bennett, and S. Palmieri. 1992. A novel oligonucleotide probe for detection of Newcastle disease virus. Avian Dis. 36:134-138. [PubMed] [Google Scholar]
- 13.Jestin, V., M. Cherbonnel, R. L'Hospitalier, and G. Bennejean. 1989. An ELISA blocking test using a peroxidase-labeled anti-HN monoclonal antibody for the specific titration of antibodies to avian paramyxovirus type 1 (PMV1). Arch. Virol. 105:199-208. [DOI] [PubMed] [Google Scholar]
- 14.Jestin, V., and A. Jestin. 1991. Detection of Newcastle disease virus RNA in infected allantoic fluids by in vitro enzymatic amplification (PCR). Arch. Virol. 118:151-161. [DOI] [PubMed] [Google Scholar]
- 15.Kant, A., G. Koch, D. J. Van Roozelaar, F. Balk, and A. Ter Huurne. 1997. Differentiation of virulent and non virulent strains of Newcastle disease virus within 24 hours by polymerase chain reaction. Avian Pathol. 26:837-850. [DOI] [PubMed] [Google Scholar]
- 16.Kho, C. L., M. L. Mohd-Azmi, S. S. Arshad, and K. Yusoff. 2000. Performance of an RT-nested PCR ELISA for detection of Newcastle disease virus. J. Virol. Methods 86:71-83. [DOI] [PubMed] [Google Scholar]
- 17.Mase, M., K. Imai, Y. Sanada, N. Sanada, N. Yuasa, T. Imada, K. Tsukamoto, and S. Yamaguchi. 2002. Phylogenetic analysis of Newcastle disease virus genotypes isolated in Japan. J. Clin. Microbiol. 40:3826-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mori, Y., K. Nagamine, N. Tomita, and T. Notomi. 2001. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289:150-154. [DOI] [PubMed] [Google Scholar]
- 19.Nagamine, K., T. Hase, and T. Notomi. 2002. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 16:223-229. [DOI] [PubMed] [Google Scholar]
- 20.Nanthakumar. T., R. S. Kataria, A. K. Tiwari, G. Butchaiah, and J. M. Kataria. 2000. Pathotyping of Newcastle disease virus by RT-PCR and restriction enzyme analysis. Vet. Res. Commun. 24:275-286. [DOI] [PubMed] [Google Scholar]
- 21.Notomi, T., H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, and T. Hase. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oberdorfer, A., and O. Werner. 1998. Newcastle Disease virus: detection and characterization by PCR of recent German isolates differing in pathogenicity. Avian Pathol. 27:237-243. [DOI] [PubMed] [Google Scholar]
- 23.Russell, P. H., and D. J. Alexander. 1983. Antigenic variation of Newcastle disease virus strains detected by monoclonal antibodies. Arch. Virol. 75:243-253. [DOI] [PubMed] [Google Scholar]
- 24.Seal, B. S., D. J. King, and J. D. Bennett. 1995. Characterization of Newcastle disease virus isolates by reverse transcription PCR coupled to direct nucleotide sequencing and development of sequence database for pathotype prediction and molecular epidemiological analysis. J. Clin. Microbiol. 33:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spradbrow, P. B. 1988. Geographical distribution, p. 247-255. In D. J. Alexander (ed.), Newcastle disease. Kluwer Academic Publishers, Boston, Mass.
- 26.Stäuber, N., K. Brechtbűhl, L. Bruckner, and M. A. Hofmann. 1995. Detection of Newcastle disease virus in poultry vaccines using the polymerase chain reaction and direct sequencing of amplified cDNA. Vaccine 13:360-364. [DOI] [PubMed] [Google Scholar]
- 27.Wise, M. G., D. L. Suarez, B. S. Seal, J. C. Pedersen, D. A. Senne, D. J. King, D. R. Kapczynski, and E. Spackman. 2004. Development of a real-time reverse-transcription PCR for detection of Newcastle disease virus RNA in clinical samples. J. Clin. Microbiol. 42:329-338. [DOI] [PMC free article] [PubMed] [Google Scholar]