Abstract
The porin gene (porB) of Neisseria gonorrhoeae encodes the major outer membrane protein identified as PI or Por. To examine the utility of por variable-region (VR) typing, porB from 206 isolates was characterized by using oligonucleotide probes in a checkerboard hybridization assay that identifies the sequence types of five VRs of both PIA and PIB porB alleles. The strains represented temporally and geographically distinct isolates, isolates from a large cluster, epidemiologically linked partner isolates, and a collection of strains from disseminated gonococcal infections. By using rigorous epidemiologic criteria for transmission of infection between sex partners, por VR typing was more discriminatory than serovar typing in classifying isolates from both members of 43 epidemiologically linked pairs: 39 of 43 pairs were classified as coinciding by por VR typing compared to 43 of 43 by serovar determination (P = 0.058). porB sequence data confirmed the accuracy of the por VR method. Relationships between VR type and serovar typing monoclonal antibodies were observed for all six PIB and three of six PIA antibodies. por VR typing is a molecular tool that appears to have broad applicability. This method can be adapted to a wide range of technologies from simple hybridization to microarray and may allow for typing from noncultured clinical specimens.
Neisseria gonorrhoeae is one of the most common communicable diseases worldwide. In the United States, 351,852 cases were reported to the Centers for Disease Control and Prevention in 2001, over six times the Healthy People 2010 objective (5). The World Health Organization estimated that in 1999, 62.35 million cases occurred worldwide (43). Reported cases vastly underestimate the prevalence of this disease (6, 36).
Gonococcal infections are usually uncomplicated genitourinary infections or are asymptomatic; however, they can result in serious medical and public health consequences such as disseminated gonococcal infection (DGI), pelvic inflammatory disease with subsequent complications of ectopic pregnancy and infertility, and increased transmission of human immunodeficiency virus infection (11). Additionally, antibiotic resistance develops rapidly, as illustrated by the spread of high-level fluoroquinolone resistance, loss of this antibiotic as first-line therapy, and the need for revised treatment guidelines (7, 44, 35).
Although N. gonorrhoeae has been described as a panmictic organism, clonal outbreaks have been described in association with disease presentation (16, 18) or antibiotic resistance (47). Within defined temporal periods and geographic regions, isolate typing can be used to examine transmission patterns, disease clusters, and antibiotic resistance outbreaks. Strain typing can assist in identifying high-prevalence, high-transmission subgroups known as core groups and in guiding focused public health interventions.
Methods available for gonococcal strain characterization include phenotypic typing such as auxotype and serovar (A/S) determination and antibiotic resistance testing. Genotypic methods include pulsed-field gel electrophoresis (45) and opa typing (29) based on restriction fragment length polymorphisms (RFLPs) and sequence typing based on one or more genes (17, 39, 40). Serovar determination is the classic phenotypic characterization of the gonococcus based on the reaction of strains with a panel of monoclonal antibodies (MAbs) directed against the porin protein (Por), historically referred to as PI (19). Serovar determination is technologically simple and has been widely performed, but problems with reproducibility and MAb availability have hampered its utility.
Characterization of the two classes of Por, PIA and PIB, has been expanded and refined by investigations of the sequence variability of the two mutually exclusive genes porB.1A and porB.1B (12, 15, 17, 31, 37). The porB gene encodes a protein that forms a homotrimer consisting of three identical barrel-shaped channels (25). Sequence diversity of porB is primarily localized to regions encoding the predicted surface-exposed loops (12). Por is essential for cell viability, does not undergo phase variation (1), and is of interest in relation to both the pathogenicity and the immunogenicity of N. gonorrhoeae (10, 14, 24, 33, 42), making Por an attractive typing target.
We have previously characterized the variability of porB by using oligonucleotide probe hybridizations to identify variable regions (VRs) (26, 34). However, this method has not been evaluated for use in diverse epidemiologic studies. In this study, we refined our method, examined the relationship between serovar, VR types and porB sequence, and applied our method to a variety of previously well-characterized strain collections to examine the potential utility of this method as a molecular epidemiologic tool.
(Part of this research was presented at the 13th International Pathogenic Neisseria Conference, 1 to 6 September 2002, Oslo, Norway, and at Diagnostic Approaches for Infectious Disease: Future Promises and Impact on Clinical Management [IDSA], Orlando, Fla., 29 April to 1 May 2001.)
MATERIALS AND METHODS
Bacterial isolates and description of infected populations.
A total of 206 study isolates and 14 control strains were characterized by por VR typing. Study isolates included the following: a panel of 18 temporally and geographically diverse strains developed to evaluate gonococcal typing methods (38); 8 strains (T13, F6, F62, N10, S12, 7122, D4, and G7) commonly used as controls in serovar determinations (kindly provided by C. Ison, Health Protection Agency, London, United Kingdom); a cluster of 14 epidemiologically linked strains (41) with 4 strains of the same serovar or auxotype; 53 DGI strains collected at Boston City and University Hospitals between 1975 and 1982 (28); and 109 previously A/S typed strains collected in the 2-year period following September 1988 in Boston, Mass. Among the 109 strains, there were 69 strains from 37 partners enrolled in a study of N. gonorrhoeae and Chlamydia transmission (21) and 12 strains from 6 partners who were classified by the same strict epidemiologic criteria but were not enrolled in the published study. The por VR typing control strains 1861, 5441, 5589, 9299, 3744, FA19, 2432, W062, PI83, S62, 256, MS11, 909, 7, 133, and 911007 have been previously described (26); strain PU186 was provided by C. Ison.
DNA preparation and porB amplification.
Genomic DNA was isolated from cultured N. gonorrhoeae cells or 200 μl of frozen culture stock by using a Wizard genomic DNA purification kit (Promega Corp., Madison, Wis.) according to the manufacturer's instructions. Amplification of the porB gene was performed as described (26) by using an Expand High-Fidelity PCR System containing Taq DNA polymerase and Tgo DNA polymerase with proofreading activity (Roche Molecular Biochemicals, Indianapolis, Ind.) and primers PIB.Fpr (5′-ATTGCCCTGACTTTGGCAGCCCTTCCT) and PIB.Rpr (5′TTGCAACCAGCCGGCAGAAACCAAGGC), complementary to the signal peptide and loop 8 coding regions, respectively. Amplifications performed equally well with either PIA or PIB porB alleles. PCR products were analyzed for size and concentration with an Agilent 2100 Bioanalyzer (Rockville, Md.).
Sequence analysis and oligonucleotide probe design.
A multiple sequence alignment analysis, using the GCG PILEUP program from the Genetics Computer Group package (GCG10.2-Unix; University of Wisconsin), was conducted by using 36 full-length PIA sequences identified in the GenBank database (accession no. AF044782 to AF044783 [17], AF090808 to AF090824 [12], AF015117 to AF015120, AF015122 [9], L19958 to L19966 [27], J03029, X58073 [4], and Z69259 [32]). Multiple sequence alignment was followed by individual alignments of the gene segments 161 to 250, 328 to 380, 437 to 530, 768 to 852, and 888 to 929 (based on J03029 [3]) encoding the predicted surface-exposed loops 1, 2, 3, 6, and 7 of the mature PIA protein. A PIB alignment (34) was followed by multiple sequence alignments of regions 251 to 316, 500 to 620, 737 to 866, 901 to 952, and 1021 to 1090 (based on M21289 [3]) encoding the predicted surface-exposed loops 1, 3, 5, 6, and 7 of the mature PIB protein. Dendrograms were generated by GCG PILEUP for regions 1, 5, and 6. Biotin 5′ end-labeled oligonucleotide probes were designed with similar melting temperatures (range, 56.6 to 61.0°C) to match the sequence variants identified for the VRs encoding the predicted surface-exposed loops. Adjustments to previously reported probes (26) (indicated in Table 1) have not altered the results, based on comparison with control strains, but improved their interpretation by increasing specific signals and/or decreasing cross-reactivity with closely related sequences.
TABLE 1.
PIA and PIB probe characteristics
| Por class and probea | Sequence (5′→3′) | Accession no.b | Control strainc |
|---|---|---|---|
| PIA | |||
| 1-1* | TTACCATGGAGCTCAGGCGGATCG | AY765454 | 177007 |
| 1-2* | CAGATCACACAGGTCGGGCGAATC | AY765457 | 252d |
| 1-3* | GAGCTCAGGCGTCTGGCGTTGAA | AY765456 | 192 |
| 2-1 | GTCAGCGGTACTGACACAGGC | J03029 | FA19 |
| 2-2 | CTAAAGAAGGCTGGGGCAGCC | AY765454 | 177007 |
| 2-3* | TACGTCAGCGGTACTCTCG | AY765459 | DG127 |
| 2-4* | TCAGCGGTACTAACGAAGGC | AY765457 | 252 |
| 3-1* | CCAACCCGAAGAACGCCA | J03029 | FA19 |
| 3-2 | CGACCCGAAGAACGCCCCA | AY765456 | 192 |
| 3-3 | CTGAACCCGAAGCACGCCACA | AY765457 | 252 |
| 6-1 | ACGCGAAATTGACTTGGAGCAACGAT | J03029 | FA19d |
| 6-2 | ACGCGAAATTGGCTTGGCCCG | AY765456 | 192 |
| 6-3* | GACGCGAAATTGGCTTTGATCG | AY765457 | 252 |
| 6-4* | CGAAATTGACTTGGCGCGACA | J03029 | FA19e |
| 7-1 | GGTTCGGTTTATGATGCAGATAACGAC | J03029 | FA19 |
| 7-2 | GGTTCGGTTTATGAGGCAAATCACG | AY765456 | 192 |
| 7-3 | TTTGGTTTATCATGCAGACTTAAGCAACGAT | AY765457 | 252 |
| PIB | |||
| 1-1* | AACATACAGACGGCAAGGTAAGTAAAGTG | M21289 | MS11 |
| 1-2* | ACATCGGGAAGGCAAAGTAGTTGGCG | AF044794 | 5589 |
| 1-3 | TGTAGAACATACAAAAGGCAAGGTAAGTAAAG | AF044788 | 9299 |
| 3-1 | GGCAATGTGCTGGAAATCAGCGG | M21289 | MS11 |
| 3-2 | CGGCGAGTTTCTGGAAATCAGCAAA | F044785 | 3744 |
| 5-1 | AAAATCGAATACGATGATCAAAATTATAGTATACCC | AF044788 | 9299 |
| 5-2 | CGAATACGATGGTCAAGCTTATAGTATGC | AY297698 | S140 |
| 5-3 | AAAAAATGGAAGGATATCTATATAATATCCCCAGT | AF044794 | 5589 |
| 5-4 | ATCGAATACGATAATCAATTTTATAGTATCCCC | AF044785 | 3744 |
| 5-5 | AAAAATGGAAGGATATACATATAATATCCCCAG | AY765442 | 255 |
| 5-6 | ATGGAAGGATATGCATATAATATCCCCAG | NAg | F62 |
| 5-7 | ATCGAATACGATAGTCAATATTATAGTATCCC | U75640 | —f |
| 5-8 | CGAATACGAACATCAAGTTTATAGTATCCC | M21289 | MS11 |
| 5-9 | AATCGAAGGCTATCAGTATAATAGCCCC | 2432h | |
| 6-1 | TATGGAGCAAGGAGGGCTAATTCG | AF044790 | 1861 |
| 6-2 | GTATGGAGCAACGAGGGTTAATTCG | AF044794 | 5589 |
| 6-3 | CCAAATTGTATGGAGCAATGAGCGG | AF044788 | 9299 |
| 6-4* | GTATGGAACATGGCGTGCTAATT | 2432h | |
| 6-5 | GCCAAATTGTATCAAAATCAAATAGTGCGTGATAATT | M21289 | MS11d |
| 6-6* | GTATAGAACATGGCATGCTAATTCG | AF044795 | 5441 |
| 7-1 | CAAAGGCACTGTTGATGATGCAAACC | 2432h | |
| 7-2 | CTCTGTTCGTAGTGCAGACTACG | AF044785 | 3744 |
| 7-3 | CTGTTGATAGTGCAGACCACGACAAT | M21289 | MS11 |
Probes marked with an asterisk (*) have minor modifications from the previously reported sequence (26) or are new.
GenBank accession numbers for porB sequences of control strains.
Control strains used in hybridizations.
Differs from probe by one base pair.
Differs from probe by two base pairs.
—, no control strain available.
NA, not available.
The porB VR5 region of this strain was identical to that of the strain with GenBank sequence accession no. NGU17235. Other VRs of this strain have not been sequenced.
Checkerboard hybridization and signal detection.
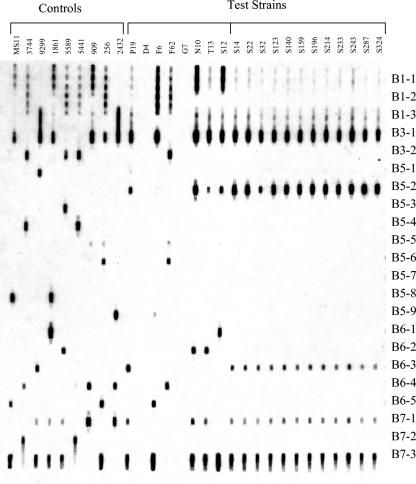
Checkerboard hybridizations were conducted as previously described (26). Briefly, 400 ng of denatured PCR-amplified porB DNA was applied to Zeta-Probe-GT nylon membranes (Bio-Rad, Hercules, Calif.) by using a 30-slot vacuum apparatus (Immunetics, Cambridge, Mass.). Hybridizations were conducted by using a 45-channel vacuum apparatus (Immunetics) at 59°C for 3 h, and bound probes were visualized by using streptavidin-horseradish peroxidase conjugate (Roche Molecular Biochemicals) and ECL chemiluminescent substrate (Amersham Pharmacia Biotech, Piscataway, N.J.). Hybridization signals were compared to those of control strains (see Fig. 2).
FIG. 2.
Checkerboard hybridization of PIB probes to serovar control strains (D4 to S12) and 12 of 14 Sheffield cluster strains (S14 to S324). The porB sequences of cluster strains S32 and S140 differed by a single base pair in VR5. S32 and S140 porB sequences were otherwise identical and differed from probes B1-1, B1-3, B 6-3, and B7-3 each by 1 bp and from B7-1 by 2 bp.
DNA sequencing.
PCR-amplified porB DNA, generated with primers GCPorBF.outer and GCPorBR.outer, was purified by QIAquick spin-columns (QIAGEN). DNA sequences were determined from both strands for each strain by using an ABI PRISM dye terminator sequencing kit with AmpliTaq DNA polymerase FS (Perkin Elmer) on a model 377 automated sequencer (Applied Biosystems). Sequences were collated and analyzed by using SEQED, (GCG10.2-Unix). The porB gene sequences of strains 280044, 280, 280042, 177, 177007, 192, 192014, 252, 255, 255034, 271, 271536, 163, 163006, DGI 17, DGI 18, DGI 19, DGI 27, DGI 29, DGI 34, DGI 37, DGI 40, DGI 43, DGI 61, DGI 70, S32, and S140 were determined. The porB sequences of seven Sheffield cluster strains were previously determined and submitted to the GenBank database under accession numbers AY297697 to AY297703.
Restriction analysis.
The transferrin-binding protein B gene (tbpB) from selected DGI strains was amplified by using primers T1 (5′ATGAACAATCCATTGGTGA) and T2 (5′TGGCGTTTCGCACCGAATAC). Amplified DNA fragments were digested with restriction endonucleases AluI, HaeIII, RsaI, and MspI (Roche Molecular Biochemicals) by using 8 μl of PCR product, 1.0 μl of 10× restriction endonuclease buffer, 10 U of restriction endonuclease, and sterile distilled water to a final volume of 10 μl. The mixture was incubated at 37°C for 1 h and then separated on 1.5% agarose gel and stained with ethidium bromide.
por VR typing nomenclature.
Hybridization results for a single VR are referred to as the “VR type,” designated by the Por class, the VR, and the probe, e.g., PIB1-1. When more than one probe is bound for a single VR, each probe is listed, separated by a comma (e.g., PIB6-4,6), and decreased signal intensity compared to homologous controls is indicated by parentheses. The “por type” includes results for all VRs tested, listed sequentially and separated by a semicolon; for example, B3;1;4;4,6;2 refers to a strain hybridizing PIB1-3, PIB3-1, PIB5-4, PIB6-4 and 6-6, and PIB7-2 probes.
Nucleotide sequence accession numbers.
The porB gene sequences of strains 280044, 280, 280042, 177, 177007, 192, 192014, 252, 255, 255034, 271, 271536, 163, 163006, DGI 17, DGI 18, DGI 19, DGI 27, DGI 29, DGI 34, DGI 37, DGI 40, DGI 43, DGI 61, DGI 70, S32, and S140 were submitted to the GenBank database under accession numbers AY765435 to AY765461.
RESULTS
The applicability of por VR typing was examined by using diverse and well-characterized collections of strains. These included temporally and geographically diverse isolates (38), serovar typing control isolates, epidemiologically linked isolates (41) with several unrelated control strains of the same A/S type, a large collection of partner isolates (21), and isolates from DGIs (28). In total, 206 isolates were por VR typed by using probes to five VRs of both PIA and PIB porB alleles. Among these isolates, 54 different por types were identified. The accuracy of por VR typing was confirmed by using 27 porB sequences determined in this study and published porB sequences of 10 control strains. The relationships between serovar and por VR type were examined by using the MAb binding patterns of strains that have been serovar typed on multiple occasions. These relationships were further investigated by examining the VR sequences of porB from PIB and PIA strains in the GenBank database that had an identified serovar.
por VR type of a panel of diverse strains.
A panel of 18 diverse strains was developed by van Looveren et al. for the purpose of evaluating gonococcal typing methods (38). The 18 strains of the panel were discriminated by por VR typing although two PIB strains (3790 and 855) differed only by the strength of the hybridization signal of probe B1-2. por VR typing was as discriminating among these strains as opa and A/S typing and was more discriminatory than serovar typing alone.
Relationship between VR type and serovar MAb binding.
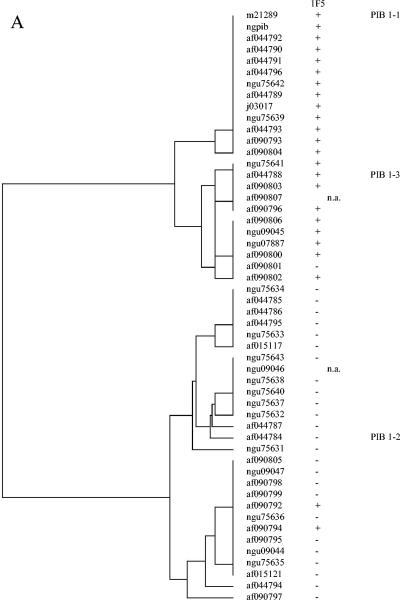
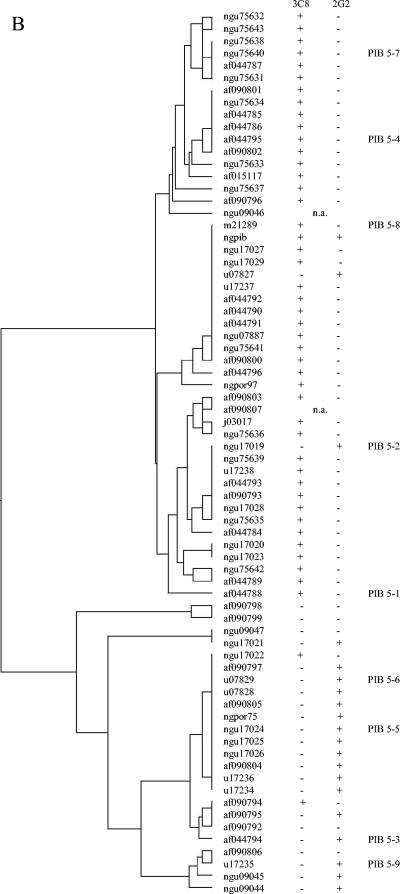
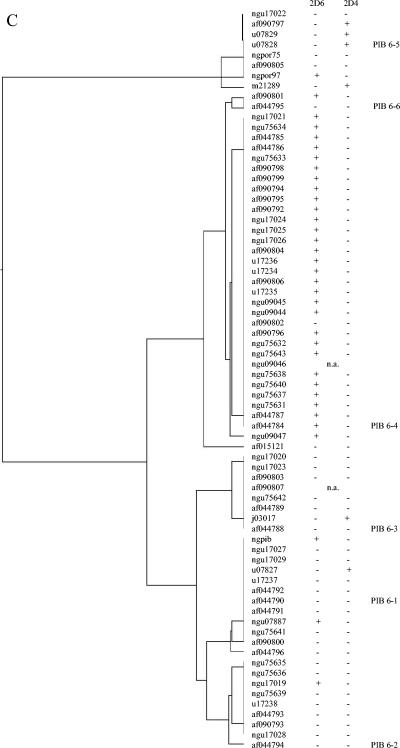
Relationships between serovar MAb binding and VR sequence were determined by examining the por VR types of the panel of strains described above, as well as eight serovar control strains that had been previously serotyped on multiple occasions (C. Ison, personal communication) and nine por VR typing control strains (Table 2 and Table 3). Associations observed between VR type, or clusters of closely related VR types, and MAb reactivities were further examined by comparing the VR sequences and serovar designation of porB genes in the GenBank database (Fig. 1A to C). MAb 1F5 was associated with PIB1-1 and some PIB1-3. MAb 3C8 was associated with PIB5-1, PIB5-2, PIB5-4, PIB5-7, and PIB5-8, and MAb 2G2 was associated with probes PIB5-3, PIB5-5, PIB5-6, and some PIB5-9. These two groups of VR5 types can be seen in the dendrogram in Fig. 1B and differ by a 6-bp deletion in the 2G2 group. Importantly, these two MAbs distinguish common serovars IB-1 and IB-2 from serovars IB-5 and IB-7. MAb 2D6 was associated with PIB6-4 or PIB6-4,6 and, in some instances, PIB6-2. MAb 2D4 was associated with PIB6-5. This MAb is one of three that define the common serovar IB-4.
TABLE 2.
PIB por VR types and serovars of gonococcal por VR and serovar control strains
| Isolate group and strain | Result of hybridization with probe(s) targeting sequence encoding loopa:
|
Binding by MAbb:
|
Serovarc | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 5 | 6 | 7 | 3C8 | 1F5 | 2D6 | 2G2 | 2D4 | 2H1 | ||
| Serovar controls | ||||||||||||
| F6 | 1-2 | 3-1 | 5-(9) | 6-5 | 7-3 | X | X | X | IB-4 | |||
| S12 | 1-1 | 3-1 | 5-(2) | 6-1 | 7-1,3 | X | X | X | IB-3 | |||
| N10 | 1-1 | 3-1 | 5-2 | 6-2 | 7-1,3 | X | X | X | X | IB-1 | ||
| T13 | 1-(1,3) | 3-1 | 5-(2) | 6-2 | 7-1,3 | X | X | X | IB-22 | |||
| F62 | 1-2 | 3-2 | 5-6 | 6-4 | NT | X | X | X | IB-7 | |||
| por VR typing controls | ||||||||||||
| MS11 | 1-1 | 3-1 | 5-8 | 6-5 | 7-3 | X | X | X | X | IB-9 | ||
| 1861 | 1-1 | 3-1 | 5-8 | 6-1 | 7-1,3 | X | X | X | IB-3 | |||
| 9299 | 1-3 | 3-1 | 5-1 | 6-3 | 7-1,3 | X | X | X | IB-3 | |||
| 3744 | 1-2 | 3-2 | 5-4 | 6-4,6 | 7-2 | X | X | X | IB-2 | |||
| 5441 | 1-2 | 3-2 | 5-4 | 6-6 | 7-2 | X | X | IB-6 | ||||
| 5589 | 1-2 | 3-2 | 5-3 | 6-2 | 7-1,3 | X | X | IB-8 | ||||
Results are given in the shortcut form VR-probe (see “por VR typing nomenclature” in the text for details). Where more than one probe was bound, probe numbers are separated by a comma. Parentheses indicate decreased signal intensity. NT, nontypeable.
X, strain was bound.
TABLE 3.
PIA por VR types and serovars of gonococcal por VR and serovar control strains
| Isolate group and strain | Result of hybridization with probe(s) targeting sequence encoding loopa:
|
Binding by MAbb:
|
Serovarc | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 6 | 7 | 4A12 | 4G5 | 2F12 | GD9 | 5G9 | 5D1 | ||
| Serovar controls | ||||||||||||
| 7122 | 1-(1) | 2-3 | 3-1 | 6-1,4 | 7-1 | +/− | X | X | X | X | X | IA-2 |
| D4 | 1-1 | 2-2 | 3-1 | 6-2 | 7-1 | X | X | X | X | IA-6 | ||
| G7 | 1-1 | 2-2 | 3-1 | 6-1,4 | 7-1 | X | X | X | X | X | ||
| por VR typing controls | ||||||||||||
| PI83 | 1-(1) | 2-1 | 3-1,2 | 6-2,3 | 7-1 | X | X | IA-8 | ||||
| PU186 | 1-1 | 2-1 | 3-1 | 6-1,4 | 7-1,(2) | X | X | X | X | IA-5 | ||
| FA19 | 1-1 | 2-1 | 3-1 | 6-1,4 | 7-1 | X | X | X | X | X | X | IA-1 |
Results are given in the shortcut form VR-probe (see “por VR typing nomenclature” in the text for details). Where more than one probe was bound, probe numbers are separated by a comma. Parentheses indicate decreased signal intensity.
±, binding result was equivocal; X, strain was bound.
Serovars were previously determined.
FIG. 1.
Dendrograms from multiple sequence alignments of PIB VR 1 (A), 5 (B), and 6 (C) were generated by using GCG PILEUP (Genetics Computer Group GCG 10.2-Unix).MAb binding is derived from the serovar stated in the GenBank database or the associated reference. +, serovar includes the identified MAb antibody; −, serovar does not include the identified antibody; n.a., a serovar designation was not available.
PIA serovar MAb 6D9 corresponded to PIA6-1,4. Antibodies 2F12 and 4G5 were bound by all strains in Table 3, suggesting an association with PIA VRs 1, 3, or 7, which were not sufficiently diverse to determine the antibody epitopes. MAbs 4A12, 5G9, and 5D1 did not appear to correspond with any single VR.
Sheffield cluster strains.
Strains identified by opa type as part of a large epidemiologically linked cluster (41) as well as four A/S-matched unrelated control strains were por VR typed in a blinded fashion. The 14 cluster strains and 1 control strain (2491) had identical por types, except for 1 cluster strain (S32) that had decreased hybridization signal of the PIB5-2 probe (Fig. 2). This was consistent with porB sequence data showing that strain S32 had a single base pair mutation in VR5. Of the three controls that had different porB sequences but the same A/S type, differences were identified by por VR type in VR1 for all three strains and in VR5 for two strains.
Boston partner strains.
The por types of 109 strains collected as part of a study of gonococcal transmission were determined. Among these strains, 28 were PIA and 81 were PIB. Four PIA por types were identified; the most common type was A1;2;1;1;1 (64% of PIA strains). Twenty-three PIB por types were identified; the three most common were B2;2;4;4;2 (18.5% of PIB strains), B2;1;6;5;3 (16% of PIB strains), and B2;2;7;4;2 (8.6% of PIB strains). In addition, eight strains (9.9%) were similar to one of these three PIB por types, differing only in the strength of the hybridization signal for one VR, which suggests that there are single nucleotide differences in those regions. No strains were completely nontypeable. Among PIB isolates, 14 strains did not bind a VR5 probe, 9 strains did not bind a VR7 probe, 4 strains did not bind a VR1 probe, and 2 strains did not bind a VR6 probe.
Among the 107 strains in which both por type and A/S type were available, there were 26 different por types, 28 different A/S types, and 15 serovar types. Although the level of discrimination was similar overall, por VR typing and A/S typing differed dramatically among PIB strains in terms of which strains were identified as having the same type (Table 4). Four different serovars (seven A/S types) were assigned to strains with the common PIB por type B2;2;4;4;2. Conversely, there were seven different por types among 18 IB1 strains: four por types among 8 IB1 proline-requiring strains and three por types among 8 IB1 prototrophic strains. Two IB1 proline- and hypoxanthine-requiring strains had a por type found among IB1 prototrophic strains. The por gene was sequenced for 14 strains and in all cases confirmed the accuracy of the por VR type. The accuracy of either typing method in distinguishing unrelated partnership pairs or in identifying extended networks cannot be accurately assessed without further analysis such as multilocus sequence typing, since extended epidemiologic data to identify links between groups are not available.
TABLE 4.
por type of Boston strains
| por type | No. of strains | Serovara | No. of strains |
|---|---|---|---|
| B2; 2; 4; 4; 2 | 15 | IB1 | 6 |
| IB2 | 5 | ||
| IB17 | 2 | ||
| IB4 | 1 | ||
| NAb | 1 | ||
| B2; 1; 6; 5; 3 | 13 | IB4 | 6 |
| IB12 | 2 | ||
| IB1 | 2 | ||
| IB8 | 2 | ||
| IB2 | 1 | ||
| B 2; 2; 7; 4; 2 | 7 | IB2 | 3 |
| IB1 | 2 | ||
| IB12 | 2 | ||
| Other PIB por typesc | 46 | IB3 | 10 |
| IB1 | 8 | ||
| IB2 | 6 | ||
| IB6 | 4 | ||
| Other (7 types) | 18 | ||
| A 1; 2; 1; 1; 1 | 18 | IA6 | 16 |
| IB3 | 2 | ||
| A 3; 1; 2; 2; 2 | 6 | IA4 | 5 |
| NA | 1 | ||
| A 3; 1; 1-2; 2-3; 2 | 3 | IB32 | 3 |
| A 2; 4; 3; 3; 3 | 1 | IB24 | 1 |
Among strains with common por types, serovar designations that are consistent with the associations between serovar MAb and VR probes are shown in bold.
NA, serovar not available.
Other PIB por types totaled 19.
Among the 109 strains, 81 were from individuals who met epidemiologic criteria for the partner transmission study: 68 strains from 34 male-female partnerships, 9 strains from three groups of one male with two identified female partners, and 4 strains from a group of one male with three female partners. Concordance of por type was found among strains from 39 of 43 defined partnerships (>90%). Of the four discordant females, two were from one partnership group in which the strain isolated from the male was different, while the strains from both women had identical por types. A third discordant female occurred in another group of one male with two identified partners, and the fourth occurred in one of the 34 male-female partnerships. In each case the A/S types were the same.
DGI strains.
The por types of 53 DGI strains were determined (Table 5). A total of 42 strains expressed PIA (79%), and among these, three PIA por types were identified: A1;3;1;1,4;1 (36 of 42, or 85.7%), A3;1;2;2;2 (4 of 42, or 9.5%), and A1;2;1;1;1 (2 of 42, or 4.8%). Eleven strains expressed PIB (21%), and seven different PIB por types were identified. The por type of 10 strains was confirmed by porB sequencing, and 10 had RFLP analysis of the tbpB gene. Five of the 10 DGI strains analyzed by tbpB RFLP were selected from the group of strains with the common A1;3;1;1,4;1 por type; the other 5 DGI strains had a variety of por types. The A1;3;1;1,4;1 strains had identical RFLP patterns with all four enzymes. Of the other 5 DGI strains, and 18 other unrelated N. gonorrhoeae strains that were not part of this study, all had unique RFLP patterns (data not shown).
TABLE 5.
por VR type of 53 disseminated gonococcal infection strains
| por type | n (%) |
|---|---|
| A 1; 3; 1; 1; 4; 1 | 36 (68) |
| A 1; 2; 1; 1; 1 | 2 (4) |
| A 3; 1; 2; 2; 3; 2 | 4 (7) |
| B (7 different por types) | 11 (21) |
DISCUSSION
por VR typing is an attractive approach for gonococcal strain typing. As a molecular method, it can be applied to nonviable bacterial cell samples (13); the reagents are easily synthesized, and the method is easy to use and highly reproducible. In order to examine the utility of por VR typing as a microbiologic and epidemiologic tool, we determined the por types of strains from several well-characterized collections. The results of this study suggest that por VR typing has broad applicability when used with the present set of 40 oligonucleotide probes. por VR typing discriminated unrelated strains and was able to accurately identify epidemiologically linked isolates; no strains were completely nontypeable, and the accuracy of por VR typing was confirmed by porB sequencing.
We explored the relationship between typing MAbs (19) and por VR types. Previous studies have identified specific binding epitopes or suggested binding regions for several antibodies. Carbonetti et al. localized MAb 1F5 to the N-terminal 60 residues and 3C8, 2H1, and 2D4 to the loop 5 or 6 regions of the PIB strain MS11 by using constructed PIA/PIB hybrids (3). Cooke et al. further determined that antibody 2D4 bound an epitope of PIB loop 6 encoded by the sequence K(L/Y)YQNQLVRD and suggested the loop 5 sequence YSIPS as the epitope for 3C8 (8). The porB sequences of naturally occurring PIA/PIB hybrids and a PIB VR5 deletion strain supported these observations and indicated that the 2H1 antibody bound a loop 5 region that was common to most PIB strains (9). Unemo et al. recently expanded and refined the comparison of the porB sequence and serovar (37). In our study, the associations of antibody 1F5 with PIB1-1 and antibody 2D4 with probe PIB6-5 are consistent with these reports; and probes PIB5-1, PIB5-2, PIB5-4, PIB5-7, and PIB5-8, corresponding to MAb 3C8, all identify sequences encoding the sequence YS(I/M)PS in the loop 5 region. Epitopes for antibodies 2G2 and 2D6 have not been previously identified, but sequence and serovar data from the GenBank and the data presented here suggest that antibody 2G2 corresponds to sequences that hybridize PIB5-3, PIB5-5, PIB5-6, and some sequences that hybridize PIB5-9 while antibody 2D6 corresponds to sequences binding probes PIB6-4 and PIB6-6 and some that bind PIB6-2.
Previous studies indicate that PIA MAbs 4A12, 5G9, and 5D1 recognize complex epitopes involving both the N- and C-terminal regions, MAb 6D9 binds the loop 6 DAKLTWRND region of strain FA19, MAb 4G5 binds the loop 3 sequence IAQPEE, and 2F12 binds the N-terminal region of strain FA19 (3, 27, 37). In this study, the PIA1-1 probe identified the N-terminal sequence required for 5D1 binding. The VR6 porB sequence of FA19 (GenBank accession number J03029 [4]) differs from the PIA6-1 probe by 1 nucleotide and from the PIA6-4 probe by 2 nucleotides, explaining the FA19 hybridization pattern PIA6-1,4 that corresponds to MAb 6D9. The 4G5 epitope corresponds to PIA3-1 and PIA3-2, consistent with the sequence analysis by Unemo et al. (37). 2F12 binding was associated with hybridization of probes PIA1-1, PIA3-1, and PIA7-1, but in the context of earlier work by Carbonetti et al. (3), the epitope corresponds to PIA1-1.
Among partner strains overall, common serovars within each por type group were consistent with the relationships described above. In many instances, though, a number of different serovars were seen within por type groups, consistent with previous reports of discrepancies between serovar determinations and porB sequence (20, 30, 37). Interestingly, based on the relationships between serovar MAbs and the previous polyclonal typing system (19), the four most common por types among these strains would also be predicted to correspond to WI (A 1;2;1;1;1), WII (B 2;2;4;4;2 and B 2;2;7;4;2), and WIII (B 2;1;6;5;3) isolates. Regardless of the differences observed between serovar and por VR type, concordance was very high among partners that met epidemiologic criteria. The few partnerships that were identified as discordant by por VR typing may have occurred because among partners that were both infected, it is possible that not all identified strains had been exchanged between partners. By using rigorous epidemiologic criteria (21), concordance rates of 97% among single male-female partnerships and >90% including males with multiple identified partners were observed. Confirmations of concordance and the ability to accurately identify strains with the same or very similar Por proteins are important for studies of transmission, acquired immunity, or pathogenesis.
PIA-expressing strains have been associated with disseminated infections (2, 28), so the predominance of PIA por types within the DGI collection was not surprising. Of interest was the identification of a single por type in 68% of the DGI strains (85.7% of PIA strains) collected over a 7-year period, as well as the suggestion that these strains represent a clonal population based on tbpB RFLP analysis. Genetic characterization of strains of the PIA-1,2 arginine-, hypoxanthine-, and uracil-requiring A/S type associated with DGI strains in Seattle also showed little genetic diversity over time (46). Further determination of the clonal nature of the DGI strains examined here, such as by multilocus sequence typing (39), is warranted in light of the potential use of this collection for studies to identify genes associated with disseminated disease.
We have previously used por VR typing to examine the diversity of Por over 10 years in a large urban community. This study and others suggest that Por diversity is restricted (12, 26). Structural, functional, or immunologic restrictions on Por diversity require further investigation, and genotypic analyses of porB will have several advantages over present serologic typing schemes. Studies have shown that strains of the same serovar may have differences in sequences encoding surface-exposed regions of Por, and, conversely, strains with different serovars may have many antigenic regions in common (12, 17, 20, 27, 34, 37). The propensity for recombination, resulting in mosaic porins, and a lack of specific antibodies for each antigenic region complicate characterization of the role that specific epitopes may play in pathogenicity or protection from disease. By identifying each VR independently, por VR typing is well suited to examining this mosaic gene.
por VR typing has some limitations. porB variation occurs through both point mutations and horizontal genetic exchange and is subject to selective pressure (12). Over time, or between geographic regions, por type may not accurately reflect strain relatedness. por VR typing may not be as discriminatory as sequencing, and minor variations in hybridization signal, suggesting single base pair differences, cannot be distinguished as synonymous versus nonsynonymous without subsequent sequence analysis. Additionally, since antigenic differences may result from single amino acid changes or may not accompany much larger changes, immunologic reactivity with sera or MAbs remains necessary for some applications.
Regardless of these limitations, molecular methods will allow for the development of rapid, accurate, and widely available typing tools. Hybridization can be utilized in a variety of methods ranging in level of technical sophistication from simple colony or dot blots to microarray or fluorescent-labeled probe detection. Analysis and interpretation of results from probe hybridization assays are technically simple. Sequencing and hybridization methods are complementary, since an approximation of sequence can be obtained from probe hybridization patterns.
Hybridization-based methods may provide an advantage in detecting mixed gonococcal infections. Coinfection with more than one gonococcal strain was recently identified in 20% of males by comparison of the opa type obtained from urethral swab specimens to the opa type of the primary culture (23). One of the goals of molecular typing is to allow for the characterization of strains directly from nucleic acid amplification test samples. We have identified mixed infections in direct clinical specimens by using por VR typing (22), and in the present study, unrecognized coinfection with more than one gonococcal strain may have contributed to the overall 9.3% (4 of 43 pairs) discrepancy observed in partners where only cultivated strains were tested rather than infected secretions. Identifying the presence of and distinguishing mixed infections in direct clinical samples may be difficult with sequence-based typing.
Rapid and widely available typing systems have the potential to provide information useful to the control and prevention of N. gonorrhoeae infections and to guide public health interventions. In this study we have shown that por VR typing is a molecular tool that is applicable to a wide variety of strain collections and is both discriminatory and accurate. It is compatible with porB sequencing methods, and it has the potential to be applied to nonculture-based clinical samples in conjunction with nucleic acid amplification diagnostic tests. por VR typing holds promise as an epidemiological tool and as a means to increase our understanding of the role of Por in neisserial pathogenicity and human immunity.
Acknowledgments
We are grateful to Cathy Ison and Iona Martin for thoughtful contributions during the development of this work, for providing strains, and for review of the manuscript, and to Carl E. Frasch, Wendy Carr, and M. S. Blake for critical comments on the manuscript.
D. McKnew was supported in part by the FDA Office of Women's Health through a postdoctoral fellowship administered by the Oak Ridge Institute for Science and Education.
REFERENCES
- 1.Blake, M. S., and E. C. Gotschlich. 1987. Functional and immunologic properties of pathogenic Neisseria surface proteins, p. 377-400. In M. Inouye (ed.), Bacterial outer membranes as model systems. John Wiley & Sons, Inc., New York, N.Y.
- 2.Brunham, R. C., F. Plummer, L. Slaney, F. Rand, and W. DeWitt. 1985. Correlation of auxotype and protein I type with expression of disease due to Neisseria gonorrhoeae. J. Infect. Dis. 152:339-343. [DOI] [PubMed] [Google Scholar]
- 3.Carbonetti, N. H., V. I. Simnad, H. S. Seifert, M. So, and P. F. Sparling. 1988. Genetics of protein I of Neisseria gonorrhoeae: construction of hybrid porins. Proc. Natl. Acad. Sci. USA 85:6841-6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carbonetti, N. H., and P. F. Sparling. 1987. Molecular cloning and characterization of the structural gene for protein I, the major outer membrane protein of Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. USA 84:9084-9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2003. Sexually transmitted disease surveillance, 2002. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, Ga.
- 6.Centers for Disease Control and Prevention. 2002. Sexually transmitted disease surveillance, 2001. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, Ga.
- 7.Centers for Disease Control and Prevention. 2000. Fluoroquinolone resistance in Neisseria gonorrhoeae, Hawaii, 1999, and decreased susceptibility to azithromycin in N. gonorrhoeae, Missouri, 1999. JAMA 284:1917-1919. [PubMed] [Google Scholar]
- 8.Cooke, S. J., H. de la Paz, C. Lapoh, C. A. Ison, and J. E. Heckels. 1997. Variation within serovars of Neisseria gonorrhoeae detected by structural analysis of outer-membrane protein PIB and by pulsed-field gel electrophoresis. Microbiology 143:1415-1422. [DOI] [PubMed] [Google Scholar]
- 9.Cooke, S. J., K. Jolley, C. A. Ison, H. Young, and J. E. Heckels. 1998. Naturally occurring isolates of Neisseria gonorrhoeae, which display anomalous serovar properties, express PIA/PIB hybrid porins, deletions in PIE or novel PIA molecules. FEMS Microbiol. Lett. 162:75-82. [DOI] [PubMed] [Google Scholar]
- 10.Edwards, J. L., E. J. Brown, S. Uk-Nham, J. G. Cannon, M. S. Blake, and M. A. Apicella. 2002. A co-operative interaction between Neisseria gonorrhoeae and complement receptor 3 mediates infection of primary cervical epithelial cells. Cell Microbiol. 4:571-584. [DOI] [PubMed] [Google Scholar]
- 11.Fleming, D. T., and J. N. Wasserheit. 1999. From epidemiological synergy to public health policy and practice: the contribution of other sexually transmitted diseases to sexual transmission of HIV infection. Sex Transm. Infect. 75:3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fudyk, T. C., I. W. Maclean, J. N. Simonsen, E. N. Njagi, J. Kimani, R. C. Brunham, and F. A. Plummer. 1999. Genetic diversity and mosaicism at the por locus of Neisseria gonorrhoeae. J. Bacteriol. 181:5591-5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giles, J. A., J. Falconio, J. D. Yuenger, J. M. Zenilman, M. dan, and M. C. Bash. 2004. Quinolone resistance-determining region mutations and por type of Neisseria gonorrhoeae isolates: resistance surveillance and typing by molecular methodologies. J. Infect. Dis. 189:2085-2093. [DOI] [PubMed] [Google Scholar]
- 14.Gorby, G. L., A. F. Ehrhardt, M. A. Apicella, and C. Elkins. 2001. Invasion of human fallopian tube epithelium by Escherichia coli expressing combinations of a gonococcal porin, opacity-associated protein, and chimeric lipo-oligosaccharide. J. Infect. Dis. 184:460-472. [DOI] [PubMed] [Google Scholar]
- 15.Gotschlich, E. C., M. E. Seiff, M. S. Blake, and M. Koomey. 1987. Porin protein of Neisseria gonorrhoeae: cloning and gene structure. Proc. Natl. Acad. Sci. USA 84:8135-8139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutjahr, T. S., M. O'Rourke, C. A. Ison, and B. G. Spratt. 1997. Arginine-, hypoxanthine-, uracil-requiring isolates of Neisseria gonorrhoeae are a clonal lineage within a non-clonal population. Microbiology 143:633-640. [DOI] [PubMed] [Google Scholar]
- 17.Hobbs, M. M., T. M. Alcorn, R. H. Davis, W. Fischer, J. C. Thomas, I. Martin, C. Ison, P. F. Sparling, and M. S. Cohen. 1999. Molecular typing of Neisseria gonorrhoeae causing repeated infections: evolution of porin during passage within a community. J. Infect. Dis. 179:371-381. [DOI] [PubMed] [Google Scholar]
- 18.Ison, C. A., J. Pepin, N. S. Roope, E. Demba, O. Secka, and C. S. F. Easmon. 1992. The dominance of a multiresistant strain of Neisseria gonorrhoeae among prostitutes and STD patients in The Gambia. Genitourin. Med. 68:356-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knapp, J. S., M. R. Tam, R. C. Nowinski, K. K. Holmes, and E. G. Sandstrom. 1984. Serological classification of Neisseria gonorrhoeae with use of monoclonal antibodies to gonococcal outer membrane protein I. J. Infect. Dis. 150:44-48. [DOI] [PubMed] [Google Scholar]
- 20.Lau, Q. C., V. T. Chow, and C. L. Poh. 1993. Polymerase chain reaction and direct sequencing of Neisseria gonorrhoeae protein IB gene: partial nucleotide and amino acid sequence analysis of strains S4, S11, S48 (serovar IB4) and S34 (serovar IB5). Med. Microbiol. Immunol. 182:137-145. [DOI] [PubMed] [Google Scholar]
- 21.Lin, J. S., S. P. Donegan, T. C. Heeren, M. Greenberg, E. E. Flaherty, R. Haivanis, X. H. Su, D. Dean, W. J. Newhall, J. S. Knapp, S. K. Sarafian, R. J. Rice, S. A. Morse, and P. A. Rice. 1998. Transmission of Chlamydia trachomatis and Neisseria gonorrhoeae among men with urethritis and their female sex partners. J. Infect. Dis. 178:1707-1712. [DOI] [PubMed] [Google Scholar]
- 22.Lynn, F., M. M. Hobbs, J. M. Zenilman, F. M. T. F. Behets, K. Van Damme, A. Rasamindrakotroka, and M. C. Bash. 2005. Genetic typing of the porin protein of Neisseria gonorrhoeae from clinical noncultured samples: strain characterization and identification of mixed gonococcal infections. J. Clin. Microbiol. 43:368-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin, I. M. C., and C. A. Ison. 2003. Detection of mixed infection of Neisseria gonorrhoeae. Sex. Transm. Infect. 79:56-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massari, P., S. Ram, H. Macleod, and L. M. Wetzler. 2003. The role of porins in neisserial pathogenesis and immunity. Trends Microbiol. 11:87-93. [DOI] [PubMed] [Google Scholar]
- 25.Mauro, A., M. Blake, and P. Labarca. 1988. Voltage gating of conductance in lipid bilayers induced by porin from outer membrane of Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. USA 85:1071-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mcknew, D. L., F. Lynn, J. M. Zenilman, and M. C. Bash. 2003. Porin variation among clinical isolates of Neisseria gonorrhoeae over a 10-year period, as determined by Por variable region typing. J. Infect. Dis. 187:1213-1222. [DOI] [PubMed] [Google Scholar]
- 27.Mee, B. J., H. Thomas, S. J. Cooke, P. R. Lambden, and J. E. Heckels. 1993. Structural comparison and epitope analysis of outer-membrane protein PIA from strains of Neisseria gonorrhoeae with differing serovar specificities. J. Gen. Microbiol. 139:2613-2620. [DOI] [PubMed] [Google Scholar]
- 28.O'Brien, J. P., D. L. Goldenberg, and P. A. Rice. 1983. Disseminated gonococcal infection: a prospective analysis of 49 patients and a review of pathophysiology and immune mechanisms. Medicine (Baltimore) 62:395-406. [PubMed] [Google Scholar]
- 29.O'Rourke, M., C. A. Ison, A. M. Renton, and B. G. Spratt. 1995. Opa-typing: a high-resolution tool for studying the epidemiology of gonorrhea. Mol. Microbiol. 17:865-875. [DOI] [PubMed] [Google Scholar]
- 30.Poh, C. L., Q. C. Lau, and V. T. Chow. 1995. Differentiation of Neisseria gonorrhoeae IB-3 and IB-7 serovars by direct sequencing of protein IB gene and pulsed-field gel electrophoresis. J. Med. Microbiol. 43:201-207. [DOI] [PubMed] [Google Scholar]
- 31.Posada, D., K. A. Crandall, M. Nguyen, J. C. Demma, and R. P. Viscidi. 2000. Population genetics of the porB gene of Neisseria gonorrhoeae: different dynamics in different homology groups. Mol. Biol. Evol. 17:423-436. [DOI] [PubMed] [Google Scholar]
- 32.Rudel, T., A. Schmid, R. Benz, H. A. Kolb, F. Lang, and T. F. Meyer. 1996. Modulation of Neisseria porin (PorB) by cytosolic ATP/GTP of target cells: parallels between pathogen accommodation and mitochondrial endosymbiosis. Cell 85:391-402. [DOI] [PubMed] [Google Scholar]
- 33.Simpson, S. D., Y. Ho, P. A. Rice, and L. M. Wetzler. 1999. T lymphocyte response to Neisseria gonorrhoeae porin in individuals with mucosal gonococcal infections. J. Infect. Dis. 180:762-773. [DOI] [PubMed] [Google Scholar]
- 34.Thompson, D. K., C. D. Deal, C. A. Ison, J. M. Zenilman, and M. C. Bash. 2000. A typing system for Neisseria gonorrhoeae based on biotinylated oligonucleotide probes to PIB gene variable regions. J. Infect. Dis. 181:1652-1660. [DOI] [PubMed] [Google Scholar]
- 35.Tompkins, J. R., and J. M. Zenilman. 2001. Quinolone resistance in Neisseria gonorrhoeae. Curr. Infect. Dis. Rep. 3:156-161. [DOI] [PubMed] [Google Scholar]
- 36.Turner, C. F., S. M. Rogers, H. G. Miller, W. C. Miller, J. N. Gribble, J. R. Chromy, P. A. Leone, P. C. Cooley, T. C. Quinn, and J. M. Zenilman. 2002. Untreated gonococcal and chlamydial infection in a probability sample of adults. JAMA 287:726-733. [DOI] [PubMed] [Google Scholar]
- 37.Unemo, M., P. Olcen, J. Albert, and H. Fredlund. 2003. Comparison of serologic and genetic porB-based typing of Neisseria gonorrhoeae: consequences for future characterization. J. Clin. Microbiol. 41:4141-4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Looveren, M., C. A. Ison, M. Ieven, P. Vandamme, I. M. Martin, K. Vermeulen, A. Renton, and H. Goossens. 1999. Evaluation of the discriminatory power of typing methods for Neisseria gonorrhoeae. J. Clin. Microbiol. 37:2183-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Viscidi, R. P., and J. C. Demma. 2003. Genetic diversity of Neisseria gonorrhoeae housekeeping genes. J. Clin. Microbiol. 41:197-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viscidi, R. P., J. C. Demma, J. Gu, and J. Zenilman. 2000. Comparison of sequencing of the por gene and typing of the opa gene for discrimination of Neisseria gonorrhoeae strains from sexual contacts. J. Clin. Microbiol. 38:4430-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward, H., C. A. Ison, S. E. Day, I. Martin, A. C. Ghani, G. P. Garnett, G. Bell, G. Kinghorn, and J. N. Weber. 2000. A prospective social and molecular investigation of gonococcal transmission. Lancet 356:1812-1817. [DOI] [PubMed] [Google Scholar]
- 42.Wen, K. K., P. C. Giardina, M. S. Blake, J. Edwards, M. A. Apicella, and P. A. Rubenstein. 2000. Interaction of the gonococcal porin P.IB with G- and F-actin. Biochemistry 39:8638-8647. [DOI] [PubMed] [Google Scholar]
- 43.World Health Organization. 2001. Global prevalence and incidence of selected curable sexually transmitted infections: overview and estimates. World Health Organization, Geneva, Switzerland.
- 44.World Health Organization.2002. Surveillance of antibiotic resistance in Neisseria gonorrhoeae in the WHO Western Pacific Region, 2001. Commun. Dis. Intell. 26:541-545. [PubMed] [Google Scholar]
- 45.Xia, M., W. L. Whittington, K. K. Holmes, F. A. Plummer, and M. C. Roberts. 1995. Pulsed-field gel electrophoresis for genomic analysis of Neisseria gonorrhoeae. J. Infect. Dis. 171:455-458. [DOI] [PubMed] [Google Scholar]
- 46.Xia, M., W. L. Whittington, K. K. Holmes, and M. C. Roberts. 1997. Genomic homogeneity of the AHU/IA-1,2 phenotype of Neisseria gonorrhoeae during its disappearance from an urban population. Sex. Transm. Dis. 24:561-566. [DOI] [PubMed] [Google Scholar]
- 47.Yagupsky, P., A. Schahar, N. Peled, N. Porat, R. Trefler, M. Dan, Y. Keness, and C. Block. 2002. Increasing incidence of gonorrhea in Israel associated with countrywide dissemination of a ciprofloxacin-resistant strain. Eur. J. Clin. Microbiol. Infect. Dis. 21:368-372. [DOI] [PubMed] [Google Scholar]