Abstract
Helicobacter pylori urease, produced in abundance, is indispensable for the survival of H. pylori in animal hosts. Urea is hydrolyzed by the enzyme, resulting in the liberation of excess ammonia, some of which neutralizes gastric acid. The remaining ammonia is assimilated into protein by glutamine synthetase (EC 6.3.1.2), which catalyzes the reaction: NH3 + glutamate + ATP→glutamine + ADP + Pi. We hypothesized that glutamine synthetase plays an unusually critical role in nitrogen assimilation by H. pylori. We developed a phenotypic screen to isolate genes that contribute to the synthesis of a catalytically active urease. Escherichia coli SE5000 transformed with plasmid pHP808 containing the entire H. pylori urease gene cluster was cotransformed with a pBluescript plasmid library of the H. pylori ATCC 43504 genome. A weakly urease-positive 9.4-kb clone, pUEF728, was subjected to nucleotide sequencing. Among other genes, the gene for glutamine synthetase was identified. The complete 1,443-bp glnA gene predicts a polypeptide of 481 amino acid residues with a molecular weight of 54,317; this was supported by maxicell analysis of cloned glnA expressed in E. coli. The top 10 homologs were all bacterial glutamine synthetases, including Salmonella typhimurium glnA. The ATP-binding motif GDNGSG (residues 272 to 277) of H. pylori GlnA exactly matched and aligned with the sequence in 8 of the 10 homologs. The adenylation site found in the top 10 homologs (consensus sequence, NLYDLP) is replaced in H. pylori by NLFKLT (residues 405 to 410). Since the Tyr (Y) residue is the target of adenylation and since the H. pylori glutamine synthetase lacks that residue in four strains examined, we conclude that no adenylation occurs within this motif. Cloned H. pylori glnA complemented a glnA mutation in E. coli, and GlnA enzyme activity could be measured spectrophotometrically. In an attempt to produce a GlnA-deficient mutant of H. pylori, a kanamycin resistance cassette was cloned into the Tth111I site of H. pylori glnA. By using the standard technique of allelic exchange mutagenesis, no verifiable glutamine synthetase double-crossover mutant of strain UMAB41 could be isolated, suggesting that the mutation is lethal. We conclude that glutamine synthetase is critical for nitrogen assimilation in H. pylori and is active under all physiologic conditions.
Helicobacter pylori, the etiologic agent of gastritis and peptic ulcer disease in humans, produces urease as one of its most abundant protein components (19). This species produces an anomalously high urease activity compared to the many other bacterial species that synthesize this enzyme (27). Urease is clearly indispensable for H. pylori, which cannot survive in an animal model of infection in its absence (5). Urea, the nitrogenous waste product of mammals, is hydrolyzed by the enzyme, resulting in the liberation of an excess of ammonia, some of which is neutralized by gastric acid. The remaining ammonia must also be dealt with.
Ammonia, a preferred nitrogen source for bacteria, is assimilated into protein and other nitrogenous compounds in bacteria by a single pathway (29). Glutamine synthetase (EC 6.3.1.2) catalyzes the reaction NH3 + glutamate + ATP→glutamine + ADP + Pi (29). Glutamine, in turn, serves as the nitrogen donor for other nitrogenous compounds including alanine, glycine, serine, histidine, tryptophan, CTP, AMP, carbamoyl-phosphate, and glucosamine 6-phosphate (29). Because of the central role of glutamine synthetase in nitrogen metabolism, most species have developed complex regulatory schemes that modulate the expression and activity of the enzyme by the use of multiple promoters (ς70 and ς54), positive activators (NRI and NRII), posttranslational adenylation, and allosteric inhibition by nitrogenous compounds (21).
During H. pylori infection of the gastric mucosa, the potential for the generation of ammonia is high. Plentiful urea is provided by the host, and plentiful urease is synthesized by the bacterium (15, 22, 24). Because of the high concentration of urease-generated ammonia produced, we hypothesized that glutamine synthetase plays an unusually critical role in nitrogen assimilation by H. pylori. The recent release of the complete nucleotide sequence of the H. pylori genome revealed a notable absence of many regulatory pathways found in Escherichia coli (34). Our work confirms this general trend for H. pylori glnA and its protein product. We describe the isolation and sequencing of the glnA gene of H. pylori and characterization of the gene product when expressed in E. coli.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. H. pylori strains were passaged on blood agar (brucella agar supplemented with 10% [vol/vol] sheep blood) at 37°C in an anaerobic jar with palladium catalyst and an activated Campypak (Becton-Dickinson). Isolates were stored at −70°C in Trypticase soy broth (BBL) supplemented with 15% (vol/vol) glycerol. For liquid culture, H. pylori strains were inoculated from fresh blood agar plates into 250 ml of Mueller-Hinton broth supplemented with 4% (vol/vol) fetal calf serum in a 500-ml flask. Cultures were incubated for 48 h with shaking (200 rpm) at 37°C in an anaerobic jar containing an activated Campypak.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype and comments | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1; used as the recipient for electroporation | 31 |
| XL1-Blue | supE44 hsdR17 recA1 endA1 gyrA46 thi relA1 F′ [proAB+ lacIqlacZΔM15 Tn10 (Tetr)] | Stratagene |
| SURE | recB recF sbcC201 uvrC umuC::Tn5 (Kanr) lacΔ(hsdRMS) endA1 gyrA96 thi relA1 supE44 F′ [proAB+ lacIq lacZΔM15 Tn10 (Tetr)] | Stratagene |
| SE5000 | araD139Δ(argF-lac)U169 rpsL150 relA1 flb-3501 deoC1 ptsF25 rbsR recA56 | F. C. Gherardini (10) |
| M5004 | M5004 [λ−trpA9825(Oc), IN(rrnD-rrnE)1 glnA3], a glutamine synthetase-deficient mutant | M. Berlyn, E. coli Genetic Stock Center, Yale University, New Haven, Conn. (23) |
| W3110 | Glutamine synthetase-sufficient parent strain of M5004 [λ− IN(rrnD-rrnE)1 rph1] | M. Berlyn, E. coli Genetic Stock Center, Yale University, New Haven, Conn. (23) |
| H. pylori | ||
| UMAB41 | Isolated from a gastric biopsy specimen taken by endoscopy from a patient with complaints of abdominal pain and a history of peptic ulcer disease | 27 |
| Leung | Gastric biopsy isolate | J. Gilbert (11) |
| ATCC 43504 | Human gastric antrum, Australia | American Type Culture Collection (12) |
| HUH1 | Gastric biopsy isolate | D. Smoot, Howard University School of Medicine |
| HUH40 | Gastric biopsy isolate | D. Smoot, Howard University School of Medicine |
| X47ZAL | Gastric biopsy isolate | H. Kleanthous, Oravax |
| Plasmids | ||
| pHP1 | 1,489-bp fragment conferring Kanr originating from streptococcal plasmid pJH1, cloned into pUC | H. Kleanthous (35) |
| pSKCAT4 | 1.5-kb EcoRI fragment derived from H. pylori NTCC 11639 plasmid pUOA26 cloned into the EcoRI site of pBluescript SK−, chloramphenicol cassette | J. Gilbert (38) |
| pHC79 | Cosmid vector, 6.43-kb Ampr Tetr derived from pBR322 | 17 (GenBank accession no. L08873) |
| pUEF728 | λZAPII genomic clone of H. pylori ATCC 43504 converted to a pBluescript-based library clone; carries entire glnA gene | This study |
| pHP808 | pHP9D11 cosmid clone derivative encoding the entire urease gene cluster | 18 |
| pUEF16-7A | Cosmid clone of H. pylori UMAB41 | This study |
| pUEF730 | 1.5-kb KpnI-SpeI glnA-containing fragment cloned into the KpnI-SpeI site of pBluescript SK+ | This study |
Gene bank preparation.
Chromosomal DNA isolated from H. pylori ATCC 43504 was used for preparation of a λ ZAPII genomic library (Stratagene) as described previously (25). Chromosomal DNA isolated from H. pylori UMAB41 was used for preparation of a pHC79-based cosmid gene bank with Sau3A partials as described previously (18).
pBluescript plasmid library.
The λ ZAPII genomic library phage suspension (107 phage) was incubated with E. coli XL1-Blue (adjusted to an optical density at 600 nm [OD600] = 1.0) along with 106 Exassist helper phage for 15 min at 37°C. The suspension was then added to 20 ml of Luria broth and incubated at 37°C for 2 h and then at 65°C for 20 min. The suspension was centrifuged (4,500 rpm [2,400 × g] for 10 min at 4°C), and the supernatant was collected. Supernatant (1 μl) was added to a suspension (OD600 = 1.0) of E. coli SURE and incubated at 37°C for 15 min. After incubation, 100 μl of the suspension was plated onto Luria agar plates containing ampicillin (200 μg/ml) and incubated for 18 h at 37°C (14). Colonies from 20 plates were pooled and used for a large-scale plasmid preparation, which represented the pooled pBluescript plasmid library.
Plasmid isolation.
Plasmid DNA was isolated by alkaline sodium dodecyl sulfate (SDS) extraction (2). DNA was purified on plasmid purification columns (Qiagen tip-100) as directed by the manufacturer.
Recombinant DNA methods.
Recombinant DNA techniques, including restriction endonuclease digestion, ligation, and transformation, were done by standard techniques (2, 31).
DNA hybridization.
DNA restriction fragments isolated from a λZAPII gene bank clone were used as gene probes to identify pHC79-based cosmid clones containing complete glnA sequences. Fragments were labeled with [γ-32P]dCTP (specific activity, 3,000 Ci/mmol; Amersham) and used to probe dot blots of the cosmid gene library. Southern blots of cosmid clones digested with various restriction enzymes were prepared by standard methods (31) and developed with the enhanced chemiluminescence system (Amersham) as specified by the manufacturer.
Nucleotide sequencing.
Double-stranded DNA was used as a template for sequencing by the dideoxy chain termination method (32). The reactions were performed with reagents from the Prism Ready Reaction Dye Deoxy Termination Kit (Applied Biosystems) and Taq polymerase. A model 373A DNA Sequencer (Applied Biosystems) was used, and the sequence was determined in each direction. DNAsis software (Hitachi, version 2.1) was used for analysis of the DNA sequence, base composition, identification of open reading frames (ORFs), restriction sites, and other basic analyses. Apparent homologies between ORFs were sought in GenBank and SwissProt data bases with Wisconsin Package programs (version 8.1; Genetics Computer Group, Inc.).
Electroporation.
Plasmid DNA was introduced into H. pylori with a Gene Pulser electroporator (Bio-Rad) in 15% glycerol–9% sucrose at 12.5 kV/cm, 25-μF capacitance, and 200 Ω by the method of Ferrero et al. (7).
PCR and oligonucleotide primer design.
PCR was performed as previously described for H. pylori (9, 37) with primers designed from aphA (35), glnA (determined in this report), and vector pBluescript SK (GenBank accession no. X52330) sequences. Oligonucleotides were synthesized by the phosphoramidite method on an Applied Biosystems automated DNA synthesizer model 380B.
Protein labeling in maxicells.
Plasmid-encoded polypeptides were labeled with [35S]methionine (specific activity, 800 to 1,000 Ci/mmol; Amersham Corp.) by using UV-treated E. coli SE5000 transformed with pBluescript or pUEF730 and the method of Gherardini et al. (10). Labeled polypeptides were solubilized in SDS-gel sample buffer, electrophoresed on an SDS–12% polyacrylamide gel, and visualized by autoradiography.
Glutamine synthetase assay.
Glutamine synthetase activity was quantitated by an assay that measures the synthesis of γ-glutamylhydroxamate from glutamine and hydroxylamine. A spectrophotometric assay that measures γ-glutamylhydroxamate formation as a gauge of the total amount of glutamine synthetase present has been described by Bender et al. (4). Soluble protein (300 μg) derived from French pressure cell lysates (bacterial suspensions passaged at 20,000 lb/in2) was assayed in a reaction volume of 8 ml. Aliquots (500 μl) were taken at 30-min intervals and placed in 1 ml of stop mix (55 g of FeCl3 · 6H2O per liter, 20 g of trichloroacetic acid per liter, 21 ml of concentrated HCl per liter). The precipitate was removed by centrifugation, and the absorbance was measured at 540 nm.
Urease assays.
H. pylori was harvested from liquid culture by centrifugation (10,000 × g for 10 min at 4°C), washed twice with 20 mM sodium phosphate (pH 6.8), suspended in 5 ml of 20 mM sodium phosphate (pH 6.8)–5 mM dithiothreitol–1 mM EDTA, and ruptured in a precooled French pressure cell at 20,000 lb/in2. Lysates were centrifuged (27,000 × g for 30 min at 4°C), and the supernatants were removed with a Pasteur pipette and used directly for the assay. The protein concentration was determined by the bicinchoninic acid method as specified by the manufacturer (Pierce) with bovine serum albumin as a standard. Rates of urea hydrolysis were measured by the spectrophotometric assay of Hamilton-Miller and Gargan (13) calibrated for the measurement of ammonia as described previously (28).
Allelic exchange mutagenesis.
A kanamycin resistance cassette (35), isolated on a 1.5-kb EcoRV fragment from pHP1 (kindly provided by H. Kleanthous, Oravax), was cloned into the Tth111I site of plasmid pUEF730 which had been rendered blunt. This construct was electroporated into H. pylori ATCC 43504 and Leung. Electrotransformants were obtained on brucella agar plates containing 4% sheep blood, kanamycin (50 μg/ml), and l-glutamine supplementation at 50 μg/ml or 1 mg/ml. Initially, growth was more substantial at the higher glutamine concentration but was not as robust as that of the wild type. A putative glutamine synthetase mutant was designated HP-GS1. With subsequent passage, the growth of HP-GS1 became hardy on brucella agar containing 4% sheep blood and kanamycin (50 μg/ml) with and without 1 mg of l-glutamine per ml. These results were more consistent with the presence of a cointegrate in H. pylori rather than the presence of a double-crossover mutation in glnA. PCR was used to evaluate the construct.
Nucleotide sequence accession numbers.
The sequences of the 1,443- and 523-bp ORFs have been deposited in GenBank under accession no. AF053357 and AF053715, respectively.
RESULTS
Urease-enhancing factor assay.
As a part of a strategy to investigate genes that contribute to the synthesis of a catalytically active urease, we developed a phenotypic screen to isolate such genes. E. coli SE5000 transformed with plasmid pHP808 containing the entire H. pylori urease gene cluster was cotransformed with a pBluescript plasmid library of the H. pylori ATCC 43504 genome. Camr Ampr colonies were plated on urea segregation agar, a medium designed to identify urease-positive colonies [E. coli SE5000(pHP808) does not produce sufficient urease on this medium to give a positive reaction]. One strongly positive clone has been identified previously as encoding NixA, a high-affinity nickel transporter (25). An additional clone was identified that turned positive for urease after 3 days of incubation. The urease activity was qualitatively positive but was not quantifiable by a spectrophotometric assay.
Nucleotide sequencing and plasmid pUEF728.
We recovered a 9.4-kb plasmid, pUEF728, in addition to pHP808 from the weakly urease-positive clone and subjected the plasmid to restriction mapping. The H. pylori chromosomal DNA insert, released by digestion with EcoRI, was estimated to be 6.5 kb. By using universal pBluescript T3 and T7 primers and additional primers based on the acquired sequence, the nucleotide sequence of approximately 1.8 kb was determined for each end of the insert. Two significant ORFs of 523 and 1,284 bp were identified, one at each end of the insert. The first ORF predicted a truncated polypeptide of 174 amino acids and a molecular weight of 20,487. The most closely related polypeptide, identified by WORDSEARCH in GenBank, was that of a GTP-binding protein of E. coli (1). The second ORF, based on very strong amino acid sequence identity to other gene bank entries, appeared to represent the H. pylori glnA gene encoding glutamine synthetase. After inspection of amino acid sequence alignments, the 3′ end of the gene did not, however, appear to be complete and was found to be fused to vector sequences.
To obtain the remaining portion of glnA, a 1,418-kb HphI-PvuII fragment of glnA (bp 527 to 1945) (Fig. 1) was isolated from pUEF728 and used as a probe to identify cosmid clones also containing these sequences. Of 2,304 pHC79-based cosmid clones screened, two hybridized strongly with the probe. One of these, designated pUEF16-7A, was isolated. A Southern blot of this cosmid DNA, digested with HindIII, was probed independently with two probes: a 1-kb AflII-Tth111I fragment (upstream sequences and the 5′ end of glnA) and a 0.4-kb Tth111I-EagI fragment (3′ end of glnA). Both probes hybridized to a 3.8-kb HindIII. This fragment was subcloned into pBluescript and subjected to nucleotide sequencing with a primer designed from sequences near the 3′ end of the truncated glnA on the previous clone. Overlapping sequences representing the end of glnA were identified and used to compile the complete glnA ORF (Fig. 1).
FIG. 1.
Nucleotide sequence of H. pylori glnA. The nucleotide sequence for glnA and the predicted amino acid sequence are shown. A Shine-Dalgarno (S.D.) sequence is underlined. ∗, stop codon.
H. pylori glnA.
The complete 1,443-bp glnA gene predicts a polypeptide of 481 amino acid residues with a molecular weight of 54,317. The G+C content of the ORF is 42.4%, slightly higher than the 39% for total genomic DNA (12, 34). An apparent Shine-Dalgarno site of AGG is located 7 bp upstream of the translational start site. No ς70 or ς54 promoter sequence is readily identifiable, nor is a rho-independent transcriptional terminator located downstream of the 3′ end of the gene. Analysis of sequences upstream (573 bp) and downstream (451 bp) of glnA did not reveal any homologs related to ammonia assimilation or nitrogen regulation. However, the sequence of the entire insert of pUEF728 was not determined. Examination of this region in the complete genome sequence of H. pylori ATCC 26695 (34) confirmed the lack of additional nitrogen regulatory homologs immediately upstream and downstream of glnA. A short ORF upstream of glnA predicts a 38-amino-acid peptide with no homologs. An ORF downstream of glnA predicts a polypeptide of 78 kDa with two homologs elsewhere in the H. pylori genome, also with no known functions. The upstream and downstream ORFs are both transcribed in the direction opposite to that of glnA, suggesting that they are not part of an operon.
H. pylori GlnA.
The predicted GlnA amino acid sequence was used to search the SwissProt data base for similar proteins by using WORDSEARCH. The top 10 matches were all bacterial glutamine synthetases from (most to least similar) Salmonella typhimurium, Azotobacter vinelandii, Methylococcus capsulatus, E. coli, Azospirillum brasilense, Synechocystis sp., Proteus vulgaris, Fremyella diplosiphon, Neisseria gonorrhoeae, and Vibrio alginolyticus. When aligned, the most similar homolog, S. typhimurium GlnA, was found to have 48.3% amino acid sequence identity and 65.3% amino acid sequence similarity (identical residues and conservative replacements) (Fig. 2). The least similar of the top 10 homologs, V. alginolyticus GlnA, has 38.7% identity and 57.4% similarity.
FIG. 2.
Alignment of H. pylori GlnA and S. typhimurium GlnA. The predicted amino acid sequences of the glutamine synthetases of H. pylori (Hp) and S. typhimurium (St) were aligned. Identical residues are denoted by a vertical line; conservative replacements are shown by a colon (:). An ATP-binding motif and the adenylation site (Tyr residue within the motif) are identified by lines drawn above the sequences.
The ATP-binding motif GDNGSG (residues 272 to 277) exactly matched and aligned with the sequence in 8 of the 10 homologs and was similar to sequences in the other two matches (five of six for Synechocystis sp. and four of six for M. capsulatus).
Lack of adenylation site.
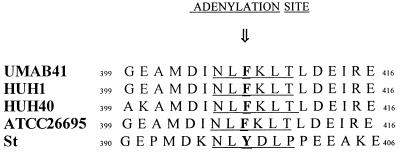
The adenylation site found in all of the 10 most similar homologs (consensus sequence, NLYDLP) (33) is replaced in H. pylori by NLFKLT (residues 405 to 410). Since the Tyr407 residue is the well-conserved target of adenylation (33) and since the H. pylori glutamine synthetase apparently lacks that residue, we concluded that no adenylation occurs for this enzyme within this motif. Another tyrosine-containing sequence, NPYLAF (residues 376 to 381), is found upstream of the consensus adenylation site and is well conserved among the homologs but has not been demonstrated as an adenylation site.
Since the change from Tyr407Phe could have resulted from a single-base-pair change (T1219A), we determined the sequence of this region of glnA from other strains. Using primers directed against sequences centered 31 bp upstream of the start codon of glnA and 21 bp downstream of glnA, we amplified a 1,522-bp sequence from two additional strains (HUH1 and HUH40) and determined the nucleotide sequence. The Tyr407Phe change is conserved in these two additional strains (Fig. 3) and also in strain 26695, for which the complete genome sequence was determined (34). This suggests that the activity of H. pylori glutamine synthetase is indeed not modulated by adenylation and thus is not regulated by posttranslational modification.
FIG. 3.
Adenylation site of H. pylori GlnA. The predicted amino acid sequences of the conserved adenylation site of GlnA from four H. pylori strains (three determined in this study and ATCC 26695 [34]) and S. typhimurium (accession no. P06201) (St) are aligned. The Tyr residue in S. typhimurium is the site of adenylation in this and most species but is replaced with Phe in H. pylori.
Protein labeling in maxicells.
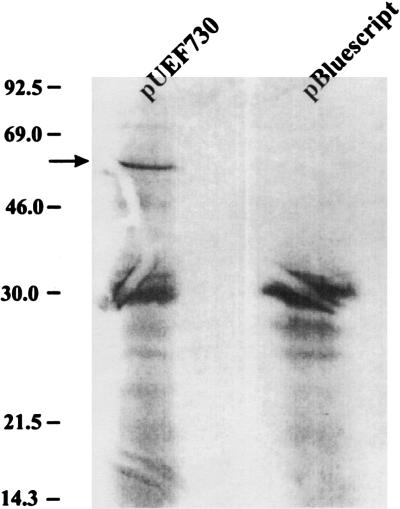
To demonstrate whether a GlnA product of the predicted size was produced by plasmid pUEF730, plasmid-encoded gene products were labeled with [35S]methionine in maxicells. Proteins encoded by this plasmid were analyzed on autoradiographs of an SDS–12% polyacrylamide gel; proteins synthesized by the pBluescript vector were included as a control (Fig. 4). In addition to vector-encoded polypeptides, pUEF730 encoded a polypeptide estimated to be 56 kDa (Fig. 4, arrow), consistent with the molecular weight of 54,317 predicted by the nucleotide sequence.
FIG. 4.
Autoradiograph of plasmid-encoded polypeptides. E. coli SE5000(pBluescript) and SE5000(pUEF730) were UV-irradiated and plasmid-encoded polypeptides were labeled with [35S]methionine and electrophoresed on an SDS–15% polyacrylamide gel, which was dried and autoradiographed. The mobilities of molecular mass markers are shown in kilodaltons at the left. The arrow points to a polypeptide synthesized by pUEF730 but not by pBluescript.
Complementation of an E. coli glnA mutant.
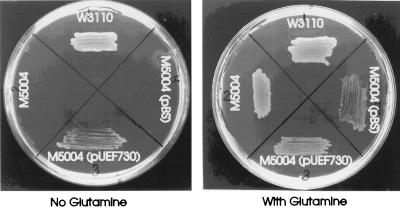
To determine whether cloned H. pylori glnA can complement a glnA mutation in E. coli, plasmid pUEF730 carrying glnA on a 1,522-bp SpeI-KpnI fragment cloned into the SpeI-KpnI site of pBluescript was constructed. Plasmid pUEF730 or vector pBluescript SK+ was transformed into E. coli W3110 and its glnA-deficient mutant M5004. These strains were plated onto minimal salts agar supplemented with tryptophan (20 μg/ml) and l-glutamine (0, 50, or 1,000 μg/ml). All strains grew within 48 h in the presence of glutamine (Fig. 5). E. coli M5004(pBluescript), however, was unable to grow in the absence of glutamine, whereas E. coli M5004(pUEF730) and W3110 grew as individual colonies after 48 h. Thus, expression of cloned H. pylori glnA is able to complement a glnA mutation of E. coli. The 48 h needed for growth of the complemented mutant is consistent with the observation that the cloned expression of H. pylori GlnA activity is weak compared to that of native E. coli.
FIG. 5.
Complementation of an E. coli glnA mutant with cloned H. pylori glnA. A glutamine synthetase mutant of E. coli, strain M5004, transformed or not with plasmid pUEF730 (contains H. pylori glnA) or pBluescript vector, and the parent strain, W3110 (produces glutamine synthetase), were plated onto minimal medium supplemented or not with glutamine (50 μg/ml). The plates were photographed after 48 h of growth at 37°C.
Glutamine synthetase activity.
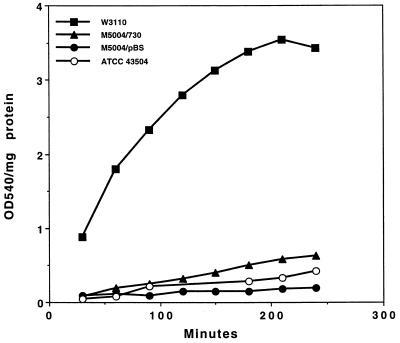
To determine whether the catalytic activity of glutamine synthetase could be detected in H. pylori and E. coli transformed with cloned H. pylori glnA, a spectrophotometric assay for glutamine synthetase was used. Use of a standard spectrophotometric assay (4) that measures total glutamine synthetase activity without regard to the adenylation status of the enzyme revealed significant activity in H. pylori and E. coli M5004(pUEF730) (Fig. 6). E. coli M5004, the glnA-deficient mutant, transformed with pUEF730 encoding H. pylori GlnA showed significantly higher rates of activity at 0.0029 OD540/min/mg of protein (P < 0.0001) than did the same strain transformed with pBluescript, which had no significant activity (0.0001 OD540/min/mg of protein). The glutamine synthetase activity of E. coli M5004(pUEF730), however, was significantly lower (P < 0.0001) than that of E. coli W3110 (0.0241 OD540/min/mg of protein), the parent strain containing the wild-type E. coli glnA gene. The weaker GlnA activity of the complemented E. coli mutant [E. coli M5004(pUEF730)] was consistent with the slower growth of this strain than of the GlnA-sufficient parent strain, E. coli W3110.
FIG. 6.
Glutamine synthetase activity of E. coli transformed with plasmids. Glutamine synthetase activity was measured in E. coli W3110 (wild type for glutamine synthetase activity) and E. coli M5004 (glutamine synthetase mutant) transformed with either vector pBluescript (pBS) or pUEF730 (730) carrying H. pylori glnA. For reference, the activity in H. pylori ATCC 43504 is also included. Values are the averages of five independent determinations. For determinations made with the E. coli strain, each curve is significantly different from the other two (P < 0.001).
Mutation of glutamine synthetase may be lethal to H. pylori.
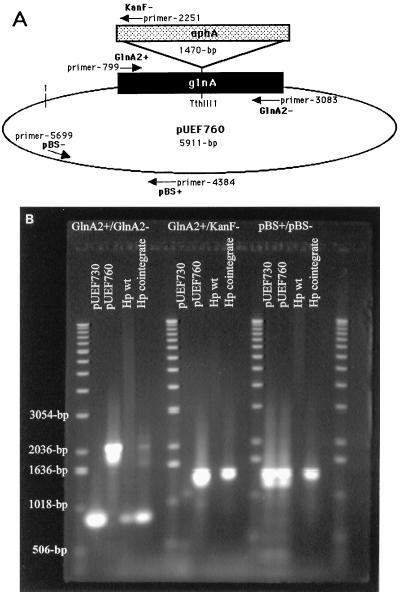
To determine the physiological consequences of mutation of glnA, we attempted to construct a GlnA-deficient mutant of H. pylori (see Materials and Methods) (Fig. 7A); four strains (UMAB41, ATCC 43504, Leung, and X47ZAL) were electroporated on approximately 30 different occasions with pUEF760. For UMAB41, no Kanr colonies were ever isolated. For the other strains, pinpoint Kanr colonies were observed but could not be successfully passaged. For example, one isolate, a putative glutamine synthetase mutant designated HP-GS1, was observed on blood agar containing l-glutamine (50 μg/ml). This mutant could not be successfully propagated, and because of the inability to obtain a sufficient number of cells, the presence of a double-crossover mutation could not be verified.
FIG. 7.
Constructs used in allelic exchange mutagenesis. (A) The H. pylori glnA gene, cloned into pBluescript, was insertionally inactivated by cloning a kanamycin resistance cassette into the Tth111I site (see Materials and Methods). The construct was electroporated into H. pylori strains and plated on blood agar containing kanamycin (50 μg/ml) supplemented or not with l-glutamine (50 μg/ml or 1 mg/ml). The location of primers used for the PCR analysis are shown along with their coordinates (given in base pairs with reference to the “1” on pBluescript vector). (B) PCR analysis of the parental strain and putative glnA cointegrate. Purified plasmid or chromosomal DNA was used as the template along with primer pairs (top) identified in panel A for PCR amplification of fragments specific for glnA, glnA::aphA, or pBluescript sequences. Plasmid pUEF730 contains intact glnA; plasmid pUEF760 carries insertionally inactivated glnA. Hp wt is wild-type strain ATCC 43504, and Hp cointegrate is the putative cointegrate. Sizes of relevant standards are given in base pairs.
In subsequent experiments, some of the Kanr isolates, obtained on blood agar supplemented with l-glutamine, were successfully passaged but regained the vigorous growth properties of the wild-type strains. These colonies retained glutamine synthetase activity. PCR analysis (Fig. 7B) of one of these strains derived from ATCC 43504 demonstrated that this isolate represented a cointegrate of the plasmid into the chromosome. When primers specific for the glnA ORF (GlnA2+ and GlnA2−) were used for PCR amplification, a 0.83-kb fragment was amplified from the parent strain; in the cointegrate, both a 0.83-kb fragment (wild-type size) and a 2.17-kb fragment (representing glnA::aphA) were amplified, indicating that both the wild-type gene and the insertionally inactivated gene were present. When one glnA (GlnA2+) and one aphA (KanF−) primer were used, a product was not amplified from the parent strain but the expected 1.48-kb product was amplified from the cointegrate. When primers specific for the pBluescript vector (pBS+ and pBS−) were used, vector sequences (1.34-kb) were amplified from the cointegrate and plasmid constructs but not from the parent strain. These results are all consistent with the cointegration of pUEF760 (the insertional inactivation construct) into the chromosome of H. pylori.
DISCUSSION
We have described how the glutamine synthetase gene of H. pylori has been cloned, sequenced, and expressed in E. coli. The amino acid sequence of the polypeptide predicted from the nucleotide sequence closely aligned with the amino acid sequences of previously described glutamine synthetases from other bacterial species (nearly 50% amino acid sequence identity to GlnA of S. typhimurium). The cloned H. pylori gene complemented a glnA mutation in E. coli, produced measurable glutamine synthetase activity, and encoded a polypeptide of the expected size (∼54 kDa) when expressed in E. coli maxicells.
The ATP-binding motif (33), required for activity, is well conserved in the H. pylori enzyme. However, the residue (Tyr407) within the adenylation motif, necessary for regulation by posttranslational modification, is absent in the H. pylori GlnA in all strains tested, as well as in strain ATCC 26695. Since adenylation is the key mechanism used to turn off enzymatic activity following translation of the enzyme, the glutamine synthetase may be catalytically active under all environmental conditions. This may be necessary because of the constant release of ammonia due to urease-mediated urea hydrolysis. The lack of such control is an exception rather than the rule among bacterial glutamine synthetases, since the enzymes from most species have rigorously conserved the Tyr-containing adenylation site (conserved in all 10 of the most closely related GlnA homologs). When the consensus adenylation site itself was used to search protein databases, 31 alignments from bacterial species were identified. Of these, only three lack the conserved Tyr residue, replacing it with Phe: Clostridium acetobutylicum (36), Lactobacillus delbrueckii (20), and Methanococcus voltae (accession no. P21154). Biochemical evidence is also provided, without the nucleotide sequence, that the GlnA of the cyanobacterium Anabaena does not undergo adenylation (8).
In most procaryotes, the ammonia assimilation pathway mediated by glutamine synthetase is regulated at every level including transcriptional regulation (ς70 and ς54 promoters, positive transcriptional activator NRI or NtrC, and catabolite activator protein-binding site), translational regulation (strength of Shine-Dalgarno sequence), posttranslational regulation (adenylation of the conserved Tyr residue), and allosteric regulation by the binding of certain nitrogenous compounds (29). For H. pylori, which has a relatively modest-sized genome of 1.7 Mb (34), one could speculate that this level of complexity of regulation may be absent or reduced. Evidence for this is the absence of NtrC or NtrB homologs encoded by sequences downstream of glnA, their usual location in procaryotes (21).
H. pylori appears to lack proteins required for transcriptional and posttranslational regulation of glutamine synthetase. Consistent with the observation that none of the H. pylori strains tested possessed the conserved Tyr407 residue, which is the target of adenylation in glutamine synthetase, homologs of proteins which are required to carry out this adenylation in other gram-negative enteric bacterial species do not appear to be present in the H. pylori genome. In E. coli, the PII protein (GlnB), uridylyltransferase/uridyl-removing enzyme (GlnD), and adenylyltransferase (GlnE) are all required for posttranslational modification of glutamine synthetase (30). Since E. coli GlnA is highly homologous (47% amino acid sequence identity) to H. pylori GlnA, E. coli GlnB, GlnD, and GlnE sequences were used to search for homologs in H. pylori. Homologs of these proteins were not identified in the recently published genome of H. pylori (no amino acid sequence identity of more than 12%) (34).
While the H. pylori genome does contain possible homologs of the transcriptional regulators NtrB (GlnL) (25% amino acid sequence identity to the E. coli protein) and NtrC (GlnG) (34% amino acid sequence identity to the E. coli protein), it is unclear whether glnA is under the transcriptional control of these proteins. First, both homologs are located distally to GlnA in the published H. pylori genome (34). Second, in other studied systems (30), a deuridylylated PII protein is required for the NtrC protein to dephosphorylate NtrB and halt transcription of glutamine synthetase. Similarly, uridylylation of PII is required for covalent modification of NtrB, leading to transcription of glnA (30). Since these modifying enzymes are absent, it is likely that transcription of glnA is not controlled in this manner.
Glutamine synthetase is intimately linked to urease, the most thoroughly investigated virulence factor of H. pylori (26). Two important roles have been proposed for the urease of H. pylori. First, hydrolysis of urea present in the host liberates ammonia, which can neutralize gastric acid, allowing the bacterium to survive its initial plunge into the gastric juice while it traverses the mucus layer to the safety of the epithelium surface. Second, ammonia that is not externalized by diffusion can serve as a preferred nitrogen source for incorporation into amino acids. This is accomplished by the action of glutamine synthetase on ammonia, glutamate, and ATP to form glutamine. Mutants of H. pylori deficient in urease cannot colonize the gastric mucosa of any animal model thus far tested (5, 6). Apparently, this inability to colonize is not solely due to acid protection mediated by urease. Gnotobiotic piglets, treated with omeprazole, a proton pump inhibitor, to raise the gastric pH to neutrality, remain resistant to colonization with a urease-negative mutant of H. pylori (6). This suggests that an additional role of urease, perhaps that of ammonia assimilation, is critical for successful colonization of the gastric mucosa. Thus, glutamine synthetase may play an unusually critical role in nitrogen metabolism in H. pylori and may be required to process the ammonia liberated by urea hydrolysis.
That glutamine synthetase is required for H. pylori viability and colonization is supported by our inability to introduce and document a double-crossover allelic exchange mutation in which a kanamycin resistance cassette was used to insertionally inactivate glnA. Mutations of nonessential H. pylori genes are carried out routinely in this manner and result in direct isolation of double-crossover mutants (3, 16). The formation of cointegrates, in which the electroporated plasmids carrying the insertionally inactivated gene is integrated into the chromosome, is in our experience a rare event unless the mutation is lethal. In this study, we observed and documented such a cointegrate formation, suggesting that glnA is an essential gene.
The isolation of glnA by the urease-enhancing assay appears to have been fortuitous. It is unlikely that synthesis of active glutamine synthetase contributed to the isolation of the clone. Indeed, upon sequence analysis and alignment with other GlnA sequences, we determined that the 3′ end of the gene was not present and thus was probably not producing active enzyme. Furthermore, if the enzyme was active, it would have reduced, not increased, the ammonia concentration and thus would not have raised the local pH, allowing the detection of an alkali-induced color change of phenol red in the urea segregation agar. This pH-elevating effect was most probably mediated by another gene product encoded on the clone, less than half of which was sequenced.
Given what we know, it is logical that GlnA remains unregulated following translation of the enzyme. H. pylori appears to be restricted to the gastric mucosa of humans. Because there are no alternate niches that would include radical changes in temperature, pH, or nitrogen availability, there would be no need for sophisticated levels of regulation for the enzyme. Also because urease does not appear to be regulated, there would be a constant supply of ammonia as long as there is a supply of urea. This pathway of nitrogen assimilation appears to be required for viability, and thus glutamine synthetase appears to represent an unusually critical enzyme in H. pylori.
ACKNOWLEDGMENT
This work was supported by Public Health Service grant AI23328 from the National Institutes of Health.
REFERENCES
- 1.Ahnn J, March P E, Takiff H E, Inouye M. A GTP-binding protein of Escherichia coli has homology to yeast RAS proteins. Proc Natl Acad Sci USA. 1986;83:8849–8853. doi: 10.1073/pnas.83.23.8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley Interscience; 1987. [Google Scholar]
- 3.Bauerfeind P, Garner R M, Mobley H L T. Allelic exchange mutagenesis of nixA in Helicobacter pylori results in reduced nickel transport and urease activity. Infect Immun. 1996;64:2877–2880. doi: 10.1128/iai.64.7.2877-2880.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender R A, Janssen K A, Resnick A D, Blumenberg M, Foor F, Magasanik B. Biochemical parameters of glutamine synthetase from Klebsiella aerogenes. J Bacteriol. 1977;129:1001–1009. doi: 10.1128/jb.129.2.1001-1009.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eaton K A, Brooks C L, Morgan D R, Krakowka S. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect Immun. 1991;59:2470–2475. doi: 10.1128/iai.59.7.2470-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eaton K A, Krakowka S. Effect of gastric pH on urease-dependent colonization of gnotobiotic piglets by Helicobacter pylori. Infect Immun. 1994;62:3604–3607. doi: 10.1128/iai.62.9.3604-3607.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrero R L, Cussac V, Courcoux P, Labigne A. Construction of isogenic urease-negative mutants of Helicobacter pylori by allelic exchange. J Bacteriol. 1992;174:4212–4217. doi: 10.1128/jb.174.13.4212-4217.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher R, Tuli R, Haselkorn R. A cloned cyanobacterial gene for glutamine synthetase functions in Escherichia coli, but the enzyme is not adenylated. Proc Natl Acad Sci USA. 1981;78:3393–3397. doi: 10.1073/pnas.78.6.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foxall P F, Hu L-T, Mobley H L T. Use of polymerase chain reaction-amplified Helicobacter pylori urease structural genes for differentiation of isolates. J Clin Microbiol. 1992;30:739–741. doi: 10.1128/jcm.30.3.739-741.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gherardini F C, Hobbs M M, Stamm L V, Bassford P J., Jr Complementation of an Escherichia coli proC mutation by a gene cloned from Treponema pallidum. J Bacteriol. 1990;172:2996–3002. doi: 10.1128/jb.172.6.2996-3002.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert J V, Ramakrishna J, Sunderman F W, Wright A, Plaut A G. Protein Hpn: cloning and characterization of a histidine-rich metal-binding polypeptide in Helicobacter pylori. Infect Immun. 1995;63:2682–2688. doi: 10.1128/iai.63.7.2682-2688.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodwin C S, Armstrong J A, Chilvers T, Peters M, Collins M D, Sly L, McConnell W, Harper W E S. Transfer of Campylobacter pylori and Campylobacter mustelae to Helicobacter gen. nov. as Helicobacter pylori comb. nov. and Helicobacter mustelae comb. nov., respectively. Int J Syst Bacteriol. 1991;41:31–38. [Google Scholar]
- 13.Hamilton-Miller J M T, Gargan R A. Rapid screening for urease inhibitors. Invest Urol. 1979;16:327–328. [PubMed] [Google Scholar]
- 14.Hay B, Short J M. ExAssist helper phage and SOLR cells for Lambda ZAP II excisions. Strategies Mol Biol. 1992;5:16–18. [Google Scholar]
- 15.Hazell S L, Borody T J, Lee A. Campylobacter pyloridis gastritis. I. Detection of urease as a marker of bacterial colonization and gastritis. Am J Gastroenterol. 1987;82:292–296. [PubMed] [Google Scholar]
- 16.Hendricks J K, Mobley H L T. Helicobacter pylori ABC transporter: effect of allelic exchange mutagenesis on urease activity. J Bacteriol. 1997;179:5892–5902. doi: 10.1128/jb.179.18.5892-5902.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hohn B, Collins J. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980;11:291–298. doi: 10.1016/0378-1119(80)90069-4. [DOI] [PubMed] [Google Scholar]
- 18.Hu L-T, Foxall P F, Russell R, Mobley H L T. Purification of recombinant Helicobacter pylori urease apoenzyme encoded by ureA and ureB. Infect Immun. 1992;60:2657–2666. doi: 10.1128/iai.60.7.2657-2666.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu L-T, Mobley H L T. Purification and N-terminal analysis of urease from Helicobacter pylori. Infect Immun. 1990;58:992–998. doi: 10.1128/iai.58.4.992-998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishino Y, Morgenthaler P, Hottinger H, Soll D. Organization and nucleotide sequence of the glutamine synthetase (glnA) gene from Lactobacillus delbrueckii subsp. bulgaricus. Appl Environ Microbiol. 1992;58:3165–3169. doi: 10.1128/aem.58.9.3165-3169.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magasanik B. Regulation of nitrogen utilization. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1344–1356. [Google Scholar]
- 22.Marshall B J, Warren J R, Francis G J, Langton S R, Goodwin C S, Blincow E D. Rapid urease test in the management of Campylobacter pyloridis-associated gastritis. Am J Gastroenterol. 1987;82:200–210. [PubMed] [Google Scholar]
- 23.Mayer E P, Smith O H, Fredricks W W, McKinney M A. The isolation and characterization of glutamine-requiring strains of Escherichia coli K 12. Mol Gen Genet. 1975;137:131–142. doi: 10.1007/BF00341679. [DOI] [PubMed] [Google Scholar]
- 24.McNulty C A M, Wise R. Rapid diagnosis of campylobacter-associated gastritis. Lancet. 1985;i:1443. doi: 10.1016/s0140-6736(85)91865-3. [DOI] [PubMed] [Google Scholar]
- 25.Mobley H, Garner R, Bauerfeind P. Helicobacter pylori nickel-transport gene nixA: synthesis of catalytically active urease in Escherichia coli independent of growth conditions. Mol Microbiol. 1995;16:97–109. doi: 10.1111/j.1365-2958.1995.tb02395.x. [DOI] [PubMed] [Google Scholar]
- 26.Mobley H L T. Structure and function of H. pylori urease. In: Ernst P B, Michetti P, Smith P D, editors. The immunobiology of H. pylori: from pathogenesis to prevention. Philadelphia, Pa: Lippincott-Raven Publishers; 1997. pp. 59–73. [Google Scholar]
- 27.Mobley H L T, Cortesia M J, Rosenthal L E, Jones B D. Characterization of urease from Campylobacter pylori. J Clin Microbiol. 1988;26:831–836. doi: 10.1128/jcm.26.5.831-836.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mobley H L T, Jones B D, Jerse A E. Cloning of urease gene sequences from Providencia stuartii. Infect Immun. 1986;54:161–169. doi: 10.1128/iai.54.1.161-169.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reitzer L J. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, l-alanine, and d-alanine. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 391–407. [Google Scholar]
- 30.Reitzer L J, Magasanik B. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, l-alanine, and d-alanine. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 302–320. [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shapiro B M, Stadtman E R. 5′-Adenylyl-O-tyrosine: the novel phosphodiester residue of adenylated glutamine synthetase from Escherichia coli. J Biol Chem. 1968;243:3769–3771. [PubMed] [Google Scholar]
- 34.Tomb J F, White O, Kerlabage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weidman J M, Fujii C, Bowman C, Watthey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 35.Trieu-Cuot P, Courvalin P. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′5′-aminoglycoside phosphotransferase type III. Gene. 1993;23:331–341. doi: 10.1016/0378-1119(83)90022-7. [DOI] [PubMed] [Google Scholar]
- 36.Usdin K P, Zappe H, Jones D T, Woods D R. Cloning, expression, and purification of glutamine synthetase from Clostridium acetobutylicum. Appl Environ Microbiol. 1986;52:413–419. doi: 10.1128/aem.52.3.413-419.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valentine J L, Arthur R R, Mobley H L T, Dick J D. Detection of Helicobacter pylori by using the polymerase chain reaction. J Clin Microbiol. 1991;29:689–695. doi: 10.1128/jcm.29.4.689-695.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Taylor D E. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene. 1990;94:23–28. doi: 10.1016/0378-1119(90)90463-2. [DOI] [PubMed] [Google Scholar]