Abstract
The taxonomic relationship of Rhinosporidium seeberi with other organisms remained controversial for over a century. Recently, molecular studies have shown R. seeberi to be a protistal microbe in the newly described class Mesomycetozoea at the animal-fungal boundary. Phylogenetic analyses of R. seeberi using 18S small-subunit (SSU) rRNA genes from several hosts suggested Rhinosporidium as a monotypic genus. To test this hypothesis, the internal transcribed spacer 1 (ITS1), 5.8S, and ITS2 from eight humans, two swans, and a dog with rhinosporidiosis were sequenced. The ITS regions were amplified by PCR using a primer designed from a unique region of R. seeberi's 18S SSU rRNA genes in combination with the ITS4 universal primer. In addition, the universal ITS4 and ITS5 primers were also used. R. seeberi's ITS sequences showed differences in the numbers of nucleotides among strains. For instance, the eight human ITS sequences were uniformly similar with only a few mismatches and ∼1,060 bp long. In contrast, sequences from one of the swans and the dog were 1,356 bp and ∼1,147 bp long, respectively. Clustal analysis of all of the ITS sequences showed multiple 50- to 60-bp gaps and several mismatches among them. Parsimony analysis placed the Rhinosporidium ITS sequences in three well-supported sister groups according to the hosts' identities. This analysis strongly suggests that the genus Rhinosporidium may possess multiple host-specific strains. No correlation was found between this finding and the phenotypic features of R. seeberi in the studied samples.
Rhinosporidiosis is a cutaneous and/or subcutaneous chronic disease of human and other animals caused by Rhinosporidium seeberi (2, 16-18). This granulomatous disease is characterized by the development of polyps primarily affecting the mucous membranes of the nostrils and the ocular conjunctivae of the infected hosts. Diagnosis is essentially based on the histological detection in tissues of R. seeberi's pathognomonic endosporulating sporangia in various stages of development. Rhinosporidiosis in not a life-threatening disease, and its treatment usually is limited to the surgical removal of the polyps (1, 4, 11).
For the last 100 years, the taxonomic affinities of R. seeberi remained obscure, principally because this unique pathogen cannot be cultured. Thus, most of its epidemiological and taxonomical characteristics were not well understood. In 1999, a team from the United States and Sri Lanka (10) using molecular tools found that R. seeberi was phylogenetically related to a novel group of protistan microbes at the divergence between animals and fungi. This finding was soon corroborated by others (8). These microbes are presently classified in the protistal class Mesomycetozoeae. R. seeberi shares several characteristics with the other members of this class; all are aquatic microbes with spherical structures containing endospores. Most of them are unculturable and were, at one point, classified as members of the fungal or protistan kingdoms.
The studies of Herr et al. (10) and later, those of Fredericks et al. (8), using the 18S SSU rRNA gene sequences of R. seeberi, showed that these regions were almost identical among mammalian hosts with rhinosporidiosis. Based on these data and those of other investigators, Rhinosporidium was considered likely to be a monotypic genus (13). Using phylogenetic analysis, we have studied the complete internal transcribed spacer 1(ITS1), 5.8S, and ITS2 sequences of eight humans, two swans, and a dog with rhinosporidiosis and have found that there are at least three well-supported groups represented by each species used in this study. This study raises the possibility that the genus Rhinosporidium may possess multiple host-specific strains.
MATERIALS AND METHODS
R. seeberi's genomic DNA isolation from fresh samples.
Freshly biopsied tissue containing R. seeberi's sporangia and endospores from two Florida swans and a dog from Georgia with rhinosporidiosis were used in this study. The tissues were aseptically collected, frozen without fixatives, and transported to our laboratory. Portions of each tissue (∼100 mg) were placed in a mortar and ground under liquid nitrogen and their genomic DNA extracted according to the protocol of Herr et al. (10). Briefly, the samples were placed in Eppendorf tubes and treated with sodium dodecyl sulfate and proteinase K. R. seeberi's genomic DNA separated from the mix after the addition of phenol and chloroform. Other portions of the tissues were used to perform direct microscopic examination in 10% KOH and hematoxylin and eosin stain to confirm the presence of R. seeberi sporangia.
Genomic DNA extraction from human paraffin-embedded tissues.
Paraffin-embedded tissues from four humans with rhinosporidiosis from India were sectioned, and 20-mg portions of the tissues were placed in Eppendorf tubes. One milliliter of xylene was added to the tubes, which were then vortexed vigorously. The mixture was incubated for 15 min at room temperature and was subsequently centrifuged at 12,000 rpm for 5 min. The supernatant was removed, and 1.0 ml of xylene was added again to the pellet. The samples were vortexed vigorously and centrifuged as before. The pellet was washed twice using 1.0 ml of absolute ethanol and centrifuged at 12,000 rpm for 5 min at room temperature each time. The supernatant was removed and the tissue dried for 30 min at room temperature. To isolate genomic DNA from the samples, the QIAmp DNA Mini Kit (QIAGEN Inc., Valencia, CA) was used. Briefly, 180 μl of ATL buffer and 20 μl of proteinase K (20 mg/ml) was added to the tubes containing the tissue samples. The tubes were vortexed vigorously for 20 s and then incubated at 56°C overnight. Two hundred microliters of AL buffer (included in the kit) were added. The samples were incubated for 15 min at 70°C, followed by the addition of 200 μl of 100% ethanol, and then centrifuged. The supernatant was loaded onto a QIAmp spin column, and the genomic DNA was extracted in accordance with the instructions of the manufacturer. In addition, genomic DNA samples from four human cases of rhinosporidiosis (Sri Lanka) obtained from previous studies (10, 13) were also used.
PCR amplification of R. seeberi's ITS1, 5.8S, and ITS2 from genomic DNA samples and sequencing.
The ITS1, 5.8S, and ITS2 sequences of R. seeberi were amplified by PCR using the specific primer RhinA2F (5′-TAGTTGCGTGATTTTTCGAA-3′) in combinations with the ITS4 universal primer (9). These sets of primers were designed to amplify approximately the last 450 bp of 18S SSU rRNA genes, the complete ITS1, 5.8S, and ITS2, and the first 60 bp of 28S large-subunit (LSU) rRNA genes. PCR consisted of 40 cycles of amplification on a Perkin-Elmer GeneAmp 9700 thermal cycler. After an initial activation of Taq Gold (Applied Biosystems) at 95°C for 10 min, each cycle consisted of 1 min of melting at 94°C, 2 min of annealing at 50°C for 18S SSU rRNA genes or 60°C for ITS, and 3 min of extension at 72°C. The last cycle was followed by an extension step at 72°C for 7 min. Amplification products were detected by electrophoresis on 0.8% agarose gels stained with ethidium bromide and visualized by using the Bio-Rad Gel Doc 1000 apparatus with the software Multi-Analyst version 1.0.2 (Bio-Rad, California). Since genomic DNA extraction from paraffin-embedded tissues often results in DNA fragmentation, the ITS sequences from these samples were amplified with the ITS5 and ITS4 universal primers, which amplified smaller fragments than those obtained using the RhinA2F and ITS4 primers.
The PCR amplicons were then cloned into pCR 2.1-TOPO plasmids (Invitrogen, Carlsbad, California) and purified, and at least 10 clones of each sample were then sequenced in both directions by using BigDye Terminator chemistry in an ABI Prism 310 genetic analyzer apparatus (Perkin-Elmer, Foster City, California).
Phylogenetic analysis.
The ITS sequences of R. seeberi from eight humans, two swans, and a dog and from Sphaerothecum destruens (accession number AY388645), were aligned using Clustal analysis software (Perkin-Elmer/Applied Biosystems) and then visually inspected. Phylogenetic analysis using the ITS sequences from the above samples was by distance methods (neighbor-joining with Kimura two-parameter multiple-hit correction) and parsimony. Support for internal branching was assessed by using 1,000 bootstrapped data sets. Parsimony analyses were conducted with the computer software programs PAUP (Phylogenetic Analysis Using Parsimony, version 3.1; D. L. Swofford, Illinois Natural History Survey, Champaign, Illinois), and MEGA version 2.1 (Sudhir Kumar, Koichiro Tamura, Ingrid B. Jakobsen, and Masatoshi Nei; MEGA2: molecular evolutionary genetics analysis software, Arizona State University, Tempe, Arizona). S. destruens' ITS sequences were used as the out-group.
Nucleotide sequence accession numbers.
The edited sequences of R. seeberi ITS1, 5.8S, and ITS 2 were deposited in GenBank under the following accession numbers (geographic origin and host in parentheses): AY610938, AY610939, AY610940, and AY610941 (Indian human); AY372367, AY378083, AY378082, and AY378081 (Sri Lankan human); AF399715 and AY486143 (Florida swans); and AY372365 (Georgia dog).
RESULTS
Light microscopy analysis.
Histological stained sections and wet prep samples showed identical phenotypic features among the human and animal strains of R. seeberi used in this study (data not shown). All examined samples showed spherical mature phenotypes with endospores and several immature forms. No obvious differences in the sizes, forms, or distributions of this pathogen in the infected tissues were noted.
Phylogenetic analysis.
The specific primers RhinA2F plus the universal primer ITS4 amplified 450 bp of the 18S SSU rRNA genes, the full length of the ITS1, 5.8S and ITS2, and 60 bp of the 28S LSU rRNA genes from each of the animal and Sri Lankan human strains. The ITS5 and ITS4 universal primers amplified only the ITS regions of R. seeberi plus 60 bp of their 28S LSU rRNA genes from the Indian human strains. This set of universal primers also amplified the human ITS regions, but fragments bearing human amplicons were not of our interest and thus not fully sequenced. R. seeberi's ITS sequences showed differences in the numbers of nucleotides among strains. For instance, R. seeberi's ITS from the eight human strains averaged ∼1,060 bp long and were all uniformly similar. In contrast, the dog's ITS was 1,147 bp and those of the two swans were 1,250 and 1,356 bp long, respectively. In the ITS1 region, the swan S2 possessed the longest sequence followed by the dog and the swan S1 sequences. The smallest sequence in the ITS2 motif was from the dog, followed by those from the eight human strains. The two swans' ITS2 sequences were the longest of all sequences analyzed (Table 1).
TABLE 1.
Characteristics of the ITS1, 5.8S, and ITS2 sequences of R. seeberi amplified from multiple host sources and of ITS sequences of the fish pathogen S. destruens
| Organism | Hosta | Accession no. | ITS1 | 5.8S | ITS2 | Total ITS |
|---|---|---|---|---|---|---|
| R. seeberi | Human-SL1 | AY372367 | 282 | 109 | 778 | 1,060 |
| R. seeberi | Human-SL2 | AY378081 | 283 | 109 | 777 | 1,060 |
| R. seeberi | Human-SL3 | AY378082 | 282 | 109 | 777 | 1,059 |
| R. seeberi | Human-SL4 | AY378083 | 283 | 109 | 776 | 1,059 |
| R. seeberi | Human-I1 | AY610938 | 275 | 109 | 777 | 1,052 |
| R. seeberi | Human-I2 | AY610939 | 280 | 109 | 780 | 1,060 |
| R. seeberi | Human-I3 | AY610940 | 282 | 109 | 780 | 1,062 |
| R. seeberi | Human-I4 | AY610941 | 280 | 109 | 780 | 1,060 |
| R. seeberi | Dog-D1 | AY372365 | 431 | 109 | 716 | 1,147 |
| R. seeberi | Swan-S1 | AY399715 | 384 | 109 | 866 | 1,250 |
| R. seeberi | Swan-S2 | AY486143 | 492 | 109 | 864 | 1,356 |
| S. destruens | Fish | AY388645 | 616 | 109 | 610 | 1,226 |
SL, Sri Lanka; I, India; D, Canis familiaris; S, Cygnus spp.
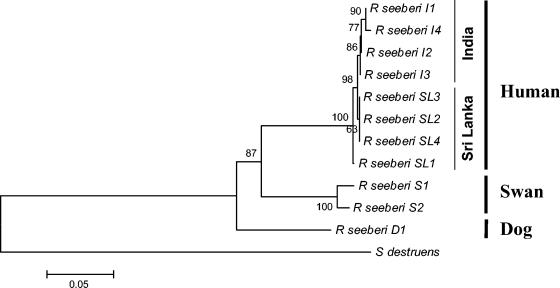
Clustal analysis of the ITS sequences showed multiple ∼50- to ∼60-bp gaps and several mismatches between the human ITS1 and ITS2 and those of the two swans and the dog (data not shown). Phylogenetic analysis by parsimony placed the R. seeberi's ITS sequences in three well-supported sister groups according to their hosts' identities (Fig. 1). In turn, the human ITS sequences grouped in two not-well-supported subgroups, the Indian and the Sri Lankan strains, respectively. The Sri Lankan SL1 strain was the most diverged of the four strains examined in this study. In this tree, the eight human ITS sequences were the sister group to Rhinosporidium's ITS from swan S1 and swan S2 (100%) and, with the inclusion of the dog's ITS sequences, to the dog (87%). The placement of the two swan strains as sister sequences was strongly supported in this analysis (Fig. 1).
FIG. 1.
Phylogenetic relationship between the ITS1, 5.8S, and ITS2 sequences of eight human strains of R. seeberi from India (four strains; I1 to I4) and Sri Lanka (four strains; SL1 to SL4), one strain of R. seeberi from a dog (D1), and two strains from swans (S1 and S2). Multiple hit correction and a 1,000-bootstrap-resampled data set were used to assess branch support. The ITS sequences of R. seeberi isolated from humans are all clustered in a solid group. In this group, the strains from India and Sri Lanka grouped in two poorly supported subgroups. In contrast, the two R. seeberi strains from swans formed a strongly supported sister group to the human strains (100% bootstrap) and with the inclusion of R. seeberi from a dog, a well-supported sister group to the dog (87% bootstrap). Numbers above the branches are percentages of bootstrap-resampled data sets obtained by neighbor-joining. The ITS sequences of S. destruens were used as out-group. The scale bar represents evolutionary distance in substitutions per nucleotide.
DISCUSSION
The use of 18S SSU rRNA gene sequences to determine the evolutionary relatedness between eukaryotic organisms with incomplete life cycles and tenuous morphological affinities has been successful (6, 14). First, Herr et al. (10) and later, Fredericks et al. (8), using the 18S SSU rRNA genes from R. seeberi in humans and a dog with rhinosporidiosis, indicated that R. seeberi was not a fungus but was related to a group of protistal mesomycetozoea at the most basal branch between the animal-fungal divergence. Based on the 18S SSU rRNA genes, these investigators believed that a single species, R. seeberi, was most likely the sole etiologic agent of rhinosporidiosis infecting mammals and birds. A rapid search with the Basic Local Alignment Search Tool (BLAST) of all R. seeberi's 18S SSU rRNA gene sequences clearly supported this idea. To challenge these investigators' data, we have studied R. seeberi's ITS sequences, including those from several strains from different hosts. The ITS alignment of R. seeberi's ITS sequences from the eight humans showed that they were uniformly similar with few mismatches. However, the ITS sequences obtained from humans differed significantly from those ITS sequences from the two swans and the dog. In turn, the sequences from the two swans' strains were also notably different from the ITS sequence obtained from the dog. The remarkable difference in the numbers of nucleotides in the two swans' ITS sequences may suggest rapid substitution in this region or perhaps indicates a different population of R. seeberi in swans.
Our phylogenetic study found that the ITS sequences from a dog, eight humans, and two swans grouped in three independent sister taxons. This molecular study suggests that the genus Rhinosporidium may possess multiple host-specific strains. Interestingly, within the group of the eight human strains, two sister subgroups, corresponding to the India and Sri Lanka strains, were also observed. Within these subgroups, the SL1 strain was found to be the most diverged of all human strains. This finding indicates that R. seeberi from humans could have diverged according to its geographical location, as it was recently found in other species inhabiting these two geographical regions (5). However, given the small number of nucleotide substitutions between these strains, the observed distribution perhaps artificial. To validate the ITS data, other regions of the genome of Rhinosporidium need to be analyzed, including protein sequences. Oddly, when histopathological sections from the tissue samples used in this study were examined, no phenotypic differences were found. Thus, this study suggests that sequencing the ITS regions of R. seeberi might be the ideal way to investigate its host relationship specificities.
The mesomycetozoeans are all aquatic microbial organisms (1, 4, 13). Some of them are well-known fish pathogens (15). So, it is quite possible that R. seeberi originated from similar aquatic environments and then evolved to become a diverged parasite in several mammals and birds. This is not a new finding; several host-restricted pathogens have exhibited similar genetic behavior (7). Based on epidemiological observations, it has been postulated that R. seeberi develops resistant spores in natural environments (2, 12, 18). The human and animal hosts acquire the infection after contacting the resistant spores through open skin. It is quite possible that the species specificity of R. seeberi was more likely acquired from an ancestor that adapted to parasitic life in response to each of its unique parasitized hosts. Thus, the genus Rhinosporidium may have evolved in each of its hosts with a degree of specificity.
Rhinosporidiosis in mammals and birds is common in the areas of endemicity. However, the experimental infection has not yet been possible (2, 4). Based on our finding of host-specific groups in R. seeberi, the failure of experimental rhinosporidiosis might be related to this trait. This could well explain why human-derived R. seeberi, used to infect several animal models, had always failed (3). It will be interesting to see whether strains recovered from Cygnus spp. (12) can infect healthy swans. However, we believe that a combination of host specificities and the fact that Rhinosporidium has resisted culture may account for the failure to produce experimental rhinosporidiosis.
Acknowledgments
We thank Sheila Bolin, Geoffrey R. Gardner, and Tracy Gieger for providing the tissue samples from two Florida swans and a Georgia dog. We also thank S. N. Arseculeratne (Sri Lanka) and Ramchandra Rao (India) for the human samples with rhinosporidiosis.
During this study, our friend and colleague Libero Ajello passed away. He is missed by many who enjoyed his friendship, wit, and wise counsel.
REFERENCES
- 1.Ajello, L. 1998. Ecology and epidemiology of hydrophilic infectious fungi and parafungi of medical mycological importance: a new category of pathogens, p. 67-73. In L. Ajello and R. J. Hay (ed.), Topley & Wilson's microbiology and microbial infections, 9th ed., vol. 4. Arnold, London, England.
- 2.Arseculeratne, S. N., and L. Ajello. 1998. Rhinosporidium seeberi, p. 596-615. In L. Ajello and R. J. Hay (ed.), Topley & Wilson's microbiology and microbial infections, 9th ed., vol. 4. Arnold, London, England.
- 3.Arseculeratne, S. N., F. N. Hussein, D. N. Atapattu, and R. Pathmanathan. 2000. Failure to infect congenitally immunodeficient SCID and NUD mice with Rhinosporidium seeberi. Med. Mycol. 30:393-395. [DOI] [PubMed] [Google Scholar]
- 4.Ashwoth, H. 1923. On Rhinosporidium seeberi (Wernicke, 1903) with special reference to its sporulation and affinities. Trans. R. Soc. Edinb. 53:301-342. [Google Scholar]
- 5.Bossuyt, F., M. Meegaskumbura, N. Beenaerts, D. J. Gower, R. Pethiyagoda, K. Roelants, A. Mannaert, M. Wilkinson, M. M. Bahir, K. Manamendra-Arachchi, P. K. L. Ng, C. J. Schneider, O. V. Oommen, and M. C. Milinkovitch. 2004. Local endemism within the western Ghats-Sri Lanka biodiversity hotspot. Science 306:479-481. [DOI] [PubMed] [Google Scholar]
- 6.Cavalier-Smith, T. 1998. Neomonada and the origin of animals and fungi, p. 375-407. In G. H. Coombs, K. Vickerman, M. A. Sleigh, and A. Warren (ed.), Evolutionary relationships among protozoa. Chapman and Hall, London, England.
- 7.Cushion, M. T. 1998. Pneumocystis carinii, p. 645-683. In L. Ajello and R. J. Hay (ed.), Topley & Wilson's microbiology and microbial infections, 9th ed., vol. 4. Arnold, London, England.
- 8.Fredericks, D. N., J. A. Jolley, P. W. Lepp, J. C. Kosek., and D. A. Relman. 2000. Rhinosporidium seeberi. A human pathogen from a novel group of aquatic protistan parasites. Emerg. Infect. Dis. 6:273-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gargas, A., and P. T. De Priest. 1996. A nomenclature for fungal PCR primers with examples from intron-containing SSU rDNA. Mycologia 88:745-748. [Google Scholar]
- 10.Herr, R. A., L. Ajello, J. W. Taylor, S. N. Arseculeratne, and L. Mendoza. 1999. Phylogenetic analysis of Rhinosporidium seeberi's 18S small-subunit ribosomal DNA groups this pathogen among members of the protoctistan mesomycetozoa clade. J. Clin. Microbiol. 37:2750-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kannan-Kutty, M., and J. B. Gomez. 1971. The ultrastructure and life history of Rhinosporidium seeberi. Southeast Asian J. Trop. Med. Public Health 2:9-16. [PubMed] [Google Scholar]
- 12.Kennedy, F. A., R. R. Buggage, and L. Ajello. 1995. Rhinosporidiosis: a description of an unprecedented outbreak in captive swans (Cygnus spp) and a proposal for revision of the ontogenic nomenclature of Rhinosporidium seeberi. J. Med. Vet. Mycol. 33:157-165. [DOI] [PubMed] [Google Scholar]
- 13.Mendoza, L., J. W. Taylor, and L. Ajello. 2002. The class Mesomycetozoea: a heterogeneous group of microorganisms at the animal-fungal boundary. Annu. Rev. Microbiol. 56:315-344. [DOI] [PubMed] [Google Scholar]
- 14.Mendoza, L., and V. Silva. 2004. The use of phylogenetic analysis to investigate uncultivated microbes in medical mycology, p. 275-298. In G. San-Blas and R. A. Calderone (ed.), Pathogenic fungi structure, biology, and taxonomy. Caister Academic Press, Norfolk, England.
- 15.Ragan, M. A., C. L. Goggin, R. J. Cawthorn, L. Cerenious, A. V. C. Jamieson, S. M. Plourdes, T. G. Tand, K. Soderhall, and R. R. Gutell. 1996. A novel clade of protistan parasites near the animal-fungal divergence. Proc. Natl. Acad. Sci. USA 93:11907-11912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savino, D. F., and C. E. Margo. 1983. Conjunctival rhinosporidiosis light and electron microscopic study. Ophthalmology 90:1482-1489. [PubMed] [Google Scholar]
- 17.Seeber, G. R. 1900. Un nuevo esporozoario parásito del hombre. Dos casos encontrados en pólipos nasales. Thesis. Universidad Nacional de Buenos Aires, Imprenta Librería “Boullosa,” Buenos Aires, Argentina.
- 18.Trianprasit, M., and K. Thagerngpol. 1989. Rhinosporidiosis. Curr. Top. Med. Mycol. 3:64-85. [DOI] [PubMed] [Google Scholar]