Abstract
Sequence analysis of the 16S rRNA gene represents a highly accurate and versatile method for bacterial classification and identification, even when the species in question is notoriously difficult to identify by phenotypic means. In this study, we evaluated the utility of 16S rRNA gene sequencing as a means of identifying clinically important Bacteroides species. We sequenced 231 clinical isolates that had been identified by a short biochemical scheme. Based on the sequence analysis, 192 clinical isolates were assigned to an established species, with the other 39 clinical strains revealing five unique sequences that may represent five novel species. This is in contrast to identification obtained from a short biochemical scheme, by which only 73.5% (172 of 231) of isolates were correctly identified to species level. Based on the solid identification obtained from 16S rRNA gene sequencing, the short biochemical scheme was modified and improved to provide clinical laboratories with an inexpensive and simple alternative for the identification of isolates of clinically significant Bacteroides species.
Anaerobic bacteria, chiefly from our indigenous flora, are involved in a variety of infections that are commonly associated with considerable morbidity and mortality (9). More than one-third of all anaerobes isolated from clinical specimens are members of the Bacteroides fragilis group. The B. fragilis group is part of the indigenous microflora of the human gastrointestinal tract, but different species in this group have been associated with different rates of mortality in bacteremic patients (19), and there are differences in susceptibility patterns as well (10). The choice of antibiotics for therapy is limited because the species of the B. fragilis group are among the most resistant of all anaerobes to antimicrobial agents, and this resistance has increased recently (8, 10, 20). Accordingly, there is a need for rapid, accurate identification of clinical isolates to permit early, effective management of infected patients.
Several identification systems are available commercially for the rapid identification of anaerobes. However, these systems are designed to identify as wide a range of anaerobes as possible, and they contain many tests of little relevance for identification of Bacteroides species. Furthermore, databases accompanying the kits are often incomplete or inaccurate, especially with a number of newly described species. A short biochemical scheme described in the Wadsworth-KTL Anaerobic Bacteriology Manual (13) was developed specifically for the identification of B. fragilis group species. It involves the minimum essential biochemical tests, thus providing clinical laboratories with an inexpensive and simple alternative for the identification of these bacteria. However, a recent study indicated that this biochemical scheme was not as accurate as a multiplex PCR identification scheme established in that study (14).
Direct sequencing of the PCR-amplified bacterial 16S rRNA gene is one of the new molecular identification methods. It has been used in identification of bacteria of many types (2, 5, 6, 11, 12, 16, 18, 21, 23, 24) and unidentified clinical bacterial isolates (7, 25). In this study, we evaluated the suitability of 16S rRNA gene sequencing for the identification of clinically significant Bacteroides species. Furthermore, using 16S rRNA gene sequencing as a standard, we have also analyzed previous errors with the biochemical scheme and modified it for more accurate identification of this group of bacteria.
MATERIALS AND METHODS
Bacterial strains and criteria for identification comparison.
Strains used in this study included 10 American Type Culture Collection (ATCC) strains and 231 Bacteroides isolates previously isolated from clinical specimens at the Wadsworth Anaerobic Bacteriology Laboratory that represent the 10 species of the B. fragilis group (Table 1). All strains were cultured anaerobically overnight on brucella blood agar (Anaerobe Systems, Morgan Hill, Calif.) at 37°C and were characterized by a combination of classical biochemical tests and a short biochemical scheme described in the Wadsworth-KTL anaerobe manual (13). Briefly, the strains were identified by means of the following reactions: bile resistance determined by growth on Bacteroides bile esculin agar or by growth in prereduced, anaerobically sterilized (PRAS) peptone yeast glucose broth containing 20% bile (Anaerobe Systems). Resistance to vancomycin (5 μg), kanamycin (1,000 μg), and colistin (10 μg) was tested by special potency disks; indole production was tested by spot indole and the PRAS tube indole test (Anaerobe Systems); and α-fucosidase or β-glucuronidase was tested with Rosco diagnostic tablets (Taastrup, Denmark). Sugar fermentation tests were performed with PRAS peptone-yeast-sugar broth tubes (Anaerobe Systems). Briefly, the PRAS tubes were inoculated from an actively growing broth culture (without carbohydrate). After incubation at 37°C until good growth was seen (of at least 2+ turbidity), the pH was measured with a pH meter. Identifications were counted as correct if both the biochemical and 16S rRNA gene sequencing methods provided the same answer. A discordant result was defined as a 16S rRNA sequence match with a species other than the species assigned by phenotypic identification. For the strains with discrepant identification, the phenotypic tests mentioned above were repeated, and the strains were also examined with commercially available API ZYM and rapid ID 32A (bioMérieux, Marcy l'Etoile, France), RapID ANA II (Remel, Inc., Lenexa, Kans.), and BBL Crystal Identification (Becton Dickinson Microbiology Systems, Cockeysville, Md.) systems, according to the respective manufacturer's instructions.
TABLE 1.
List of strains used in this study
| Speciesa | Strain or isolates | No. of strains or isolates |
|---|---|---|
| Bacteroides fragilis | ATCC 25285T | 1 |
| Clinical isolates | 32 | |
| Bacteroides thetaiotaomicron | ATCC 29148T | 1 |
| Clinical isolates | 28 | |
| Bacteroides stercoris | ATCC 43183T | 1 |
| Clinical isolates | 21 | |
| Bacteroides uniformis | ATCC 8492T | 1 |
| Clinical isolates | 22 | |
| Bacteroides caccae | ATCC 43185T | 1 |
| Clinical isolates | 30 | |
| Bacteroides distasonis | ATCC 8503T | 1 |
| Clinical isolates | 33 | |
| Bacteroides eggerthii | ATCC 27754T | 1 |
| Clinical isolates | 3 | |
| Bacteroides merdae | ATCC 43184T | 1 |
| Clinical isolates | 15 | |
| Bacteroides ovatus | ATCC 8483T | 1 |
| Clinical isolates | 26 | |
| Bacteroides vulgatus | ATCC 8482T | 1 |
| Clinical isolates | 21 |
Initial identification by phenotypic tests.
16S rRNA gene sequencing.
Genomic DNA was extracted and purified from cells in the mid-logarithmic-growth phase with a QIAamp DNA Mini kit (QIAGEN, Inc., Chatsworth, Calif.). The 16S rRNA gene fragments were amplified as previously described (3). Briefly, two subregions of the 16S rRNA gene were amplified with two pairs of primers. The two subregions were defined as follows: region A was an 899-bp sequence between primer 8UA and 907B, and region B was a 711-bp sequences between primer 774A and 1485B. PCR was performed for 35 cycles, each cycle consisting of 30 s at 95°C, 30 s at 45°C, and 1 min at 72°C; this was followed by a final extension at 72°C for 5 min. PCR products were excised from a 1% agarose gel after electrophoresis, purified with a QIAquick Gel Extraction kit (QIAGEN, Inc.), and sequenced directly with a Biotech Diagnostic Big Dye sequencing kit (Biotech Diagnostics, Laguna Niguel, Calif.) on an ABI 377 sequencer (Applied Biosystems, Foster City, Calif.).
Sequence data analysis.
The sequencing data were analyzed by (i) assembly of the reverse and forward sequences into a consensus sequence, (ii) editing of the consensus sequence to resolve discrepancies between the two strands by evaluation of the electropherograms, and (iii) comparison of the consensus sequences with GenBank sequences by using the Ribosomal Database Project (RDP-II) (Michigan State University, East Lansing, Mich.) (15) and the Basic Local Alignment Search Tool (BLAST) (1). Analysis of Bacteroides clinical isolates was performed by comparing the sequences against the sequences of type strains determined in this study as well as related sequences retrieved from GenBank.
RESULTS
16S rRNA gene sequencing for identification of clinical isolates.
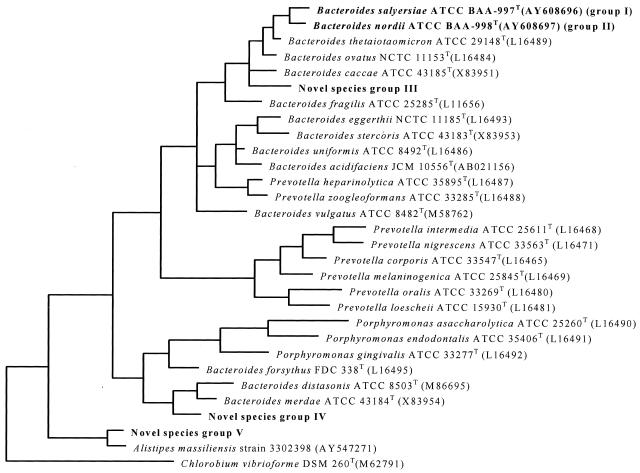
A total of 241 strains including the 10 ATCC type strains and 231 clinical isolates representing the 10 B. fragilis group species were subjected to 16S rRNA sequence analysis. The 10 type strains we analyzed had a good match (similarity of ≥99%) with sequences of their corresponding strains from GenBank as determined by both BLAST and RDP-II. Of the 231 clinical strains, 192 strains had a sequence with high similarity (≥99%) to an established species. Twenty strains had high sequence similarities to two uncharacterized strains in GenBank; these included 7 isolates of B. merdae and 13 isolates of B. stercoris, originally identified by phenotypic testing, that were determined to be 99% similar to 16S rRNA gene sequences of two uncharacterized strains, Bacteroides sp. ASF519 and bacterium strain 31285, respectively. They had only 94.0 and 95% similarities to B. merdae and B. stercoris, respectively (novel species groups IV and II) (Table 2). The remaining 19 strains had three unique sequences that were distinct (similarity, ≤96%) from any sequence in the GenBank database (novel species groups I, III, and V) (Table 2). These were assigned a phylogenetic position by construction of a phylogenetic tree with their related sequences (Fig. 1).
TABLE 2.
Comparison of 16S rRNA sequencing identification with phenotypic identification
| Phenotypic identification | No. of isolates | 16S rRNA gene sequencing identification
|
||
|---|---|---|---|---|
| Result (no. of isolates identified)a | % similarity with reference sequence | Reference sequence | ||
| B. caccae | 30 | B. caccae (19) | ≥99 | B. caccae ATCC 43185T (X83951) |
| B. ovatus (4) | ≥99 | B. ovatus ATCC 8483T (X83952) | ||
| Novel species group V (7) | 96 | A. massiliensis strain 3302398 (AY547271) | ||
| B. distasonis | 33 | B. distasonis (33) | ≥99 | B. distasonis ATCC 8503T (X86695) |
| B. eggerthii | 3 | B. eggerthii (3) | ≥99 | B. eggerthii NCTC 11185T (L16485) |
| B. fragilis | 32b | B. fragilis (11) | ≥99 | B. fragilis ATCC 25285T (X83935) |
| B. fragilis (21) | ≥99 | B. fragilis strain 2393 (X83948) | ||
| B. merdae | 15 | B. merdae (8) | ≥99 | B. merdae ATCC 43184T (X83954) |
| Novel species group IV (7) | ≥99 | Bacteroides sp. ASF519 (AF157056) | ||
| 94 | B. merdae ATCC 43184T (X83954) | |||
| B. ovatus | 26 | B. ovatus (17) | ≥99 | B. ovatus ATCC 8483T (X83952) |
| B. thetaiotaomicron (7) | ≥99 | B. thetaiotaomicron ATCC 29148T (X83952) | ||
| Novel species group III (2) | 93 | B. ovatus ATCC 8483T (X83952) | ||
| B. stercoris | 21 | B. stercoris (8) | ≥99 | B. stercoris ATCC 43183T (X83953) |
| Novel species group II (13) | ≥99 | Bacterium strain 31285 (AF227834) | ||
| 96 | B. thetaiotaomicron ATCC 29148T (X83952) | |||
| B. thetaiotaomicron | 28 | B. thetaiotaomicron (19) | ≥99 | B. thetaiotaomicron ATCC 29148T (X83952) |
| B. uniformis (9) | ≥99 | B. uniformis ATCC 8492T (L16486) | ||
| B. uniformis | 22 | B. uniformis (12) | ≥99 | B. uniformis ATCC 8492T (L16486) |
| Novel species group I (10) | 96 | B. thetaiotaomicron ATCC 29148T (X83952) | ||
| B. vulgatus | 21 | B. vulgatus (21) | ≥99 | B. vulgatus ATCC 8482T (M58762) |
Novel species groups IV and II had sequence similarity of ≥99% to Bacteroides sp. ASF519 and bacterium strain 31285, respectively, and the closest valid species to them are B. merdae (94.0%) and B. stercoris (95%), respectively. Novel species groups I, III, and V had a relatively low sequence similarity to B. thetaiotaomicron (96%), B. ovatus (93%), A. massiliensis (96%), respectively.
A total of 32 isolates of B. fragilis fell into two clusters, with approximately 1.5% difference between them.
FIG. 1.
The phylogenetic tree indicates the phylogenetic relationship of the five possible novel species with their related established species, including one representative strain for each species. The tree was rooted using Chlorobium vibrioforme DSM 260T (M62791) as the outgroup sequence. The closest phylogenetically valid species to novel species groups I and II is B. thetaiotaomicron (96%). The closest valid species to novel species groups III, IV, and V are B. ovatus (93%), B. merdae (94.0%), and Alistipes massiliensis (96%), respectively. NOvel species groups I and II have recently been proposed as B. sulyersiae and B. nordii, respectively.
Comparison of phenotypic and 16S rRNA gene sequencing identification.
Comparison between sequence-based identification and phenotypic identification showed that only 172 (74.5%) isolates had concordant results between the original identification and the 16S rRNA gene sequencing identification, while the other 59 (25.5%) isolates had a molecular identification that was discordant with the original identification (Table 2). B. fragilis (n = 32), B.distasonis (n = 33), B. eggerthii (n = 3), and B. vulgatus (n = 21) were consistently identified correctly by both phenotypic and genotypic methods: 63.3% (19 of 30) of B. caccae isolates, 53.3% (8 of 15) of B. merdae isolates, and 65.4% (17 of 26) of B. ovatus isolates. A total of 38.1% (8 of 21) of B. stercoris isolates, 67.9% (19 of 28) of B. thetaiotaomicron isolates, and 54.5% (12 of 22) of B. uniformis isolates were also identified correctly by both methods. Among the 59 discrepant isolates, 20 strains were reidentified as a different valid Bacteroides species by 16S rRNA gene sequencing: four strains initially identified as B. caccae, nine strains initially identified as B. thetaiotaomicron, and seven strains initially identified as B. ovatus were reidentified as B. ovatus, B. uniformis, and B. thetaiotaomicron, respectively. The other 39 strains that were misidentified to species level by phenotypic tests may be novel species, as they shared low sequence similarities against the sequences of valid species in the GenBank database (Table 2).
Analysis of failures and modification of the biochemical scheme.
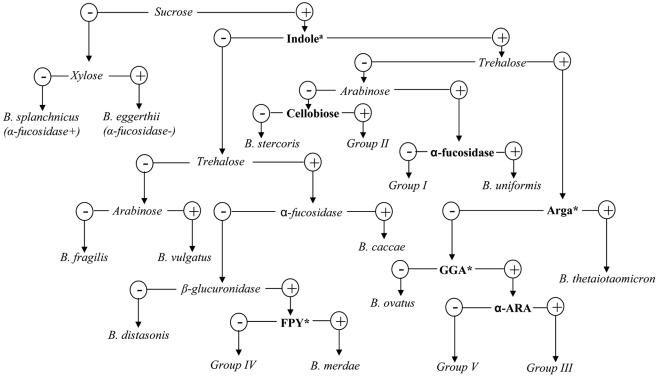
The established biochemical scheme for identification of B. fragilis group species was reevaluated by comparison with 16S rRNA gene sequence analysis of 231 clinical isolates that were previously identified to the species level by this scheme. The key phenotypic characteristics on the flow chart of the biochemical scheme were retested for the 59 strains that had discordant identifications between 16S rRNA gene sequencing and the biochemical scheme; the results are shown in Table 3. For the 20 strains that were reidentified as a different valid Bacteroides species by 16S rRNA gene sequencing, 4 were strains of B. ovatus misidentified as B. caccae because of a false-negative spot indole test; they were indole positive only by the tube test. For the sugar fermentation tests, a pH of ≤5.5 in the PRAS tubes was interpreted as acid (positive), 5.6 to 5.8 was interpreted as weakly acid (weak positive), and ≥5.9 was interpreted as negative fermentation. Seven strains of B. thetaiotaomicron were misidentified as B. ovatus because of the unmatched xylan fermentation profile; they had pHs near 5.5 (range, 5.3 to 5.6) in the xylan reaction. They were interpreted as xylan positive and therefore were misidentified as B. ovatus. Similarly, nine strains of B. uniformis were misidentified as B. thetaiotaomicron because of the unmatched trehalose fermentation profile; their reactions with trehalose were interpreted as positive (pH range, 5.4 to 5.6), in contrast to the negative status in the flow chart. Further characterization of these strains by biochemical kits showed that arginine arylamidase and leucine arylamidase could be used to separate xylan-positive B. thetaiotaomicron (both arginine arylamidase and leucine arylamidase are positive) from B. ovatus (both are negative), and trehalose-positive B. uniformis (both are negative) from B. thetaiotaomicron. For the other 39 strains that may represent five novel species by 16S rRNA gene sequence analysis, phenotypic tests were carried out to characterize and distinguish them from the species they resembled. For example, α-fucosidase, α-arabinosidase, and α-chymotrypsinase could be used to distinguish group I novel species from B. uniformis; glutamic acid decarboxylase, arginine dihydrolase, and p-nitrophenyl-β,d-disaccharidase could differentiate group II novel species from B. stercoris. Glutamyl glutamic acid arylamidase and pyrrolidonyl arylamidase could be used to separate group III and IV species from the biochemically similar B. ovatus and B. merdae, respectively (Table 3). According to the results we obtained from this study, the biochemical scheme was modified and is shown in Fig. 2.
TABLE 3.
Key characteristics to distinguish misidentified species or novel species from the species they resemble phenotypicallya
| Characteristic | Species and response
|
|
|---|---|---|
| Novel species group I (B. salyersiae) | B. uniformis | |
| α-Fucosidase | − | + |
| α-Arabinosidase | − | + |
| α-Chymotrypsinase | − | + |
| Novel species group II (B. nordii) | B. stercoris | |
| Glutamic acid | + | − |
| Decarboxylase | ||
| Arginine dihydrolase | − | + |
| p-Nitrophenyl-β,d-disaccharidase | + | − |
| Novel species group III | B. ovatus | |
| Glutamyl glutamic acid arylamidase | + | − |
| α-Arabinosidase | Novel species group III | Novel species group V |
| + | − | |
| Novel species group IV | B. merdae | |
| Pyrrolidonyl arylamidase | − | + |
| Novel species group V | B. caccae | |
| B. ovatus | B. thetaiotaomicron | |
| B. unformis | B. thetaiotaomicron | |
| Arginine arylamidase | − | + |
| Leucine arylamidase | − | + |
All the key characteristics except pyrrolidonyl arylamidase can be tested by using Key tablets (Key Scientific, Round Rock, Tex.). Pyrrolidonyl arylamidase can be tested with the BBL Crystal identification system (Becton Dickinson Microbiology Systems).
FIG. 2.
Flow chart illustrating modified scheme for identification of B. fragilis group and related Bacteriodes spp. Adapted from the flow chart in reference 13 with permission of the publisher. Boldface type indicates the changes made in this study from our original flow chart (13). Indolea, the indole test should be performed by the PRAS tube test; Arga, arginine arylamidase (tested by rapid ID 32A); GGA, glutamyl glutamic acid arylamidase (tested by rapid ID 32A); FPY, pyrrolidonyl arylamidase (tested with BBL Crystal Identification systems); α-ARA, α-arabinosidase (tested by RapidID 32A).
DISCUSSION
The B. fragilis group is commonly associated with a variety of human infections such as intra-abdominal abscess, wound infections, and bacteremia. B. fragilis group bacteremia contributes significantly to morbidity and mortality (19). There is a tendency for clinicians to choose an antimicrobial drug for empirical treatment of a suspected anaerobic or mixed infection because many clinical microbiology laboratories do not provide identification rapidly or reliably. The widespread application of this type of therapy leads to increased resistance to antimicrobial agents, including the most active ones, among anaerobes. The species of the B. fragilis group are among the most resistant of all anaerobes to antimicrobial agents, and this resistance has increased recently (8, 10, 20). As we apply genetic techniques more and more in the study of anaerobic bacteria, it is likely that we may find that certain presumed relationships between taxa are different, that the habitats of various species will prove to be different, that certain types of infections and the severity of these infections will be found to involve organisms different from the ones originally described in these processes, and that species other than those originally thought to be involved will account for resistance to certain antimicrobial agents. Since different types and severities of infections produced and antimicrobial susceptibility patterns vary according to the particular species involved, the accurate differentiation of species is important to both clinicians and microbiologists so that everyone knows just what organism is being encountered, how virulent it is, and what therapy should be chosen.
16S rRNA gene sequencing has been widely used for bacterial identification. The large rRNA sequence databases at GenBank and at RDP-II allow for a quick comparison of 16S ribosomal DNA (rRNA gene) sequences and to provide phylogenetically useful information, even if a particular sequence has no match in the database. However, its use for species identification is not without limitations. For example, there is no accepted cutoff value of 16S rRNA sequence similarity for species definition (17, 22). Studies of numerous diverse taxa clearly indicated that the majority of recognized species that have been examined to date differ in their 16S rRNA sequences from related species of the same genus in at least 1% of the sequence positions, and typically by more. Our 16S rRNA sequence data showed that the recognized species within the Bacteroides phylogenetic cluster (which excludes B. merdae and B. distasonis) has up to 7.0% or more average divergence. This indicates clearly that the species of the Bacteroides genus are typically separated by good evolutionary distances. All the isolates within the same species shared ≥99% sequence similarity with their corresponding type strains except for 32 isolates of the B. fragilis species, which fell into two clusters with approximately 1.5% difference between them. One cluster shared ≥99% sequence similarity with that of B. fragilis strain ATCC 25285T, and the other cluster had a high similarity (≥99%) with that of B. fragilis strain 2393. These variations might be attributed to intraspecies heterogeneity, but it also suggests that they may in fact be new species. Since there is no clear cutoff value for species assignment, in this study we decided to use stringent criteria (≥99% similarity) for species identification. If an isolate showed a genetic difference of ≥1.00 and ≤2.00%, we reported it as closely related to its best match; thus, the 21 isolates that had a 98.6% sequence similarity with the B. fragilis type strain might be the same species or a subspecies of B. fragilis. In cases where the genetic difference was >2%, the isolate was reported as a unique isolate that may represent a novel taxon (Table 2). The reason that we used only type strains was to eliminate any possible errors of species identification due to initial strain misidentification.
The other issue is the quality of public databases such as GenBank. It is commonly known that sequences are deposited in these databases largely independent of their quality, e.g., the number of ambiguous nucleotides, sequence gaps and errors, and the length of the sequence. Therefore, caution was exercised when we carried out the sequence analysis for species identification. Analysis of Bacteroides clinical isolates was performed by comparing the sequences against related sequences retrieved from GenBank as well as against the sequences of type strains determined in this study. In addition, for an accurate determination of species similarities, all 5′ and 3′ ends of the sequences were cut at identical positions along the gene, at Escherichia coli bp 49 to 1450, and the sequences determined in this study and their corresponding sequences in GenBank were aligned and manually analyzed.
Despite some unresolved issues in 16S rRNA gene sequencing for microbial identification, the advantages of this technique outweigh its limitations, in general. It provides more accurate, more objective, and comprehensive species concepts. Signature nucleotides of 16S rRNA genes allow the determination of phylogenetic relationships among the bacteria that would not have resulted from phenotypic data alone. In this study, we encountered five groups of clinical isolates that may represent five novel taxa. Two of them (novel species groups I and II) have been described recently as B. salyersiae and B. nordii, respectively; further characterization of the other three groups will be carried out. Systematic description of new bacterial species recovered from patients may contribute to the description of emerging infections. Therefore efforts should be made to report the possible novel species, even if only very few strains have been isolated. The strains should be deposited in an international collection and the sequence should be accessible through public databases. This will allow further definition of their clinical significance.
Although the idea that 16S rRNA gene sequencing could be used for bacterial identification of cultured bacteria has become increasingly realistic, the cost is still a critical issue in the evaluation of 16S rRNA gene sequence analysis as a diagnostic tool in clinical laboratories. Therefore, in this study, based on solid identification obtained from sequencing, we reevaluated and modified the biochemical flow chart.
Citron et al. (4) have developed a short biochemical scheme for identification of Bacteroides species. This scheme involves the minimum essential biochemical tests and only requires overnight incubation; therefore, it provides a rapid, cost-effective, and simple alternative for the identification of these bacteria. However, this scheme possesses the limitations of all biochemical identification schemes, such as the fact that phenotypic characteristics of clinical isolates are not always typical and may lead to ambiguous results; interpretation of test results involves substantial subjective judgment. Especially with those newly encountered novel species, the scheme is not sufficient to distinguish them from the others. For example, we found that 7 of 26 B. thetaiotaomicron and 9 of 21 B. uniformis isolates had pHs near 5.5 with reactions to xylan and trehalose, respectively, and were thus interpreted as xylan or trehalose positive and misidentified as B. ovatus and B. thetaiotaomicron, respectively. This indicated that xylan and trehalose fermentation tests are not reliable for making distinctions among these species. Further tests showed that arginine arylamidase and leucine arylamidase could be used to distinguish these species (Table 3 and Fig. 2). Furthermore, since 4 of 21 B. ovatus strains gave a false-negative spot indole test, we suggest that the tube indole test should be used instead of the spot indole test. The key characteristics distinguishing the possible novel species from the other Bacteroides species were added to a biochemical flow chart that we have previously published (Fig. 2) (13).
In summary, the overall performance of 16S rRNA gene sequencing for Bacteroides species identification was excellent. However, since 16S rRNA gene sequencing is not used routinely in clinical microbial identification, we have improved the short biochemical scheme based on the solid identification we obtained from sequencing to provide clinical laboratories with an inexpensive and simple alternative.
Acknowledgments
This work was carried out, in part, with financial support from Veterans Administration Merit Review funds.
REFERENCES
- 1.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, B. A. Rapp, and D. L. Wheeler. 2002. GenBank. Nucleic Acids Res. 30:17-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosshard, P. P., S. Abels, R. Zbinden, E. C. Bottger, and M. Altwegg. 2003. Ribosomal DNA sequencing for identification of aerobic gram-positive rods in the clinical laboratory (an 18-month evaluation). J. Clin. Microbiol. 41:4134-4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brosius, J., M. L. Palmer, P. J. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Citron, D. M., E. J. Baron, S. M. Finegold, and E. J. Goldstein. 1990. Short prereduced anaerobically sterilized (PRAS) biochemical scheme for identification of clinical isolates of bile-resistant Bacteroides species. J. Clin. Microbiol. 28:2220-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cloud, J. L., H. Neal, R. Rosenberry, C. Y. Turenne, M. JAMA, D. R. Hillyard, and K. C. Carroll. 2002. Identification of Mycobacterium spp. by using a commercial 16S ribosomal DNA sequencing kit and additional sequencing libraries. J. Clin. Microbiol. 40:400-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cloud, J. L., P. S. Conville, A. Croft, D. Harmsen, F. G. Witebsky, and K. C. Carroll. 2004. Evaluation of partial 16S ribosomal DNA sequencing for identification of Nocardia species by using the MicroSeq 500 system with an expanded database. J. Clin. Microbiol. 42:578-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drancourt, M., C. Bollet, A. Carlioz, R. Martelin, J. P. Gayral, and D. Raoult. 2000. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J. Clin. Microbiol. 38:3623-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falagas, M. E., and E. Siakavellas. 2000. Bacteroides, Prevotella, and Porphyromonas species: a review of antibiotic resistance and therapeutic options. Int. J. Antimicrob. Agents 15:1-9. [DOI] [PubMed] [Google Scholar]
- 9.Finegold, S. M. 1977. Anaerobic bacteria in human disease. Academic Press, New York, N.Y.
- 10.Finegold, S. M. 1995. Anaerobic infections in humans: an overview. Anaerobe 1:3-9. [DOI] [PubMed] [Google Scholar]
- 11.Gorkiewicz, G., G. Feierl, C. Schober, F. Dieber, J. Kofer, R. Zechner, and E. L. Zechner. 2003. Species-specific identification of campylobacters by partial 16S rRNA gene sequencing. J. Clin. Microbiol. 41:2537-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall, L., K. A. Doerr, S. L. Wohlfiel, and G. D. Roberts. 2003. Evaluation of the MicroSeq system for identification of mycobacteria by 16S ribosomal DNA sequencing and its integration into a routine clinical mycobacteriology laboratory. J. Clin. Microbiol. 41:1447-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jousimies-Somer, H. R., P. Summanen, D. M. Citron, E. J. Baron, H. M. Wexler, and S. M. Finegold. 2002. Wadsworth-KTL anaerobic bacteriology manual, 6th ed. Star Publishing Co., Belmont, Calif.
- 14.Liu, C., Y. Song, M. McTeague, A. W. Vu, H. M. Wexler, and S. M. Finegold. 2003. Rapid identification of the species of the Bacteroides fragilis group by multiplex PCR assays using group- and species-specific primers. FEMS Microbiol Lett. 222:9-11. [DOI] [PubMed] [Google Scholar]
- 15.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mellmann, A., J. L. Cloud, S. Andrees, K. Blackwood, K. C. Carroll, A. Kabani, A. Roth, and D. Harmsen. 2003. Evaluation of RIDOM, MicroSeq, and GenBank services in the molecular identification of Nocardia species. Int. J. Med. Microbiol. 293:359-370. [DOI] [PubMed] [Google Scholar]
- 17.Palys, T., L. K. Nakamura, and F. M. Cohan. 1997. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int. J. Syst. Bacteriol. 47:1145-1156. [DOI] [PubMed] [Google Scholar]
- 18.Patel, J. B., D. G. B. Leonard, X. Pan, J. M. Musser, R. E. Berman, and I. Nachamkin. 2000. Sequence-based identification of Mycobacterium species using the MicroSeq 500 16S rDNA bacterial identification system. J. Clin. Microbiol. 38:246-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redondo, M. C., M. D. Arbo, J. Grindlinger, and D. R. Snydman. 1995. Attributable mortality of bacteremia associated with the Bacteroides fragilis group. Clin. Infect. Dis. 20:1492-1496. [DOI] [PubMed] [Google Scholar]
- 20.Snydman, D. R., N. V. Jacobus, L. A. McDermott, R. Ruthazer, E. J. C. Goldstein, S. M. Finegold, L. J. Harrell, D. W. Hecht, S. G. Jenkins, C. Pierson, R. Venezia, J. Rihs, and S. L. Gorbach. 2002. National survey on the susceptibility of Bacteroides fragilis group: report and analysis of trends for 1997-2000. Clin. Infect. Dis. 35:S126-S134. [DOI] [PubMed] [Google Scholar]
- 21.Song, Y., C. Liu, M. McTeague, and S. M. Finegold. 2003. 16S ribosomal DNA sequence-based analysis of clinically significant gram-positive anaerobic cocci. J. Clin. Microbiol. 41:1363-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.Song, Y. L. C. X. Liu, M. McTeague, and S. M. Finegold. 2004. “Bacteroides nordii” sp. nov. and “Bacteroides salyersae” sp. nov. isolated from clinical specimens of human intestinal origin. J. Clin. Microbiol. 42:5565-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stackebrandt, E., and B. M. Goebel. 1994. A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 23.Tang, Y. W., A. Von Graevenitz, M. G. Waddington, M. K. Hopkins, D. H.Smith, H. Li, C. P. Kolbert, S. O. Montgomery, and D. H. Persing. 2000. Identification of coryneform bacterial isolates by ribosomal DNA sequence analysis. J. Clin. Microbiol. 38:1676-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang, Y. W., N. M. Ellis, M. K. Hopkins, D. H. Smith, D. E. Dodge, and D. H. Persing. 1998. Comparison of phenotypic and genotypic techniques for identification of unusual aerobic pathogenic gram-negative bacilli. J. Clin. Microbiol. 36:3674-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woo, P. C., K. H. Ng, S. K. Lau, K. T. Yip, A. M. Fung, K. W. Leung, D. M. Tam, T. L. Que, and K. Y. Yuen. 2003. Usefulness of the MicroSeq 500 16S ribosomal DNA-based bacterial identification system for identification of clinically significant bacterial isolates with ambiguous biochemical profiles. J. Clin. Microbiol. 41:1996-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]