Abstract
The apparent genetic homogeneity of Mycobacterium ulcerans contributes to the poorly understood epidemiology of M. ulcerans infection. Here, we report the identification of variable number tandem repeat (VNTR) sequences as novel polymorphic elements in the genome of this species. A total of 19 potential VNTR loci identified in the closely related M. marinum genome sequence were screened in a collection of 23 M. ulcerans isolates, one Mycobacterium species referred to here as an intermediate species, and five M. marinum strains. Nine of the 19 loci were polymorphic in the three species (including the intermediate species) and revealed eight M. ulcerans and five M. marinum genotypes. The results from the VNTR analysis corroborated the genetic relationships of M. ulcerans isolates from various geographical origins, as defined by independent molecular markers. Although these results further highlight the extremely high clonal homogeneity within certain geographic regions, we report for the first time the discrimination of the two South American strains from Surinam and French Guyana. These findings support the potential of a VNTR-based genotyping method for strain discrimination within M. ulcerans and M. marinum.
Buruli ulcer (BU) is an emerging necrotic skin disease caused by Mycobacterium ulcerans. Several rural communities in West Africa have recorded dramatic increases in disease incidence over the past decade (7, 12). In some areas in West Africa, BU has replaced tuberculosis and leprosy as the most prevalent mycobacterial disease (1, 7). The foci of BU disease are usually associated with communities living near slow-flowing waters and swamps. On the basis of epidemiological observations it is therefore suggested that aquatic environs could be sources of M. ulcerans. Although several attempts to recover M. ulcerans in pure culture from the environment have remained unsuccessful (14), the identification of M. ulcerans-specific DNA sequences in water and detritus in Australia (18, 23) and in aquatic insects in Ghana and Benin (15), along with recent evidence for association of this mycobacterium with aquatic plants in Ivory Coast (11), suggests that M. ulcerans is an environmental pathogen.
Other molecular techniques have been applied with relatively modest gains in further unraveling aspects of the epidemiology of the disease. The genetic relationships among M. ulcerans strains from different geographical regions have been inferred from comparisons of insertion sequences and 16S rRNA, housekeeping, and structural gene sequences. These markers have indicated restricted genetic variation in M. ulcerans, especially among strains from the same geographic origin (5, 16, 21, 22), strongly hampering the determination of the sources of infection at a local level.
Tandem repeat (TR) DNA sequences are important sources of polymorphism in the genome of many eukaryotes and prokaryotes. Genomic regions showing polymorphism due to different numbers of TR motifs in different strains or individuals are described as variable number tandem repeat (VNTR) loci. Allele-length polymorphism associated with VNTR loci can be indexed by PCR amplification. Especially in highly monomorphic and clonal species such as Mycobacterium tuberculosis (8, 13, 17, 19, 20, 25), Bacillus anthracis (9), and Yersinia pestis (10), VNTR arrays represent polymorphism islets in the otherwise highly conserved genomic backbones and have therefore been very useful in differentiating and studying the genetic relationships of strains of these species.
The main objective of this study was to investigate the existence of VNTR loci in M. ulcerans and to assess their utility for the discrimination of strains of this organism.
MATERIALS AND METHODS
Bacterial strains and DNA preparation.
A total of 23 M. ulcerans isolates, 5 M. marinum isolates, and one mycobacterial strain (ITM 00-1026) referred to as an intermediate species (4) were used in this study (Table 1). All isolates were from the culture collection of the Institute of Tropical Medicine, Antwerp (Antwerp, Belgium). The M. ulcerans isolates were recovered from tissue fragments of patients suffering from BU. Among the M. marinum isolates, two were obtained from granulomatous lesions of patients from Belgium, one was obtained from a fish from Nicaragua, and one (ITM 7732) was obtained from a fish and one (ITM 1717) was obtained from an armadillo in the United States. The isolate from the intermediate species was recovered from a granulomatous lesion of a French patient (4).
TABLE 1.
Strain information
| Species | Straina | Originb | Received fromc (other strain designation) | RFLP typed | 2426 typee | MLST typef | VNTR type |
|---|---|---|---|---|---|---|---|
| M. ulcerans | ITM 5142 | Australia | ATCC19423 | Australian | Victorian | Victorian | Australian |
| ITM 5147 | Australia | J. L. Stanford | Australian | ND | ND | Australian | |
| ITM 94-1324 | Australia | 176862 | ND | ND | ND | SE Asian | |
| ITM 9537 | PNG | D. Dawson (11878/70) | ND | PNG I | SE Asian | PNG | |
| ITM 94-1331 | PNG | D. Dawson (186395) | S. Asia | PNG II | SE Asian | SE Asian | |
| ITM 94-1328 | Malaysia | D. Dawson (186510) | S. Asia | Malaysian | SE Asian | SE Asian | |
| ITM 98-912 | China | W. R. Faber | Asian | Asian | Asian | Asian | |
| ITM 8756 | Japan | ATCC 33728 | Asian | Asian | Asian | Asian | |
| ITM 5114 | Mexico | P. Lavalle | ND | Mexican | Mexican | Mexican | |
| ITM 5143 | Mexico | F. Portaels | Mexican | Mexican | Mexican | Mexican | |
| ITM 7922 | F. Guyana | F. Portaels | S. American | ND | ND | F. Guyana | |
| ITM 842 | Surinam | F. Portaels | S. American | Surinamese | Surinamese | Surinamese | |
| ITM 96-658 | Angola | F. Portaels | ND | African | African | African | |
| ITM 97-111 | Benin | F. Portaels | ND | African | African | African | |
| ITM 97-483 | Benin | F. Portaels | African | ND | African | African | |
| ITM 97-104 | Benin | F. Portaels | African | ND | ND | African | |
| ITM 5152 | DRC | F. Portaels | ND | African | African | African | |
| ITM 5155 | DRC | F. Portaels | ND | ND | ND | African | |
| ITM 97-610 | Ghana | F. Portaels | ND | African | African | African | |
| ITM 97-684 | Benin | F. Portaels | ND | ND | ND | African | |
| ITM 97-680 | Togo | F. Portaels | African | African | African | African | |
| ITM 94-662 | Iv. Coast | F. Portaels | ND | ND | ND | African | |
| ITM 94-511 | Iv. Coast | F. Portaels | African | ND | ND | African | |
| M. marinum | ITM 8022 | Belgium | F. Portaels | NA | NA | ND | 1 |
| ITM 94-56 | Belgium | F. Portaels | NA | NA | ND | 2 | |
| ITM 1320 | Nicaragua | F. Portaels | NA | NA | ND | 3 | |
| ITM 1717 | USA | F. Portaels | NA | NA | ND | 4 | |
| ITM 7732 | USA | ATCC 927 | NA | NA | ND | 5 | |
| Intermediate | ITM 00-1026 | France | F. Portaels | ND | ND | ND | MI |
ITM, Institute for Tropical Medicine.
PNG, Papua New Guinea; DRC, Democratic Republic of Congo; Iv. Coast, Ivory Coast; USA, United States; F. Guyana, French Guyana.
F. Portaels, Institute of Tropical Medicine, Antwerp, Belgium; J. L. Stanford, School of Pathology, London, United Kingdom; D: Dawson, Laboratory of Microbiology and Pathology, Queensland Health, Brisbane, Australia; P. Lavalle, Centro Dermatologico Pascua, Mexico City, Mexico.
RFLP, genotype designation determined by IS2404 restriction fragment length polymorphism typing (6). S. Asian, South Asian; S. American, South American. NA, not applicable. ND, not done.
2426 type, genotype designation determined by 2426-PCR (22). PNG I and PNG II, Papua New Guinea 2426 types I and II, respectively.
MLST, genotype designation determined by multilocus sequence typing (21). SE Asian, Southeast Asian.
DNA was extracted as described previously (26). Briefly, bacterial suspensions were prepared by scraping three to four loopfuls of colonies in TE (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]) and digestion with lysozyme (1 mg/ml). After proteinase K (0.1 mg/ml) and sodium dodecyl sulfate (1%) treatment, the suspensions were incubated with 0.6 M NaCl and 0.27 M N-acetyl-N,N,N-trimethyl ammonium bromide. The DNA was extracted with chloroform-isoamyl alcohol and precipitated with isopropanol. Alternatively, the DNA was obtained by resuspending bacteria into 100 to 200 μl of TE followed by heat inactivation at 100°C for 10 min and by subsequent centrifugation (10,000 × g for 20 min at 4°C) to remove cellular debris.
Location of tandem repeats in M. marinum and M ulcerans.
The unassembled M. marinum genome sequence available as of August 2003 at the Sanger Centre website (http://www.sanger.ac.uk/Projects/M_marinum/) was screened for the presence of TR loci by using the Tandem Repeat Finder program of the Department of Biomathematical Science of Mount Sinai School of Medicine (http://c3.biomath.mssm.edu/trf.html).
BLAST searches were performed on the M. ulcerans genome sequences available as of August 2003 on the BURULIST Web server of the Pasteur Institute (http://genopole.pasteur.fr/Mulc/BuruList.html) to establish the presence of homologous TR loci in the M. ulcerans genome sequence. The sequences were also aligned with the M. tuberculosis genome sequence available on the TUBERCULIST Web server (http://genolist.pasteur.fr/TubercuList/).
VNTR PCR.
The primers for PCR amplification (Table 2) were designed on the basis of the flanking sequences of each TR locus of M. marinum by the use of Oligo 5.0 software (National Biosciences). The PCR was carried out using a Hotstar Taq DNA polymerase kit (QIAGEN). Sample DNA (purified or crude extract) (3 μl) was added to 27 μl of a PCR mix containing 15.5 μl of water, 3 μl of 10× PCR buffer (containing 1.5 mM MgCl2 at the final concentration), 0.4 μM of each primer, 0.2 mM of each deoxynucleotide triphosphate (Roche), 1× Q-solution, and 0.1 μl of HotstarTaq DNA polymerase (0.5 U).
TABLE 2.
Characteristics of VNTR loci in M. marinum and M. ulcerans
| Locus | Primer sequence (5′ to 3′)a | Repeat length (bp)b | ORFc | Functiond |
|---|---|---|---|---|
| 1 | F GGCAGTGGGTGACGTCTCAGT | 57 | Rv2188c | Glycosyltransferase |
| R TCGAGGCGATCTACACCAAGGATTA | FadD15 | Long-chain fatty acid coenzyme A ligase | ||
| 4 | F GCCTTGCTTACCGTCGTGCCAA | 61 | Rv1864c | Conserved hypothetical protein |
| R CGAGCCAAGTTGGACCGTCAACACAT | NF | NA | ||
| 6 | F GACCGTCATGTCGTTCGATCCTAGT | 56 | PfkA | Phosphofructokinase |
| R GACATCGAAGAGGTGTGCCGTCT | gatB | Glutamyl-tRNA amidotransferase | ||
| 8 | F CGGATGACGTCGGAACTCTGA | 58 | cobB | Cobalamine biosynthesis |
| R GGACGCGGTAGCACGTTTTGT | cysG | Cobalamine biosynthesis | ||
| 9 | F GGTGGATCTCCGCGTCATTTG | 57 | Rv3200c | Transmembrane cation transporter |
| R CGACCGCCCTCGAGACAG | nudC | NADP pyrophosphate | ||
| 10 | F ACAAGCCACGGCGAGATATAG | 71 | NF | NA |
| R GCGGGGCTTTTATCTGCTTA | NF | NA | ||
| 13 | F CAGGTATTCCAGGAGATCAAA | 53 | RegX3 | TCS regulator |
| R GGCGACAAGGCTCGTT | SenX3 | TCS sensory protein | ||
| 14 | F CCTTGTATCCGAGTTTCAGTT | 54 | fgd1 | Glucose-6 phosphate dehydrogenase |
| R GTCGACCAGATATGAGCAAT | Rv0406c | β-lactamase-like protein | ||
| 15 | F GCCACCGGTCAGGTCAGGTT | 54 | MgtE | Mg2+ transmembrane transporter |
| R TCACCAACTACGACGGCGTTC | fbA | Fructose biphosphate aldolase | ||
| 16 | F CCAACGCTCCCCCAACCAT | 59 | NF | NA |
| R GCTCACAGGCCTTCGCTCAGA | Rv0238 | Transcriptional regulator | ||
| 18 | F CCCGGAATTGCTGATCGTGTA | 63 | NF | NA |
| R GGTGCGCAGACTGGGTCTTA | NF | NA | ||
| 19 | F CCGACGGATGAATCTGTAGGT | 56 | DeaD | Cold shock protein |
| R TGGCGACGATCGAGTCTC | IprE | Lipoprotein |
F, forward primer, R, reverse primer.
For a full length consensus repeat in M. marinum. Lengths could not be determined in several loci of the M. ulcerans sequence strain that only include a single copy of repeat element.
ORF, homologous open reading frame in M. tuberculosis H37Rv genome. NF, not found.
As predicted from Tuberculist. TCS, two-component system; NA, not applicable.
The PCRs were run on a PTC 100 thermocycler (MJ Research, Waltham, Mass.) at 95°C for 15 min, followed by 40 cycles of 94°C for 30 s, 59°C for 1 min, and 72°C for 1 min 30 s and a final extension at 72°C for 10 min. A total of 3 μl of the PCR products was electrophoretically separated using a 3% small-fragment agarose gel (Eurogentec, Seraing, Belgium) in 0.5× TAE (20 mM Tris-acetate, 0.5 mM EDTA at the final concentration) buffer at 100 V, and the gel was then stained with ethidium bromide. The sizes of the amplicons were estimated by comparison with a 50- and 100-bp stepladder (Promega, Leiden, The Netherlands).
Genetic distance analysis.
The analysis of the genetic relationships on the basis of the VNTR amplicon size profiles was performed using the neighbor-joining algorithm included in the PAUP software package (D. Swofford, phylogenetic analysis using parsimony, 4.0 beta version; Sinauer Associates, Inc., Sunderland, Mass.).
RESULTS
Location of tandem repeat loci in M. marinum and M ulcerans.
The presence of TR loci was initially investigated in sequences of the closely related M. marinum genome (21), because the M. ulcerans genome sequences were not directly accessible for Tandem Repeat Finder analysis. Numerous tandem repeat loci of different sizes were found in the M. marinum genome. We focused on TRs of the minisatellite category, classically defined by a repeat unit size between 10 and 100 bp. This choice was motivated by the relative ease with which allelic differences can be resolved by agarose gel electrophoresis and by the relatively large range of variability of sequences of this type in M. tuberculosis (13, 25). Among the 59 TR loci with period size ranges between 40 and 100 bp identified, 19 loci with more than 95% nucleotide identity between individual repeat units and with two or more copies of repeats were selected for experimental analysis. These two criteria were used on the basis of the observation that the presence of at least two identical or nearly identical repeats is necessary and sufficient to generate TR variability in the case of M. tuberculosis minisatellites (25).
The loci were given numerical labels 1 to 19. A total of 13 of them were found to be polymorphic in a small collection of M. marinum strains. Table 2 shows their locus information. Most of them, with the exception of locus 18, contained repeats with complete consensus sizes plus one partial repeat, with lengths ranging from half to nearly complete units. An alignment of the sequences of each locus with the homologous sequences in the M. ulcerans genome revealed complete identity of TR unit sequences among both mycobacterial species at loci 8, 9, and 14. In the other loci, there were nucleotide deletions of from 2 to 17 bp and/or from 2 to 14 nucleotide substitutions in the M. ulcerans units compared to those of M. marinum. The sequences flanking the repeat sequences were found to be highly conserved in both species and homologous to M. tuberculosis open reading frames encoding products of diverse functional categories (Table 2). Most of the corresponding TR sequences were found in intergenic positions in these regions. At least one of these homologous regions (locus 13) corresponds to a known VNTR locus in M. tuberculosis, namely, the senX3-regX3 locus (25).
Typing of strains and cluster analysis.
The size polymorphism of the 19 TR loci was analyzed by PCR using a collection of 23 M. ulcerans isolates, 5 M. marinum isolates, and an intermediate isolate. This latter strain contains a low copy number of the M. ulcerans-specific insertion sequence IS2404, while many other of its phenotypic and genetic characteristics are similar to those of M. marinum (4). The amplification was robust and yielded reproducible results for all loci and all isolates. A total of 13 of these loci were polymorphic among the M. marinum control strains; nine were also polymorphic in M. ulcerans (Fig. 1). (Only the results obtained for the nine polymorphic loci in both species are shown).
FIG. 1.
Dendrogram showing the genetic relationships of the M. marinum and M. ulcerans isolates determined using nine VNTR loci. The dendrogram was built using the neighbor-joining algorithm and ITM 1717 and 7732 as outgroups, as described in Material and Methods. The VNTR amplicon sizes and the corresponding genotype designations are indicated at the right. The linkage distance scale and the VNTR locus numbers are indicated at the bottom. MM, M. marinum; MU, M. ulcerans; MI, intermediate isolate.
For the M. ulcerans reference strain (ATCC 19423), the experimentally determined sizes were consistent with the predicted sizes determined from the sequence, except for loci 4, 8, and 14. These discrepancies in the amplicon sizes could be due to inaccuracies in the incomplete M. ulcerans genome sequence available. In view of these discrepancies, the differences in repeat unit sizes between M. ulcerans and M. marinum, and the apparent involvement of incomplete repeat units in the variation in some loci (see below), we used the experimentally determined amplicon sizes (Fig. 1) instead of repeat copy numbers to generate profiles for indexing strain differences. For each locus, amplicon sizes of the different strains were found in most cases to differ by nearly exact multiples of complete repeat unit lengths predicted for the respective loci in M. ulcerans and M. marinum. However, smaller differences in amplicon sizes were also observed in some loci, suggesting the involvement of shorter repeat variants in the variability of these loci. These features are consistent with polymorphisms associated with VNTRs.
Eight different M. ulcerans genotypes, five M. marinum genotypes, and a unique intermediate strain genotype were found by VNTR typing (Fig. 1). Although the test panel is of limited size, the results suggest differential variability among the nine loci. Loci 18 and 19 were the most variable loci, with a total of seven and six alleles overall, respectively. Locus 19 was the most discriminatory at the intraspecies level, showing four different alleles in M. ulcerans and four in M. marinum (Fig. 2A and B, respectively). At the other extremity, locus 6 was among the least discriminatory, with only three alleles in total, of which two and three were found in M. ulcerans and M. marinum, respectively. Nevertheless, only loci 6 and 14 could be used to differentiate between the Surinam and French Guyana M. ulcerans isolates (Fig. 1).
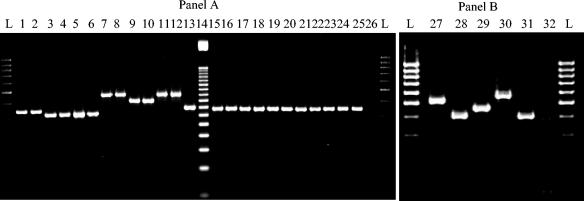
FIG. 2.
PCR analysis of VNTR locus 19. (A) M. ulcerans and the intermediate strain. Lanes: L, 100-bp DNA ladder; 1, ITM 5142; 2, ITM 5147; 3, ITM 94-1324; 4, ITM 9537; 5, ITM 94-1331; 6, ITM 94-1328; 7, ITM 98-912; 8, ITM 8756; 9, ITM 842; 10, ITM 7922; 11, ITM 5114; 12, ITM 5143; 13, ITM 00-1026; 14, 50-bp ladder (locus 19); 15, ITM 96-658; 16, ITM 97-111; 17, ITM 97-483; 18, ITM 97-104; 19, ITM 5152; 20, ITM 5155; 21, ITM 97-610; 22, ITM 97-648; 23, ITM 97-680; 24, ITM 94-662; 25, ITM 94-511; 26, negative control. (B) M. marinum. Lanes: L, 100-bp DNA ladder; 27, ITM 8022; 28, ITM 1717; 29, ITM 94-56; 30, ITM 1320; 31, ITM 7732; 32, negative control.
Genetic relationships of the M. marinum and M. ulcerans isolates.
A dendrogram (Fig. 1) of the VNTR profiles of the 29 isolates was built using the neighbor-joining algorithm.
A first group in the dendrogram included one Australian genotype (alias Victoria) (22), two genotypes designated Southeast Asian and Papua New Guinea of M. ulcerans, and one African genotype. The Australian genotype included two isolates from Australia (Victoria), while the Southeast Asian genotype comprised the third isolate from Australia (Queensland), the Malaysian isolate, and one Papua New Guinea isolate (ITM 94-1331). The genotype of the other Papua New Guinea isolate (ITM 9537) was distinct from the Southeast Asian genotype by only one locus. Strikingly, the African genotype included the 11 M. ulcerans isolates from the seven different African countries tested; these isolates did not display any differences among the nine VNTR loci investigated. Similarly, the isolates from Japan and China had identical profiles at all the nine loci and constituted the Asian genotype. The two Mexican isolates had also identical profiles at all nine loci and were designated the Mexican genotype. This Mexican type was found to be closely related to the Asian type. The two South American isolates from Surinam and French Guyana both gave unique but closely related genotypes, referred to as the Surinam and Guyana genotypes, respectively. These strains appeared to be more distant from the other M. ulcerans strains.
The five M. marinum isolates had five unique genotypes but could be classified into two subgroups, showing some correlation with their source of isolation. These two groups included the two isolates from Belgium and the one from Nicaragua as one group and the two isolates from the United States as the other group. The isolate of the intermediate species (ITM 00-1026) was found to be distantly related to the M. ulcerans Asian-Mexican genotype.
DISCUSSION
M. ulcerans features low variability in house-keeping gene sequences and other genetic elements, especially among isolates from the same geographic region (5, 16, 21, 22). Given the informative value of VNTRs for many other genetically homogeneous organisms, the identification of VNTRs in M. ulcerans may be useful for phylogenetic and epidemiological studies of this pathogen. In this study, we have identified 9 VNTR loci in M. ulcerans and 13 in M. marinum and assessed their usefulness as new molecular targets to study genetic relationships in these two species.
The set of nine VNTR loci revealed eight genotypes among the 23 M. ulcerans reference isolates tested. Using largely overlapping sets of isolates, six genotypes have previously been found by multilocus sequence typing (MLST) analysis of eight structural and house-keeping genes (21) and by restriction fragment length polymorphism (RFLP) of IS2404 (6), whereas nine genotypes were found with 2426-PCR, a repetitive-element PCR targeting IS2404 and IS2606 (22) (Table 1). In contrast, amplified fragment length polymorphism (AFLP) revealed only three M. ulcerans subtypes (6). Although the different typing methods were not applied to exactly the same strain collections, these results suggests that the typing method used on the basis of the set of nine VNTR loci described here is at least as discriminatory as the previous typing methods.
VNTR typing groups M. ulcerans isolates according to their geographic origin similarly to MLST results. Both methods cluster the Malaysian and Papua New Guinean isolates ITM 94-1331 as the Southeast Asian type. VNTR analysis provided additional resolution by discriminating one Papua New Guinean (strain 11878/70 or ITM 9537) genotype, but this genotype was consistently closely related by a single-locus difference to the South East Asian genotype. Numerical analyses of the profiles generated by both methods indicate the closest evolutionary link between strains from Southeast Asia, Australia, and Africa and a comparatively more distant relation among genotypes from other regions. However, at variance with 2426-PCR results (22) but consistent with MLST results (21), multilocus VNTR analysis shows the Asian genotype (Japan-China) to be much more closely related to the Mexican genotype than to the Surinam genotype. Considering that the most reliable method for determining genetic relatedness is sequence comparison, the congruence between VNTR-based and MLST-based (21) groupings indicates that reliable phylogenetic inferences can be made from this set of markers over the scale of evolutionary divergence considered. Furthermore, such congruence is consistent with the clonal population structure of M. ulcerans apparent from MLST analysis (21).
It is particularly noteworthy that despite the resolution power of these markers, we did not find any size differences in any of nine VNTR loci tested among the 11 African isolates coming from the seven different countries. Consistent with the absence of any sequence variation among African strains identified so far (21, 22), these findings highlight the extremely high clonal homogeneity of M. ulcerans in Africa and lend support to the hypothesis of a recent distribution of the organism across this continent (21). The level of genetic conservation is not so extreme within other geographic regions. In addition to discriminating the isolates from Papua New Guinea by one locus (see above), VNTR analysis could discriminate between the isolates from French Guyana and Surinam by two loci, in contrast to IS2404 RFLP results (5) and AFLP results (6). This finding identifies for the first time two closely related but different M. ulcerans genotypes in South America. Furthermore, the difference between these two isolates and the Mexican isolates, also apparent from MLST analysis (21), was much greater (at seven loci out of nine) than that between the two South American isolates. This is the most extensive difference among M. ulcerans strains originating from a same continent observed so far by VNTR analysis and suggests that the corresponding genotypes may have arisen independently in the Americas.
Beyond this comparison with the Mexican isolates, the South American isolates appeared distantly related to the M. ulcerans strains from the other geographic regions in general. Consistently, the isolate from Surinam appeared as the most distal geographical genotype among the M. ulcerans isolates tested by MLST (21) and was distinct from M. ulcerans isolates from the other geographic regions by the sequence of the 3′-terminal end of its 16S rRNA gene, which is identical to that of M. marinum strains (16). Similarly, the isolate of the intermediate species (ITM 00-1026) was found to be distantly related to the Asian-Mexican M. ulcerans genotypes and one of the M. marinum subgroups. However, the analysis of more deep-rooted relationships should be considered with some caution because of the limited degree of confidence inherent in investigations conducted using a set presently restricted to nine VNTR loci.
In conclusion, this study supported the potential of a VNTR-based genotyping method for M. ulcerans and M. marinum. Since VNTR genotyping has also been successfully applied to the M. tuberculosis (8, 17, 19, 20, 25) and M. avium (2, 3) complexes, it is likely that they can be used for the typing of other species of the mycobacterial genus as well. This method offers several practical advantages over AFLP- or RFLP-based typing methods, such as easier DNA extraction (from boiled colonies) suitable for typing and easier interpretation of the banding patterns. The panel of nine markers identified here permitted the discrimination of M. ulcerans isolates within some but not all geographic regions and offers a resolution power comparable to or slightly higher than that of these methods and of MLST. The identification of these nine loci can therefore be considered as a first promising step towards the potential use of VNTRs as markers for molecular epidemiological studies of BU within given geographic regions. This set of loci most likely represents only a fraction of the VNTR patrimony of M. ulcerans, as numerous and diverse arrays of TRs identified in M. marinum have homologs in M. ulcerans which have not been explored yet. Conversely, it is possible that in M. ulcerans, some VNTR loci, such as in the recently identified giant plasmid encoding its macrolide toxin (24), exist that have no counterparts in the M. marinum genome and thus could not be detected by our homology-based strategy. Hence, the publication of the complete M. ulcerans genome sequence will provide an opportunity for the discovery of additional tandem repeat loci, which should enhance strain resolution. Higher resolution will be especially useful to further assess the genetic homogeneity of African M. ulcerans isolates.
Acknowledgments
We thank P. Lavalle, W. R. Faber, and P. H. G. van Keulen for the donation of mycobacterial isolates. We are grateful to Juan Carlos Palomino, Leen Rigouts, Anandi Martin, and Pieter Stragier for their assistance and advice. We thank Pim de Rijk, Krista Fissette, and Cecile Uwizeye for the excellent technical work. We also thank the Sanger Center and The Pasteur Institute for making unpublished genome sequences available.
This work was partly supported by a grant from the Fund for Scientific Research, Flanders (Flanders, Belgium) (F. W. O. Vlaanderen; contracts G. 0301.01 and G. 0471.03N), by the Institut Pasteur de Lille, and by INSERM. A.A. was supported by a grant from the Damien Foundation (Brussels, Belgium). P.S. is a Researcher of the Centre National de la Recherche Scientifique (CNRS).
REFERENCES
- 1.Amofah, G. K., C. Sagoe-Moses, C. Adjei-Acquah, and E. H. Frimpong. 1993. Epidemiology of Buruli ulcer in Amansie West district, Ghana. Trans. R. Soc. Trop. Med. Hyg. 87:644-645. [DOI] [PubMed] [Google Scholar]
- 2.Amonsin, A., L. L. Li, Q. Zhang, J. P. Bannantine, A. S. Motiwala, S. Sreevatsan, and V. Kapur. 2004. Multilocus short sequence repeat sequencing approach for differentiating among Mycobacterium avium subsp. paratuberculosis strains. J. Clin. Microbiol. 42:1694-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bull, T. J., K. Sidi-Boumedine, E. J. McMinn, K. Stevenson, R. Pickup, and J. Hermon-Taylor. 2003. Mycobacterial interspersed repetitive units (MIRU) differentiate Mycobacterium avium subspecies paratuberculosis from other species of the Mycobacterium avium complex. Mol. Cell. Probes 17:157-164. [DOI] [PubMed] [Google Scholar]
- 4.Chemlal, K., G. Huys, F. Laval, V. Vincent, C. Savage, C. Gutierrez, M.-A. Laneelle, J. Swings, W. M. Meyers, M. Daffe, and F. Portaels. 2002. Characterization of an unusual mycobacterium: a possible missing link between Mycobacterium marinum and Mycobacterium ulcerans. J. Clin. Microbiol. 40:2370-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chemlal, K., K. de Ridder, P. A. Fonteyne, W. M. Meyers, J. Swings, and F. Portaels. 2001. The use of IS2404 restriction fragment length polymorphisms suggests the diversity of Mycobacterium ulcerans from different geographic areas. Am. J. Trop. Med. Hyg. 64:270-273. [DOI] [PubMed] [Google Scholar]
- 6.Chemlal, K., G. Huys, P. A. Fonteyne, V. Vincent, A. G. Lopez, L. Rigouts, J. Swings, W. M. Meyers and F. Portaels. 2001. Evaluation of PCR-restriction profile analysis and IS2404 restriction fragment length polymorphism and amplified fragment length polymorphism fingerprinting for identification and typing of Mycobacterium ulcerans and Mycobacterium marinum. J. Clin. Microbiol. 39:3272-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Debacker, M., J. Aguiar, C. Steunou, C. Zinsou, W. M. Meyers, A. Guédénon, J. T. Scott, M. Dramaix, and F. Portaels. 2004. Trends in Mycobacterium ulcerans disease (Buruli ulcer) patients as seen in a rural hospital of Southern Benin, 1997 through 2001. Emerging Infect. Dis. 10:1391-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frothingham, R., and W.A. Meeker-O'Connell. 1998. Genetic diversity in Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144:1189-1196. [DOI] [PubMed] [Google Scholar]
- 9.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus-variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klevytska, A., L. B. Price, J. M. Schupp, P. L. Worsham, J. Wong, and P. Keim. 2001. Identification and characterization of variable-number tandem repeats in Yersinia pestis genome. J. Clin. Microbiol. 39:3179-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marsollier, L., R. Robert, J. Aubry, J. P. Saint Andre, H. Kouakou, P. Legras, A. L. Manceau, C. Mahaza, and B. Carbonnelle. 2002. Aquatic insects as a vector for Mycobacterium ulcerans. Appl. Environ. Microbiol. 68:4623-4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marston, B. J., M. O. Diallo, C. R. Horsburgh, Jr., I. Diomande, M. Z. Saki, J. M. Kanga, G. Patrice, H. B. Lipman, S. M. Ostroff, and R. C. Good. 1995. Emergence of Buruli ulcer disease in the Daloa region of Côte d'Ivoire. Am. J. Trop. Med. Hyg. 52:219-224. [DOI] [PubMed] [Google Scholar]
- 13.Mazars, E., S. Lesjean, A. Banuls, M. Gilbert, V. Vincent, B. Gicquel, M. Tibayrenc, C. Locht, and P. Supply. 2001. High resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. USA 98:1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Portaels, F. 1995. Epidemiology of mycobacterial diseases. Clin. Dermatol. 13:207-222. [DOI] [PubMed] [Google Scholar]
- 15.Portaels, F., P. Elsen, A. Guimares-Peres, P. A. Fonteyne, and W. M. Meyers. 1999. Insects in the transmission of Mycobacterium ulcerans infection. Lancet 353:986. [DOI] [PubMed] [Google Scholar]
- 16.Portaels, F., P. A. Fonteyne, H. de Beehouwer, P. de Rijk, A. Guedenon, J. Hayman, and W. M. Meyers. 1996. Variability in the 3′ end of 16S rRNA sequence of Mycobacterium ulcerans is related to the geographic origin of isolates. J. Clin. Microbiol. 34:962-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roring, S., A. Scott, D. Brittain, I. Walker, G. Hewinson, S. Neill, and R. Skuse. 2002. Development of variable-number tandem repeat typing of Mycobacterium bovis: comparison of results with those obtained by using existing exact tandem repeats and spoligotyping. J. Clin. Microbiol. 40:2126-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ross, B. C., P. D. R. Johnson, F. Oppedisano, L. Marino, A. Sievers, T. Stinear, J. Hayman, M. G. K. Veitch, and R. M. Robins-Browne. 1997. Detection of Mycobacterium ulcerans in environmental samples during an outbreak of ulcerative disease. Appl. Environ. Microbiol. 63:4135-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skuce, R. A., T. P. MacCorry, J. F. McCarroll, S. M. M. Roring, A. N. Scott, D. Brittain, S. L. Hughes, R. G. Hewinson, and S. D. Neill. 2002. Discrimination of Mycobacterium tuberculosis complex bacteria using novel VNTR-PCR targets. Microbiology 148:519-528. [DOI] [PubMed] [Google Scholar]
- 20.Smittipat, N., and P. Pallittapongarnpim. 2000. Identification of possible loci of variable number of tandem repeats in Mycobacterium tuberculosis. Tuber. Lung Dis. 80:69-74. [DOI] [PubMed] [Google Scholar]
- 21.Stinear, T. P., G. A. Jenkins, P. D. R. Johnson, and J. K. Davis. 2000. Comparative genetic analysis of Mycobacterium ulcerans and Mycobacterium marinum reveals evidence of recent divergence. J. Bacteriol. 182:6322-6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stinear, T. P., J. K. Davis, G. A. Jenkins, F. Portaels, B. C. Ross, F. Oppedisano, M. Purcell, J Hayman, and P. D. R. Johnson. 2000. A simple PCR method for rapid genotype analysis of Mycobacterium ulcerans. J. Clin. Microbiol. 38:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stinear, T. P., J. K. Davis, G. A. Jenkins, J. A. Hayman, F. Oppedisano, and P. D. R. Johnson. 2000. Identification of Mycobacterium ulcerans in the environment from regions in Southeast Australia in which it is endemic with sequence capture-PCR. Appl. Environ. Microbiol. 66:3206-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stinear, T. P., A. Mve-Obiang, P. L. C. Small, W. Frigui, M. J. Pryor, R. Brosch, G. A. Jenkin, P. D. R. Johnson, J. K. Davies, R. E. Lee, S. Adusumilli, T. Garnier, S. F. Haydock, P. F. Leadlay, and S. T. Cole. 2004. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc. Natl. Acad. Sci. USA 101:1345-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 39:3563-3571. [DOI] [PubMed] [Google Scholar]
- 26.van Embden, J. D. A., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]