Abstract
Mycobacterium tuberculosis complex isolates (n = 248) collected during a 1-year period in Tuscany, Italy, were genotyped for the katG463 and gyrA95 polymorphisms and by standard spacer oligonucleotide typing (spoligotyping) and IS6110 restriction fragment length polymorphism (RFLP) assays. Most of the isolates (n = 212; 85.5%) belonged to genotypic groups 2 and 3, which included most isolates from Italian-born patients. The remaining isolates were genotypic group 1 organisms, which were prevalent among foreign-born patients (29 of 36; 80.6%). Spoligotype analysis detected 116 unique patterns and 34 clusters including 166 isolates. The combination of spoligotyping and IS6110 RFLP analyses yielded 28 distinct clusters including 65 identical isolates (26.2%)—22 clusters with 2 isolates, 4 clusters with 3 isolates, 1 cluster with 4 isolates, and 1 cluster with 5 isolates—thus proving a low transmission rate in the community. Predominant spoligotypes representing 50% of clustered isolates were found in six clusters that included widespread type ST53 (clade T1) with 29 isolates (11.7% of total isolates); types ST50 and ST47 (Haarlem family) with 18 isolates (7.3%) and 8 isolates (3.2%), respectively; type ST42 (Latino-American and Mediterranean clade) with 13 isolates (5.2%); new type ST1737 (named “Tuscany”) with 8 isolates (3.2%); and type ST1 (W-Beijing family) with 7 isolates (2.8%). Other spoligotype families, such as the Mycobacterium africanum, East African-Indian (EAI2/Manila), and central Asia 1 (CAS1/Delhi) families (all including organisms of genotypic group 1) and the Cameroun family (genotypic group 2), were detected especially among immigrant patients. The occurrence of genotypes originally found in distant geographic areas with a high prevalence of tuberculosis may represent a hallmark for changes in the dynamics of transmission of tuberculosis in the region in the near future.
Since the discovery of polymorphic DNA in Mycobacterium tuberculosis, molecular typing of strains has become a valuable tool in the study of the epidemiology of tuberculosis (TB) by allowing investigators to track epidemics, detect new outbreaks, and achieve better knowledge of moving and expanding strains (1).
Analysis of restriction fragment length polymorphisms (RFLPs), based on the variability of the chromosomal location and copy number of insertion sequence IS6110, is considered the standard approach for the genotyping of M. tuberculosis isolates and has allowed the construction of large databases of IS6110-based genotypes (24). IS6110 fingerprinting has been shown to have great potential for the clonal differentiation of M. tuberculosis isolates (11); however, IS6110 RFLP analysis is a labor-intensive and time-consuming methodology, and its use is hampered for strains with fewer than six IS6110 insertions that have a limited degree of IS6110 polymorphisms or when rapid results are needed, such as in cases of nosocomial transmission of TB or laboratory cross-contaminations. In this context, PCR-based spacer oligonucleotide typing (spoligotyping), based on the variability of spacer sequences interspersed with repeat sequences in the polymorphic chromosomal direct repeat (DR) locus (10), has emerged as a rapid and reliable supplementary tool for molecular epidemiological analysis of TB (5, 6), even though it is endowed with less power to discriminate among M. tuberculosis strains with high IS6110 copy numbers than is IS6110-based genotyping (11). Moreover, spoligotyping has been proven useful for differentiating mycobacterial species within the M. tuberculosis complex and for identifying peculiar genotypes, such as those of the W-Beijing family (9), as well as for investigating the evolutionary genetics of the tubercle bacilli (4, 21) when used in conjunction with other genotyping assays (26).
M. tuberculosis isolates can be divided into three principal genotypic groups on the basis of single nucleotide polymorphisms at codon 463 of the katG gene, encoding the catalase-peroxidase enzyme, and at codon 95 of the gyrA gene, encoding the A subunit of DNA gyrase. The potential value of this typing system is derived from the finding that isolates belonging to groups 1 and 2 are more frequently associated with clustered cases of TB than are isolates belonging to group 3; because clustering is considered a marker for transmission, it has been suggested to indicate a higher level of virulence of group 1 and group 2 organisms (22).
Italy is a country with a low prevalence of TB, where reported notification rates for bacteriologically confirmed cases in 1980 to 2001 ranged from 6 to 10 per 100,000 population (7 per 100,000 in 2001) (27). In the last few years, however, Italy has faced sustained immigration, mostly from Asia, Africa, and eastern Europe. The aim of the present investigation is to provide a description of the population structure of M. tuberculosis in a region of central Italy, Tuscany (with a population of 3.5 × 106 persons in 2001, 3.1% of whom were foreign born), in order to achieve knowledge of the circulating strains and to monitor the dynamics of transmission of TB in the region.
MATERIALS AND METHODS
Bacterial strains, drug resistance assays, and patients.
A total of 248 M. tuberculosis complex strains were collected during a 1-year study period, from 1 January to 31 December 2002, from as many TB patients hospitalized in Tuscany, Italy. All isolates were identified by use of DNA molecular probes (Accuprobe [Gen-Probe] and/or InnoLipa [Innogenetics]). A total of 156 isolates were from Italian-born patients, and 92 were from foreign-born patients. The age of the patients was known for 75% of the study population; Italian patients were 61.1 ± 21.4 (mean ± standard deviation) years old, and immigrant patients were 30.8 ± 7.4 years old. A total of 136 isolates were from human immunodeficiency virus (HIV)-negative patients, and 17 were from HIV-positive patients; HIV serology was not known for 95 patients. The site of TB was pulmonary for 179 patients, extrapulmonary for 45, and unknown for 24.
The drug susceptibility of isolates was determined by use of the radiometric BACTEC 460 TB system (Becton Dickinson, Towson, Md.) according to the proportional method in accordance with the manufacturer's recommendations. The mutations responsible for rifampin and isoniazid resistance were searched for in selected regions of gene rpoB (rifampin) by real-time PCR (23) and in genes katG, kasA, and ndh and in the promoter regions of inhA and ahpC (isoniazid) by real-time PCR (17, 23) and/or nucleotide sequencing.
katG463 and gyrA95 polymorphisms.
Isolates of M. tuberculosis were assigned to one of the three genotypic groups delineated by Sreevatsan et al. (22) on the basis of the combination of polymorphisms at codon 463 of the katG gene and at codon 95 of the gyrA gene. Polymorphisms were evaluated by a real-time PCR assay with primers and probes designed by use of LightCycler probe design software (Roche Applied Science, Mannheim, Germany) in a single reaction tube (18). Codon sequences were obtained by analysis of melting curves derived from the continuous monitoring of probe hybridization status during slow heating.
Spoligotyping.
Spoligotyping analysis of strains was performed basically as described by Kamerbeek et al. (10). Briefly, genomic mycobacterial DNA was extracted from bacteria by the cetyltrimethylammonium bromide (CTAB) method. Oligonucleotides DRa and DRb were used as primers to amplify the whole DR region by PCR. The amplified biotinylated products were hybridized to a set of 43 immobilized oligonucleotides, each corresponding to one of the unique spacer DNA sequences within the DR locus. After hybridization, the membrane was washed, incubated with streptavidin-peroxidase conjugate (Boehringer), and finally developed by chemiluminescence. Spoligotyping results were analyzed as previously described by use of the Excel (Microsoft, Cupertino, Calif.) and Taxotron (P. A. D. Grimont, Taxolab, Institut Pasteur, Paris) software packages. The dendrogram was constructed with the unweighted-pair group method using average linkages (UPGMA) after pairwise comparison of strains by calculation of the Jaccard index.
IS6110 RFLP analysis.
Extraction of DNA from mycobacterial strains and DNA fingerprinting with IS6110 as a probe were performed by standardized methods (24). In brief, after 3 to 4 weeks of growth on Lowenstein-Jensen medium, bacteria were harvested and heat killed (80°C for 20 min). DNA was extracted by the CTAB method and digested with PvuII. After electrophoresis of the digested DNA on agarose gels, restriction fragments were blotted onto nylon sheets, which were then probed with a peroxidase-labeled 245-bp IS6110-specific sequence and finally developed by chemiluminescence (Amersham). The films (Hyperfilm ECL; Amersham) were scanned, and the patterns were compared by the UPGMA clustering method with the Dice coefficient by use of the Gelcompar 4.1 software package (Applied Maths, Kortrijk, Belgium). Clinical isolates with identical IS6110 RFLP fingerprints or those that differed in one IS6110 band were considered to constitute one cluster.
RESULTS AND DISCUSSION
A total of 248 M. tuberculosis complex isolates obtained during a 1-year period from patients hospitalized in Tuscany, Italy, were independently genotyped by the standard spoligotyping and IS6110 RFLP assays and assigned to the genotypic groups defined by the polymorphisms of the katG and gyrA genes as described by Sreevatsan et al. (22). As shown in Table 1, most of the isolates (n = 212; 85.5%) belonged to groups 2 and 3, which also included the vast majority of isolates from Italian-born patients; this finding is in agreement with prevalence data recently reported from northeastern Italy (2). Group 1 isolates were largely prevalent in foreign-born patients (29 of 36; 80.6%).
TABLE 1.
Distribution of M. tuberculosis genotypic groups with respect to patient's place of birth and IS6110 and spoligotype clustering
| Genotypic group | Total no. of isolates | No. (%) of isolates with respect to:
|
|||
|---|---|---|---|---|---|
| Patient's place of birth
|
Clustering
|
||||
| Italy | Foreign country | Spoligotyping | IS6110 RFLP analysis | ||
| 1 | 36 | 7 (19.4) | 29 (80.6) | 22 (61.1) | 14 (38.9) |
| 2 | 153 | 102 (66.7) | 51 (33.3) | 103 (67.3) | 55 (35.9) |
| 3 | 59 | 47 (79.7) | 12 (20.3) | 41 (69.5) | 13 (22.0) |
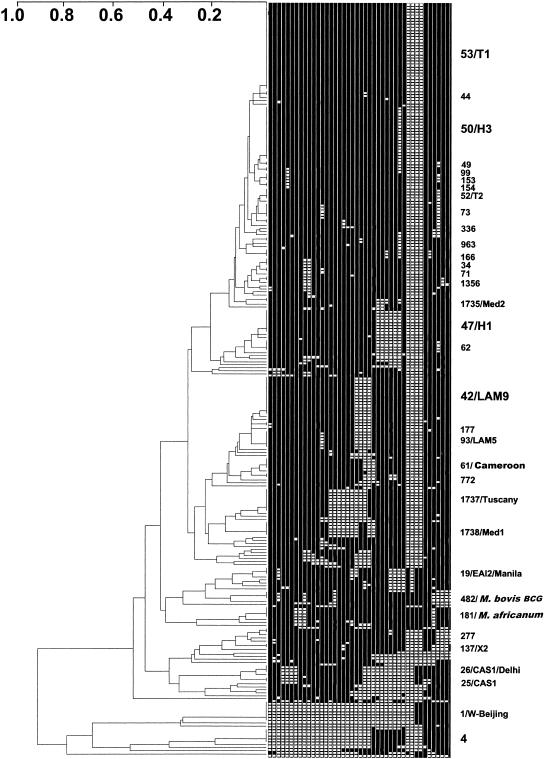
Spoligotyping detected a total of 116 unique patterns and 34 clusters including 166 isolates (67% of total isolates). The spoligotypes were compared to those in the SpolDB4 database containing, at the time of the matching analysis, 30,924 spoligotype patterns from more than 120 countries (unpublished data; see SpolDB3 at www.pasteur-guadeloupe.fr/tb/spoldb3 for the last public release). Twelve new shared types were created when the patterns were not found in the database either for new observed shared types (ST1735, ST1737, and ST1738; see below) or when an orphan isolate from this study matched an orphan isolate in the SpolDB4 database (ST1736, ST1739, ST1740, ST1741, ST1742, ST1743, ST1744, ST1745, and ST1746). After comparison with the spoligotypes in the SpolDB4 database, the total number of isolates for which no matching pattern was detected was reduced to 33. A dendrogram showing relatedness among the spoligotypes detected in our survey is shown in Fig. 1. It must be made clear, however, that matching to the international database does not necessarily indicate a clonal relationship among the isolates. Genetic convergence may explain the existence of some spoligotype-based clusters, as demonstrated by Warren et al. (26); however, previous demonstrations of congruence among various polymorphic genetic markers as well as a clonal population structure for M. tuberculosis suggest that events of genetic convergence are rare for spoligotyping (21).
FIG. 1.
Dendrogram and schematic representation of spoligotyping results obtained for 248 clinical isolates of M. tuberculosis complex from Tuscany, Italy. The phylogenetic tree was constructed with the UPGMA method by calculation of the Jaccard index by use of the Taxotron software package; shared type/family (e.g., 1/W-Beijing) designations appear at the right after the binary spoligotype. The size of the font is proportional to the size of the cluster. Newly described Tuscany, Med1, and Med2 clones are shown with their shared type numbers.
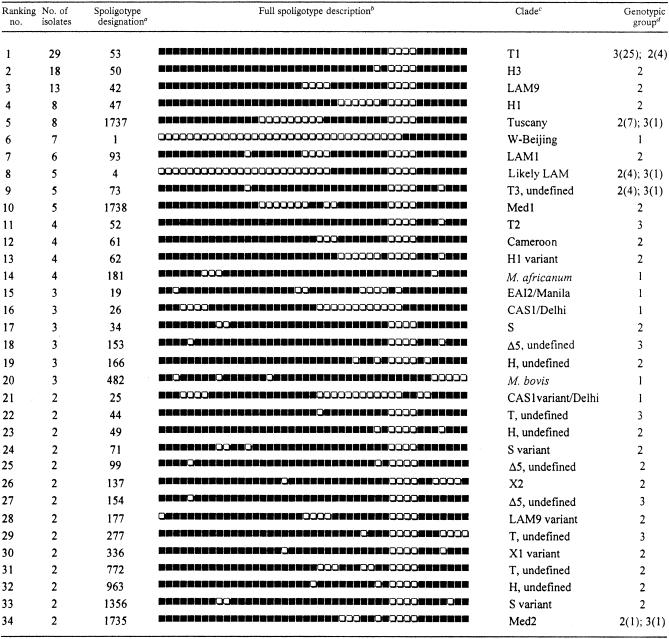
The distribution of the clustered isolates and their assignment to the katG- and gyrA-based genotypic groups are shown in Table 2. In general, clustered isolates with a given spoligotype belonged to the same genotypic group; however, in five cases spoligotypes were shared by organisms of genotypic groups 2 and 3 (ST4, ST53, ST73, ST1735, and ST1737). No sharing of spoligotypes among organisms belonging to groups 1 and 2 or among organisms belonging to groups 1 and 3 was found. Moreover, all isolates of genotypic groups 2 and 3 failed to hybridize with spoligotype probes 33 to 36, as previously reported in the Houston study by Soini et al. (20). These data confirm previous observations indicating that group 1 organisms can be regarded as evolutionarily old and may have further evolved into group 2 and group 3 organisms (20, 22). No association of genotypic groups with spoligotype clusters was found (Table 1). As shown in Table 2, 50% of the clustered isolates were found in only six types. These predominant spoligotypes included the ubiquitous clade T1 (ST53, with 29 isolates), which likely represents relatively old genotypes and is prevalent in Europe (5); the Haarlem family, clades H3 and H1 (ST50 and ST47, with 18 and 8 isolates, respectively) (11); the Latino-American and Mediterranean (LAM9) clade (ST42, with 13 isolates) (5); new spoligotype ST1737, with 8 isolates (see below); and the W-Beijing family (ST1, with 7 isolates), whose members are becoming prevalent worldwide (9).
TABLE 2.
Distribution of clustered isolates determined by spoligotyping
Spoligotype designations were assigned according to the definitions in the SpolDB4 database(see the text).
Black and white boxes indicate the presence and absence, respectively, of the specific spacer at positions 1 to 43 in the DR locus.
Clades are defined mainly as described by Filliol et al.(5) and this study and should be considered subject to change.
When a clade included isolates belonging to different genotypic groups, the number of isolates in each group is given in parentheses.
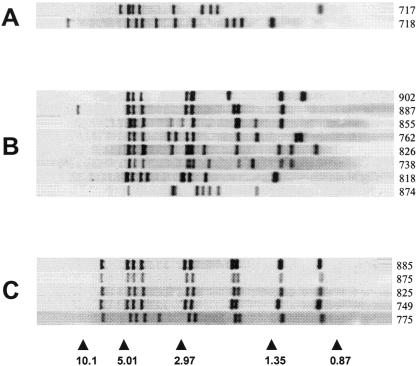
Notably, as anticipated, spoligotyping revealed three new shared types, designated ST1735, ST1737, and ST1738. In particular, ST1737 included eight isolates characterized by the absence of spacers 15 to 23 and 33 to 36. These isolates had distinct IS6110 fingerprints (Fig. 2), but seven of them had some bands at conserved positions. Seven patients were Italian born; the eighth patient was a foreign-born medical postdoctoral fellow attending the Infectious Diseases Unit of a Tuscan hospital. As these isolates seemed to be autochthonous, we propose the designation “Tuscany” for the ST1737 family. ST1738 (proposed designation: Med1 [Mediterranean]) included five isolates characterized by the absence of spacers 15 to 21, 24 to 25, and 33 to 36. Three isolates were from Italian-born patients; the others were from patients who were born in Algeria and Morocco and had recently emigrated to Italy. The IS6110 fingerprints of the ST1738 isolates were basically identical (one isolate had one extra band) (Fig. 2), indicating that there was a possible epidemiological link between the TB cases or that the isolates shared a common ancestor. Isolates of the third new type (ST1735; proposed designation: Med2), characterized by the absence of spacers 26 to 28, 31, and 33 to 36, included two isolates with distinct IS6110 patterns (Fig. 2). These isolates were from an Italian-born patient and an Albanian-born patient.
FIG. 2.
IS6110 fingerprints of M. tuberculosis complex isolates showing new shared spoligotype patterns ST1735 (A), ST1737 (B), and ST1738 (C). The arrowheads and numbers at the bottom indicate the positions and sizes (in kilobase pairs), respectively, of standard DNA fragments. Numbers at right indicate the codes of the isolates.
Besides these types, it is worth mentioning the following families detected in the region: (i) the Mycobacterium africanum family, reported, to our knowledge, for the first time in Italy, including five isolates which lacked spacers 8, 9, and 39 and which were isolated from patients who were born in Senegal and had emigrated to Italy in 2001 to 2002 (the isolates belonged to genotypic group 1 and showed basically the same IS6110 RFLP profiles—one had an extra band; four shared spoligotype profile ST181, characterized by the absence of spacers 7 to 9 and 39; and the fifth one had an orphan profile characterized by the absence of spacers 7 to 9, 12, 19, 20, and 39); (ii) the Mycobacterium bovis family (ST482), including three isolates which had a single IS6110 insertion, which belonged to genotypic group 1, and which were isolated from Italian-born patients in three distinct city hospitals; (iii) the central Asia 1 (CAS1) family (ST25 and ST26), which was characterized by the absence of spacers 4 to 7 and 23 to 34 and, in IS6110 fingerprinting, by two high-molecular-weight bands at 12.1 and 10.1 kb and which was present in India, Pakistan, Sudan, and elsewhere (7, 19, 25); (iv) the East African-Indian 2 (EAI2/Manila) family (ST19) (3); and (v) the Cameroun family (ST61), including four isolates, all from African-born patients (16).
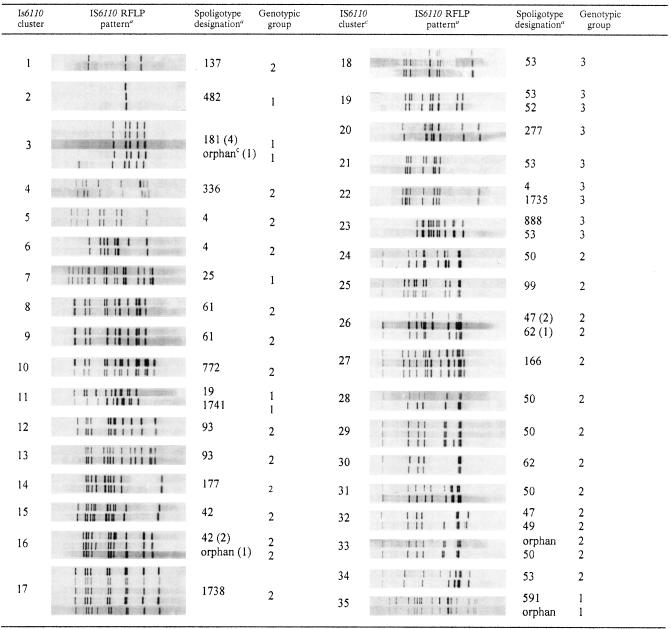
In the IS6110 RFLP analysis, the 248 isolates yielded a total of 201 distinct IS6110 patterns; 82 isolates (33.1%) occurred in 35 distinct clusters. Of these 35 clusters, 27 contained two isolates, 6 contained three isolates, and 2 contained five isolates. Table 3 shows the molecular traits of the isolates in the IS6110 clusters with respect to spoligotypes and katG and gyrA polymorphisms. The isolates included in each IS6110 cluster belonged to the same genotypic group; unexpectedly, however, a number of clusters (i.e., 3, 11, 16, 19, 22, 23, 26, 32, 33, and 35) included isolates with different spoligotypes. This is an unusual finding, since the general belief is that IS6110 RFLP analysis is generally more discriminatory than spoligotyping. Two nonexclusive interpretations may be suggested. On the one hand, IS6110 genetic convergence in a different spoligotyping background, although unlikely, may explain this result. Preferential IS6110 insertion sites have already been described (12). On the other hand, this investigation may have provided an exceptional environment in which to detect true molecular evolution of the DR locus. With the exception of IS6110 cluster 22, whose spoligotyping structures (ST4 and ST1735) cannot be directly linked by a single molecular evolutionary mechanism, all other IS6110 clusters harboring different spoligotypes are easily explained by such a mechanism (Table 3); this explanation favors the “historical transmission” interpretation of IS6110 clustering or the independent introduction of strains harboring more diverse spoligotypes than IS6110 patterns. For the Haarlem and LAM clusters of strains (clusters 16, 26, 32, and 33 in Table 2), the first interpretation is more likely, whereas for the M. africanum and EAI clusters (clusters 3, 11, and 35 in Table 3), the second hypothesis is more likely.
TABLE 3.
Spoligotypes and katG and gyrA polymorphisms of isolates in IS6110 clusters
Each panel shows the IS6110 banding patterns of the isolates included in the cluster.
See Table 2, footnote a. When an IS6110 cluster included different spoligotypes, the number of isolates with a given spoligotype or orphan isolates is given in parentheses.
“Orphan” indicates a spoligotype not matching any existing pattern in the database.
From the epidemiological point of view, the combination of IS6110 RFLP typing results and spoligotyping results for the study population yielded 28 distinct clusters including 65 identical isolates (26.2%). These isolates were included in 22 clusters with two isolates, 4 clusters with three isolates, 1 cluster with four isolates, and 1 cluster with five isolates. Assuming that a cluster includes one reactivation case, the “active transmission rate” in the study area is (65 − 28)/248, that is, 14.9%. Although an accurate evaluation of the active transmission rate requires a survey of a longer duration (8), this preliminary low rate indicates that in the study area TB cases attributed to recent transmission are rare and that disease caused by reactivation is more prevalent. The rate of TB transmission is in overall agreement with that in previous studies performed with isolates collected in northeastern Italy (2) and southern Italy (15); however, it is lower than that found by Moro et al. (14) in the area of Milan, where 28.1% of TB cases in a population with a higher percentage of AIDS patients were attributed to recent transmission. With regard to the association of IS6110 clusters with katG- and gyrA-based genotypic groups (Table 1), organisms of groups 1 and 2, considered together, were more likely to be found in IS6110 clusters than were group 3 organisms (P value determined by Fisher's exact test, <0.05).
Finally, in agreement with the low TB prevalence in the study area, the overall prevalence of drug resistance of the study isolates to first-line anti-TB drugs (Table 4) was generally low, and multidrug resistance was observed in only five isolates (2%) (one isolate resistant to two drugs, three isolates resistant to three drugs, and one isolate resistant to five drugs). It is worth mentioning, however, that at least for isoniazid and rifampin, the prevalences of resistant isolates (9.1 and 3.5%, respectively) were higher than those reported in a recent national survey (3.5 and 0.8%, respectively) (13). Mutations associated with drug resistance were rpoB531 in 6 of 8 rifampin-resistant isolates and katG315 and −15 inhA in 13 and in 4 of 21 isoniazid-resistant isolates, respectively; 2 isoniazid-resistant isolates harbored both the katG315 and the −15 inhA mutations. No mutations associated with isoniazid resistance were found in the ahpC, kasA, and ndh genes. The genetic basis of drug resistance remained undetermined for six isoniazid-resistant isolates (data not shown). The drug resistance of most studied isolates probably reflects primary resistance because, in a small proportion of patients receiving anti-TB drug treatment for at least 30 days, the prevalence of drug resistance of the isolates was approximately two- to eightfold higher (data not shown).
TABLE 4.
Patterns of drug resistance of M. tuberculosis complex isolates in Tuscany, Italy, in 2002
| Drug(s)a | Total no. of isolates tested | No. (%) of drug-resistant isolatesb |
|---|---|---|
| INH | 232 | 21 (9.1) |
| RIF | 232 | 8 (3.5) |
| STR | 110 | 4 (3.6) |
| ETH | 226 | 5 (2.2) |
| PZA | 231 | 5 (2.2) |
| INH + RIF | 1 | |
| INH + RIF + STR | 1 | |
| INH + RIF + ETH | 2 | |
| INH + RIF + STR + ETH + PZA | 1 |
INH, isoniazid; RIF, rifampin; STR, streptomycin; ETH, ethambutol; PZA, pyrazinamide.
Determined by use of the radiometric BACTEC 460 TB system.
In conclusion, molecular epidemiological analysis provides a description of the population structure of M. tuberculosis in Tuscany, Italy. The marked heterogeneity of molecular characteristics of the isolates involved in microoutbreaks of TB testifies to the low transmission rate occurring in the study area and indicates that most cases are caused by reactivation of latent TB rather than recent transmission; moreover, multidrug resistance is still rare. However, the occurrence, mostly among immigrant patients, of organisms such as those belonging to the families W-Beijing, M. africanum, EAI2/Manila, and CAS1 (genotypic group 1) as well as to the family Cameroun (genotypic group 2), originally found in distant geographic areas with a high prevalence of TB, may represent a hallmark for changes in the dynamics of transmission of TB in the region in the near future. Preliminary data on M. tuberculosis strains isolated in the region during 2003 in fact indicate a significant increase in multidrug resistance (unpublished data). Further and continuous molecular epidemiological surveillance is needed in order to trace and monitor trends in prevalence and interhuman dissemination of moving and expanding M. tuberculosis clones for a proper TB control program.
Acknowledgments
This work was supported by funds from MIUR (PRIN 2002-04) and in part by Istituto Superiore di Sanità (National Research Program on AIDS).
We acknowledge S. Brignali, R. Mattei, and S. Valentini for providing mycobacterial strains isolated in their laboratories.
REFERENCES
- 1.Barnes, P. F., and M. D. Cave. 2003. Molecular epidemiology of tuberculosis. N. Engl. J. Med. 349:1149-1156. [DOI] [PubMed] [Google Scholar]
- 2.Dolzani, L., M. Rosato, B. Sartori, E. Banfi, C. Lagatolla, M. Predominato, C. Fabris, E. Tonin, F. Gombac, and C. Monti-Bragadin. 2004. Mycobacterium tuberculosis isolates belonging to katG gyrA group 2 are associated with clustered cases of tuberculosis in Italian patients. J. Med. Microbiol. 53:155-159. [DOI] [PubMed] [Google Scholar]
- 3.Douglas, J. T., L. Qian, J. C. Montoya, J. M. Musser, J. D. van Embden, D. van Soolingen, and K. Kremer. 2003. Characterization of the Manila family of Mycobacterium tuberculosis. J. Clin. Microbiol. 41:2723-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duchene, V., S. Ferdinand, I. Filliol, J. F. Guegan, N. Rastogi, and C. Sola. 2004. Phylogenetic reconstruction of Mycobacterium tuberculosis within four settings of the Caribbean region: tree comparative analyses and first appraisal on their phylogeography. Infect. Genet. Evol. 4:5-14. [DOI] [PubMed] [Google Scholar]
- 5.Filliol, I., J. R. Driscoll, D. Van Soolingen, B. N. Kreiswirth, K. Kremer, G. Valétudie, D. D. Anh, R. Barlow, D. Banerjee, P. J. Bifani, K. Brudey, A. Cataldi, R. C. Cooksey, D. V. Cousins, J. W. Dale, O. A. Dellagostin, F. Drobniewski, G. Engelmann, S. Ferdinand, D. Gascoyne-Binzi, M. Gordon, M. C. Gutierrez, W. Haas, H. Heersma, G. Källenius, E. Kassa-Kelembho, T. Koivula, H. M. Ly, A. Makristathis, C. Mammina, G. Martin, P. Moström, I. Mokrousov, V. Narbonne, O. Narvskaya, A. Nastasi, S. Ngo Niobe Eyangoh, J. W. Pape, V. Rasolofo-Razanamparany, M. Ridell, M. L. Rossetti, F. Stauffer, P. N. Suffys, H. Takiff, J. Texier-Maugein, V. Vincent, J. de Waard, C. Sola, and N. Rastogi. 2002. Global distribution of Mycobacterium tuberculosis spoligotypes. Emerg. Infect. Dis. 8:1347-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filliol, I., J. R. Driscoll, D. van Soolingen, B. N. Kreiswirth, K. Kremer, G. Valetudie, D. A. Dang, R. Barlow, D. Banerjee, P. J. Bifani, K. Brudey, A. Cataldi, R. C. Cooksey, D. V. Cousins, J. W. Dale, O. A. Dellagostin, F. Drobniewski, G. Engelmann, S. Ferdinand, D. Gascoyne-Binzi, M. Gordon, M. C. Gutierrez, W. H. Haas, H. Heersma, E. Kassa-Kelembho, M. L. Ho, A. Makristathis, C. Mammina, G. Martin, P. Mostrom, I. Mokrousov, V. Narbonne, O. Narvskaya, A. Nastasi, S. N. Niobe-Eyangoh, J. W. Pape, V. Rasolofo-Razanamparany, M. Ridell, M. L. Rossetti, F. Stauffer, P. N. Suffys, H. Takiff, J. Texier-Maugein, V. Vincent, J. H. de Waard, C. Sola, and N. Rastogi. 2003. Snapshot of moving and expanding clones of Mycobacterium tuberculosis and their global distribution assessed by spoligotyping in an international study. J. Clin. Microbiol. 41:1963-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gascoyne-Binzi, D. M., R. E. Barlow, A. Essex, R. Gelletlie, M. A. Khan, S. Hafiz, T. A. Collyns, R. Frizzell, and P. M. Hawkey. 2002. Predominant VNTR family of strains of Mycobacterium tuberculosis isolated from South Asian patients. Int. J. Tuberc. Lung Dis. 6:492-496. [DOI] [PubMed] [Google Scholar]
- 8.Glynn, J. R., E. Vynnycky, and P. E. M. Fine. 1999. Influence of sampling on estimates of clustering and recent transmission of Mycobacterium tuberculosis derived from DNA fingerprinting techniques. Am. J. Epidemiol. 149:366-371. [DOI] [PubMed] [Google Scholar]
- 9.Glynn, J. R., J. Whiteley, P. J. Bifani, K. Kremer, and D. van Soolingen. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agderveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. D. A. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. Hermans, C. Martin, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D. van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 37:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McHugh, T. D., and S. H. Gillespie. 1998. Nonrandom association of IS6110 and Mycobacterium tuberculosis: implications for molecular epidemiological studies. J. Clin. Microbiol. 36:1410-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Migliori, G. B., R. Centis, L. Fattorini, G. Besozzi, C. Saltini, C. Scarparo, D. Cirillo, A. Gori, A. Cassone, and C. Piersimoni. 2004. Mycobacterium tuberculosis complex drug resistance in Italy. Emerg. Infect. Dis. 10:752-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moro, M. L., G. Salamina, A. Gori, V. Penati, R. Sacchetti, F. Mezzetti, A. Infuso, and L. Sodano. 2002. Two-year population-based molecular epidemiological study of tuberculosis transmission in the metropolitan area of Milan, Italy. Eur. J. Clin. Microbiol. Infect. Dis. 21:114-122. [DOI] [PubMed] [Google Scholar]
- 15.Nastasi, A., and C. Mammina. 1999. Epidemiological study of tuberculosis in Palermo, Italy: IS6110 fingerprinting of Mycobacterium tuberculosis strains isolated in the years 1994-1998. Infection 27:318-322. [DOI] [PubMed] [Google Scholar]
- 16.Ngo Niobe-Eyangoh, S., C. Kuaban, P. Sorlin, P. Cunin, J. Thonnon, C. Sola, N. Rastogi, V. Vincent, and M. C. Gutierrez. 2003. Genetic biodiversity of Mycobacterium tuberculosis complex strains from patients with pulmonary tuberculosis in Cameroon. J. Clin. Microbiol. 41:2547-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rindi, L., L. Bianchi, E. Tortoli, N. Lari, D. Bonanni, and C. Garzelli. 2003. A real-time PCR assay for detection of isoniazid resistance in Mycobacterium tuberculosis clinical isolates. J. Microbiol. Methods 55:797-800. [DOI] [PubMed] [Google Scholar]
- 18.Rindi, L., N. Lari, D. Bonanni, and C. Garzelli. 2004. Detection of Mycobacterium tuberculosis principal genetic groups by a duplex real-time PCR targeting the katG and gyrA genes. J. Microbiol. Methods 59:283-287. [DOI] [PubMed] [Google Scholar]
- 19.Sharaf-Eldin, G. S., N. S. Saeed, M. E. Hamid, A. M. Jordaan, G. D. van der Spuy, R. M. Warren, P. D. van Helden, and T. C. Victor. 2002. Molecular analysis of clinical isolates of Mycobacterium tuberculosis collected from patients with persistent disease in the Khartoum region of Sudan. J. Infect. 44:244-251. [DOI] [PubMed] [Google Scholar]
- 20.Soini, H., X. Pan, A. Amin, E. A. Graviss, A. Siddiqui, and J. M. Musser. 2000. Characterization of Mycobacterium tuberculosis isolates from patients in Houston, Texas, by spoligotyping. J. Clin. Microbiol. 38:669-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sola, C., I. Filliol, E. Legrand, I. Mokrousov, and N. Rastogi. 2001. Mycobacterium tuberculosis phylogeny reconstruction based on combined numerical analysis with IS1081, IS6110, VNTR, and DR-based spoligotyping suggests the existence of two new phylogeographical clades. J. Mol. Evol. 53:680-689. [DOI] [PubMed] [Google Scholar]
- 22.Sreevatsan, S., X. Pan, K. Stockbauer, N. Connell, B. Kreiswirth, T. Whittam, and J. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torres, M. J., A. Criado, J. C. Palomares, and J. Aznar. 2000. Use of real-time PCR and fluorimetry for rapid detection of rifampin and isoniazid resistance-associated mutations in Mycobacterium tuberculosis. J. Clin. Microbiol. 38:3194-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Embden, J. D. A., M. D. Cave, J. T. Crowford, J. W. Dale, K. D. Eisenhach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. S. Shinnick, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vijaya-Bhanu, N., D. van Soolingen, J. D. A. van Embden, L. Dar, R. M. Pandey, and P. Seth. 2002. Predominance of a novel Mycobacterium tuberculosis genotype in the Delhi region of India. Tuberculosis 82:105-112. [DOI] [PubMed] [Google Scholar]
- 26.Warren, R. M., E. M. Streicher, S. L. Sampson, G. D. Van Der Spuy, M. Richardson, D. Nguyen, M. A. Behr, T. C. Victor, and P. D. Van Helden. 2002. Microevolution of the direct repeat region of Mycobacterium tuberculosis: implications for interpretation of spoligotyping data. J. Clin. Microbiol. 40:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.World Health Organization. 2003. W. H. O. report. Global tuberculosis control. http://www.who.int/gtb/publications/globrep.