Abstract
Simple Summary
Grade 2 and 3 and dedifferentiated chondrosarcomas represent rare malignant bone neoplasms. These tumors are often associated with isocitrate dehydrogenase (IDH) mutations. Unfortunately, treatment options are limited for advanced disease and at the present time both IDH mutant and WT tumors are treated similarly. This study compares differential gene expression in IDH mutant and WT chondrosarcomas with RNA sequencing and stratifies clinical outcome by IDH status and tumor grade.
Abstract
Background: Grade 2 and 3 and dedifferentiated chondrosarcomas (CS) are frequently associated with isocitrate dehydrogenase (IDH) mutations and often exhibit a poor clinical outcome. Treatment is limited mainly to surgery. Defining IDH status (wild type (WT) and mutant) and the associated transcriptome may prove useful in determining other therapeutic options in these neoplasms. Methods: Formalin-fixed paraffin-embedded material from 69 primary and recurrent grade 2, 3 and dedifferentiated CS was obtained. DNA sequencing for IDH1 and IDH2 mutations (n = 47) and RNA sequencing via Nextseq 2000 (n = 14) were performed. Differentially expressed genes (DEGs) were identified and used to predict aberrant biological pathways with Ingenuity Pathway Analysis (IPA) software (Qiagen). Gene Set Enrichment Analyses (GSEA) using subsets C3, C5 and C7 were performed. Differentially expressed genes were validated by immunohistochemistry. Outcome analysis was performed using the Wilcoxon test. Results: A set of 69 CS (28 females, 41 males), average age 65, distributed among femur, pelvis, humerus, and chest wall were identified from available clinical material. After further selection based on available IDH status, we evaluated 15 IDH WT and 32 IDH mutant tumors as part of this dataset. Out of 15 IDH WT tumors, 7 involved the chest wall/scapula, while 1 of 32 mutants arose in the scapula. There were far more genes overexpressed in IDH WT tumors compared to IDH mutant tumors. Furthermore, IDH WT and IDH mutant tumors were transcriptomically distinct in the IPA and GSEA, with IDH mutant tumors showing increased activity in methylation pathways and endochondral ossification, while IDH WT tumors showed more activity in normal matrix development pathways. Validation immunohistochemistry demonstrated expression of WT1 and AR in IDH WT tumors, but not in IDH mutants. SATB2 was expressed in IDH mutant tumors and not in WT tumors. Outcome analysis revealed differences in overall survival between mutant and WT tumors (p = 0.04), dedifferentiated mutant and higher-grade (2, 3) mutant tumors (p = 0.03), and dedifferentiated mutant and higher-grade (2, 3) WT tumors (p = 0.03). The longest survival times were observed in patients with higher-grade WT tumors, while patients with dedifferentiated mutant tumors showed the lowest survival. Generally, patients with IDH WT tumors displayed longer survival in both the higher-grade and dedifferentiated groups. Conclusions: Grade 2, 3 and dedifferentiated chondrosarcomas are further characterized by IDH status, which in turn informs transcriptomic phenotype and overall survival. The transcriptome is distinct depending on IDH status, and implies different treatment targets.
Keywords: chondrosarcoma, RNA sequencing, isocitrate dehydrogenase, genomics
1. Introduction
Conventional chondrosarcoma is the second most common primary malignancy of bone [1,2,3,4]. Approximately 50% to 70% of these tumors harbor isocitrate dehydrogenase (IDH) 1 or IDH2 mutations [5,6,7]. IDH mutations are thought to represent an early driver event of oncogenesis in conventional chondrosarcoma [5,8].
Clinical outcome is stratified by grade, based on cellularity, increasing nuclear pleomorphism, and mitoses. Dedifferentiated chondrosarcoma (DDCS) is characterized by a biphasic histology; an area with cartilage matrix is juxtaposed to a highly cellular, pleomorphic sarcoma devoid of matrix. Similar mutations are present in both components, supporting a common origin [9]. However, histologic features do not inform IDH status in these malignancies. Approximately 10% of conventional chondrosarcomas undergo dedifferentiation and are associated with the worst prognosis [10]. Treatment options are limited because chondrosarcoma is resistant to adjuvant therapies and represents a mainly surgically treated disease [11].
National Comprehensive Cancer Network (NCCN 1.2024) guidelines for chondrosarcoma recommend wide excision with negative margins in surgically resectable cases [12]. However, unresectable, locally advanced, and widely metastatic chondrosarcoma require systemic therapy in an effort to control disease progression. Post-operative radiation therapy may be offered for conventional chondrosarcomas in select situations. Dedifferentiated chondrosarcoma may be treated with cytotoxic drug regimens similar to those used in osteosarcoma [12,13]. Conventional chondrosarcoma is typically resistant to chemotherapy and there are no uniformly established protocols [12,14]. Other novel therapies include tyrosine kinase inhibition by agents such as dasatinib [15] and pazopanib [16], and IDH1 inhibition with application of ivosidenib in patients with susceptible mutations [11]. Comprehensive molecular testing may be considered to determine potential treatment targets in individual patients [12].
In this work, we compared the genomic and transcriptomic characteristics of IDH mutant to wild type dedifferentiated and higher-grade (2, 3) chondrosarcomas. The results obtained may impact potential therapeutic options for these aggressive neoplasms.
2. Methods
2.1. Case Material
Formalin-fixed paraffin-embedded (FFPE) material from 69 primary and recurrent conventional higher-grade (2, 3) and dedifferentiated chondrosarcomas from 1999–2021 was obtained from the UPMC Department of Pathology under IRB approval (IRB 20050109). Hematoxylin and eosin slides were reviewed by a senior musculoskeletal pathologist (KS) and regions of interest were selected for tissue microarray construction (2 mm cores).
2.2. IDH Analysis
DNA was extracted from FFPE material and targeted amplification Sanger sequencing was performed for IDH1 and IDH2 mutations using ampliTAQ Gold360 PCR Master Mix (Applied Biosystems, Waltham, MA, USA) and capillary gel electrophoresis on the ABI3730xl (Applied Biosystems) [17].
Interpretable results were obtained in 47 cases.
2.3. RNA Sequencing
RNA was extracted from FFPE material (8 IDH-mutant, 6 IDH wild-type) after quality control (Tape Station HSD1000). Samples were sequenced via Nextseq 2000. The reverse-stranded paired-end RNA-Seq reads were checked for the presence of adapters and high-quality bases using FastQC (v 0.11.7). These high-quality reads were trimmed for the TruSeq adapters using Cutadapt (v 1.18). The trimmed reads were later mapped against the Ensembl human reference genome (GRCh38 v 107) using the STAR (v 2.7.9a) mapping tool. For better mapping outcomes, the STAR parameters were modified to utilize outFilterScoreMinOverLread and outFilterMatchNminOverLread, where both parameters were set to 0.3 instead of the standard. The output file from STAR was converted from SAM format to BAM format using SAMtools (v 1.9). Counts for expressed genes were generated using HT-Seq (v 0.11.2) and output was generated in text format. These count text files were then imported into the Bioconductor R package, edgeR (v 3.38.4). After importing the counts text files, ComBat seq was performed from the sva package (v 3.44.0) to compensate for the different sequencing batches without removing biological differences between samples. The edgeR package was then again utilized to identify differentially expressed genes based on the criteria of the genes having an expression count of absolute value log base 2 greater than 1 between two experimental conditions and a false discovery rate of less than 0.05 using an Exact test. Based on this standard, a single comparison of 6 wild type vs. 8 IDH mutant samples produced 743 differentially expressed genes.
After the differentially expressed genes were identified each list of genes along with their differential expression values were uploaded to Ingenuity Pathway Analysis (IPA) and used to identify aberrant biological pathways (FDR 0.05). Gene Set Enrichment Analyses (GSEA) were performed using the GSEA software (v 4.2.1 [build 5]) from the Broad Institute. Subsets C3, C5 and C7 were utilized in the GSEA. Immunohistochemistry: Differentially expressed genes (Wilms Tumor 1, WT1, Androgen Receptor, AR) and (Special AT-rich sequence-binding protein 2, SATB2 suggested by the GSEA) were validated on tissue microarrays (TMAs), and whole tumor sections using monoclonal antibodies (WT1 predilute, Ventana-Roche Oro, Valley, AZ, USA,; AR 1:100, Dako Santa Clara, CA, USA; SATB2 predilute, Cell Marque, Rocklin, CA, USA).
2.4. RT-qPCR Analysis
A subset of 5 IDH mutant samples of fresh frozen tissue were analyzed for the presence of SATB2, MMP13, and COL10A1 using commercially available primers. Briefly, RNA was extracted from a subset of 5 IDH mutant fresh frozen tissue samples according to the manufacturer’s protocol (Qiagen RNeasy Mini Kit, 74106 Germantown, MD, USA). RNA was analyzed using a one-step RT-qPCR protocol (Bio-Rad iTaq Universal SYBR Green One-Step Kit, 1725151) at a total reaction volume of 20 µL. GAPDH and SYMPK were used as housekeeping gene references. Each reaction was setup according to the manufacturer’s protocol using 150 ng RNA input for each respective sample reaction. Reactions were set up in a 384-well hard-shell plate and loaded on a CFX Opus 384 RT-qPCR instrument (Bio-Rad, Cat #12011452 Hercules, CA, USA). Expression data were analyzed using CFX Maestro Software (version 2.3).
2.5. Outcome Analysis
Clinical outcome data were obtained from UPMC electronic medical records and the UPMC Network Cancer Registry. Kaplan–Meier plots were generated and statistical analyses were performed using Wilcoxon tests and Prism9.
3. Results
3.1. Patients
Sixty-nine primary and recurrent conventional higher-grade (2, 3) and dedifferentiated conventional chondrosarcomas from 28 females and 41 males, average age 65 (range 14–91), were collected from the UPMC Department of Pathology archives. The primary sites included mainly the femur, pelvis, humerus, and chest wall (Table 1). All patients were treated surgically. Histologically, the dedifferentiated chondrosarcomas appeared similar to each other with an absent chondroid matrix, spindled-to-epithelioid and occasionally rhabdoid cells, mitotic activity, and necrosis.
Table 1.
Demographics of Grade 2, 3 and dedifferentiated chondrosarcomas with site, size and IDH status (- indicates data not available).
| IDH | ||||||
|---|---|---|---|---|---|---|
| Case | Age | Sex | Site | Size cm | Grade | Status |
| 1 | 37 | M | Arm | 22.5 | dd | Mut |
| 2 | 81 | M | Prox femur | 17.6 | dd | Mut |
| 3 | 82 | F | Thorax | 24.1 | 2 | WT |
| 4 | 57 | M | Pelvis | 9.8 | 3 | Mut |
| 5 | 70 | M | Sternum | - | 2 | WT |
| 6 | 85 | F | Humerus | - | dd | Mut |
| 7 | 72 | M | Femur | - | dd | Mut |
| 8 | 83 | F | Humerus | 7.2 | dd | - |
| 9 | 77 | M | Humerus | 9.2 | dd | Mut |
| 10 | 56 | M | Femur | - | 2 | Mut |
| Scapula | ||||||
| 11 | 62 | F | Chest | 5.8 | dd | Mut |
| 12 | 44 | M | Prox femur | 21.2 | dd | Mut |
| 13 | 76 | M | Chest wall | 12.5 | 3 | WT |
| 14 | 65 | M | Pelvis | 6.3 | 3 | - |
| 15 | 76 | M | Pelvis | - | 2 | Mut |
| 16 | 91 | M | Chest wall | 10 | dd | WT |
| 17 | 58 | M | Femur | - | 2 | Mut |
| 18 | 38 | M | Neck | - | 3 | Mut |
| 19 | 56 | F | Femur | 33 | dd | Mut |
| 20 | 74 | M | Pelvis | 13.2 | dd | WT |
| 21 | 61 | F | Pelvis | - | 3 | Mut |
| 22 | 84 | F | Hand | - | 2 | Mut |
| 23 | 54 | F | Femur | 9 | 2 | - |
| 24 | 79 | M | Prox femur | 17.5 | dd | Mut |
| 25 | 70 | F | Femur | - | 2 | Mut |
| 26 | 72 | F | Rib | 7 | 2 | WT |
| 27 | 62 | M | Pelvis | 9 | dd | Mut |
| 28 | 72 | F | Pelvis | 13.5 | 2 | - |
| 29 | 67 | M | Femur | 27 | dd | WT |
| 30 | 74 | M | Sacrum | 6.2 | dd | - |
| 31 | 57 | M | Humerus | 9.7 | 2 | - |
| 32 | 71 | M | Chest wall | 4.5 | 2 | - |
| 33 | 88 | F | Humerus | - | 2 | Mut |
| 34 | 14 | F | Pelvis | 6 | 2 | - |
| 35 | 62 | F | Chest wall | - | 2 | - |
| 36 | 83 | F | Femur | 6 | 3 | Mut |
| 37 | 80 | M | Pelvis | - | dd | WT |
| 38 | 63 | F | Talus | 11 | dd | - |
| 39 | 70 | M | Humerus | 7 | dd | - |
| 40 | 18 | F | Femur | - | 2 | - |
| 41 | 74 | M | Chest wall | 2 | 2 | - |
| 42 | 83 | M | Rib | 6 | dd | WT |
| 43 | 88 | M | Pelvis | 5.1 | 2 | - |
| 44 | 64 | M | Pelvis | 10.6 | dd | Mut |
| 45 | 65 | F | Femur | 16 | dd | - |
| 46 | 66 | M | Sternum | 8 | 2 | - |
| 47 | 70 | F | Humerus | - | dd | Mut |
| 48 | 50 | M | Femur | 5.5 | 2 | Mut |
| 49 | 88 | F | Femur | 11 | dd | Mut |
| 50 | 51 | M | Scapula | - | 2 | WT |
| 51 | 34 | F | Humerus | 5.5 | 2 | - |
| 52 | 49 | F | Thigh | - | 3 | - |
| 53 | 43 | M | Pelvis | 25 | 2 | WT |
| 54 | 74 | F | Femur | - | dd | WT |
| 55 | 56 | M | Pelvis | 18 | 2 | - |
| 56 | 84 | M | Pelvis | 9 | 2 | Mut |
| 57 | 64 | M | Femur | 11.5 | dd | WT |
| 58 | 82 | F | Femur | 3.5 | 2 | WT |
| 59 | 68 | M | Pelvis | 5.5 | 2 | Mut |
| 60 | 70 | M | Tibia | 9.5 | dd | WT |
| 61 | 67 | F | Humerus | 14 | dd | Mut |
| 62 | 56 | M | Femur | 10 | dd | Mut |
| 63 | 71 | F | Femur | 20.5 | dd | Mut |
| 64 | 32 | M | Pelvis | 18 | dd | Mut |
| 65 | 74 | F | Femur | 6 | dd | - |
| 66 | 49 | M | Pelvis | 10.5 | dd | - |
| 67 | 75 | M | Humerus | 9.5 | dd | - |
| 68 | 57 | M | Pelvis | 12 | dd | Mut |
| 69 | 56 | F | Pelvis | - | dd | Mut |
Abbreviations: dd, dedifferentiated; Mut, IDH mutant; WT, wild type; Prox, proximal.
3.2. IDH Analysis
DNA analysis of FFPE tissue from 47 high-grade and dedifferentiated chondrosarcomas revealed 15 WT and 32 mutant tumors. Of the mutants, 20 were IDH1, and 12 were IDH2. Correlation with the site of disease demonstrated chest wall/scapula involvement in 7 of 15 WT tumors, while only 1 of 32 mutant tumors was located in the scapula, and none in the chest wall (Table 1).
3.3. RNA Sequencing
Differentially expressed genes were entered into the Qiagen IPA to predict associated pathways (IPA, FDR 0.05) and showed that IDH WT and mutant tumors were transcriptomically distinct (Figure 1). Furthermore, IDH mutant tumors were associated with DNA methyltransferase pathways. Genes implicated in malignant behavior included increased expression of collagen 10 alpha 1 (COL10A1), matrix metalloproteinase 13 (MMP13), TP53, and WRAP53. By contrast, the IPA implicated long non-coding RNAs such as Braveheart (BvHT) in WT tumors. RNA-sequencing exhibited higher differentially expressed genes (DEGs) in WT tumors than in the mutants. Several development-associated genes such as SRY-box 2 (SOX2), insulin-like growth factor-2 (IGF2) and the Homeobox family of genes revealed increased expression in the WT tumors. The data were also interrogated via GSEA using subsets C3, C5 and C7. Selected differentially expressed genes (DEGs) correlating with pathways highlighted in the IPA are listed in Table 2. The complete lists of DEGs are included in the Supplementary Materials (Data S1 and S2).
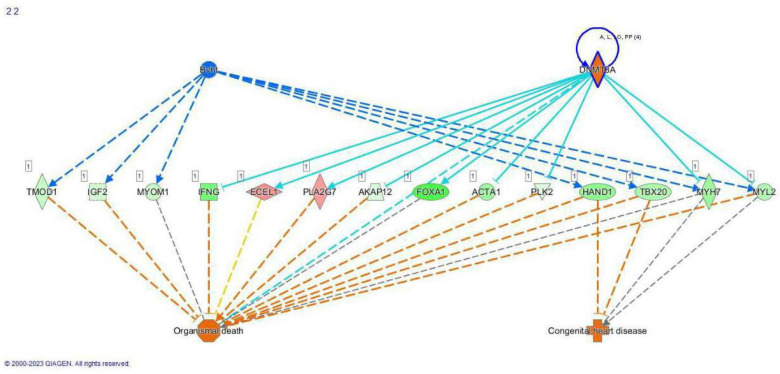
Figure 1.
Qiagen IPA diagram demonstrating differences in gene expression between IDH WT (indicated by BvHT in blue) and IDH mutant (indicated by DNA methyltransferase 3 alpha, DNMT3A in an orange diamond rimmed by purple) tumors. Implicated pathways are generated by Qiagen IPA software based on DEG data input and suggest that endothelin-converting enzyme-like 1 (ECEL1) and phospholipase A2 group VII (PLA2G7) may mediate cell death (solid arrows) in the IDH mutant tumors. Dashed lines are suggested pathways.
Table 2.
Selected differentially expressed genes in IDH WT vs. mutant chondrosarcomas (grades 2, 3 and dedifferentiated).
| DEG Higher in IDH WT | DEG Higher in IDH Mut |
|---|---|
| GREM1 | COL10A1 |
| TMEM52 | MMP13 |
| FOXA1 | HHIP |
| ALX Homeobox 1, 3 | IBSP |
| HOXA2 | COL26A1 |
| WT1 | WRAP53 |
| BMP7 | TP53 |
| SOX2 | |
| IGF2 | |
| MAGEA1 | |
| NES | |
| ERG | |
| FGF18 | |
| DKK2 |
Immunohistochemical stains were performed for WT1, AR, and SATB2. Immunohistochemical stains were chosen based on gene expression or highlighted in the GSEA and ease of interpretation (nuclear staining). WT1 and AR were associated with the WT tumors. Although SATB2 did not meet defined cutoffs for a differentially expressed gene, it was identified in the GSEA and immunohistochemistry revealed positivity only in the mutant tumors (Figure 2 and Figure 3 and Supplementary Materials Data S3). Additionally, a subset of IDH mutant tumors was subjected to RT-qPCR testing for MMP13, Col10A1 and SATB2 expression (Table 3).
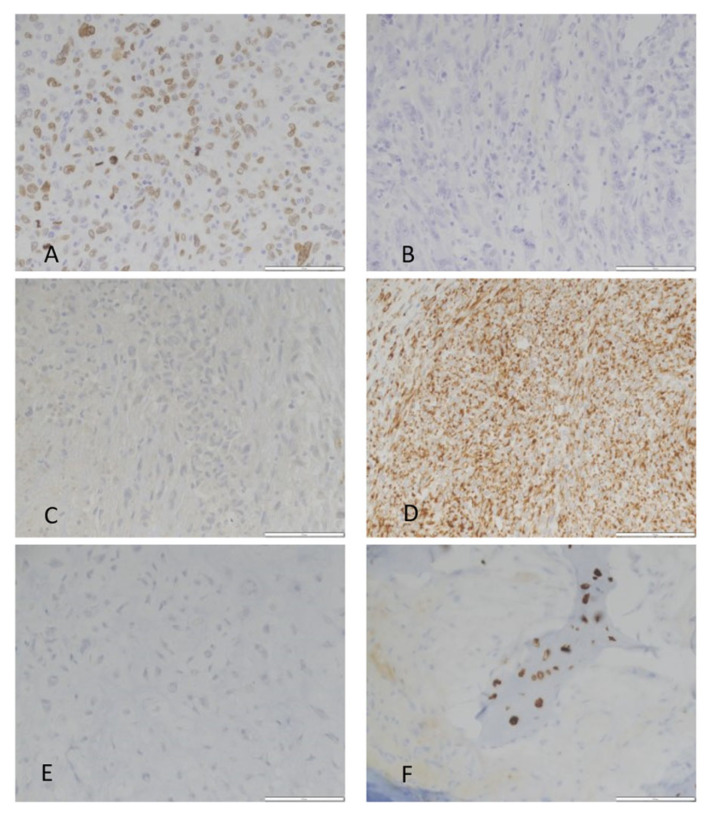
Figure 2.
Immunohistochemical validation of SATB2, WT1 and AR in IDH mutant and wild type chondrosarcoma: (A) SATB2 shows nuclear positivity in IDH mutant CS. (B) SATB2 immunostaining is negative in WT CS. (C) WT1 immunostaining is negative in dedifferentiated IDH mutant CS; (D) WT1 cytoplasmic and focal nuclear positivity in an IDH WT dedifferentiated CS; (E) AR negative IDH mutant CS; and (F) AR positive IDH WT CS. All photomicrographs 400×.
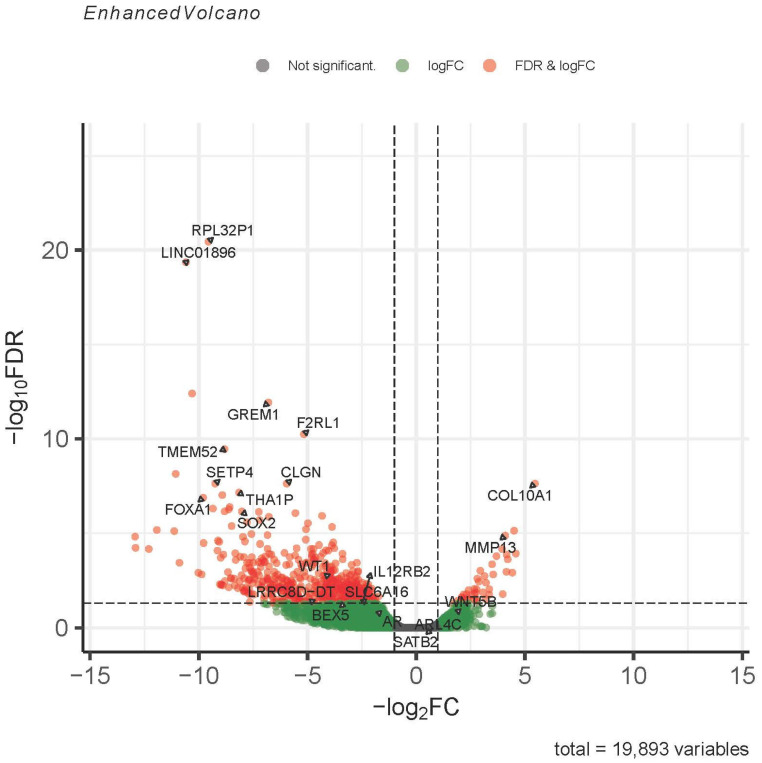
Figure 3.
Volcano plot comparing gene expression in IDH mutant/WT CHS. Expression of development-related genes are decreased in IDH mutant CHS compared to wild type whereas transcripts related to endochondral ossification are increased in the IDH mutant population compared to WT. Transcripts highlighted in red pass abs log2FC > 1 and FDR < 0.05; genes in the green zone pass only abs log2FC > 1; and transcripts highlighted in grey zone do not meet either FC or FDR thresholds. The gene expression used for validation by immunohistochemistry includes WT1 (red zone), AR (green zone) and SATB2 (grey zone). SATB2 was highlighted by a GSEA gene set (C3) in the IDH mutant tumors (see Supplementary Materials Data S3).
Table 3.
Relative expression of genes of interest (compared to housekeeping genes) in IDH mutant fresh frozen chondrosarcomas using qPCR.
| Case | COL10A1 | MMP13 | SATB2 | IDH Mutation | Grade |
|---|---|---|---|---|---|
| 9 | Expressed | Expressed | Expressed | IDH1 R132S | Dedifferentiated |
| 6 | Expressed | Low Expression | Expressed | IDH2 R172S | Dedifferentiated |
| 17 | Expressed | Expressed | Expressed | IDH1 R132L | 2 |
| 4 | High Expression | High Expression | High Expression | IDH1 R132C | 3 |
| 2 | High Expression | High Expression | High Expression | IDH1 R132C | Dedifferentiated |
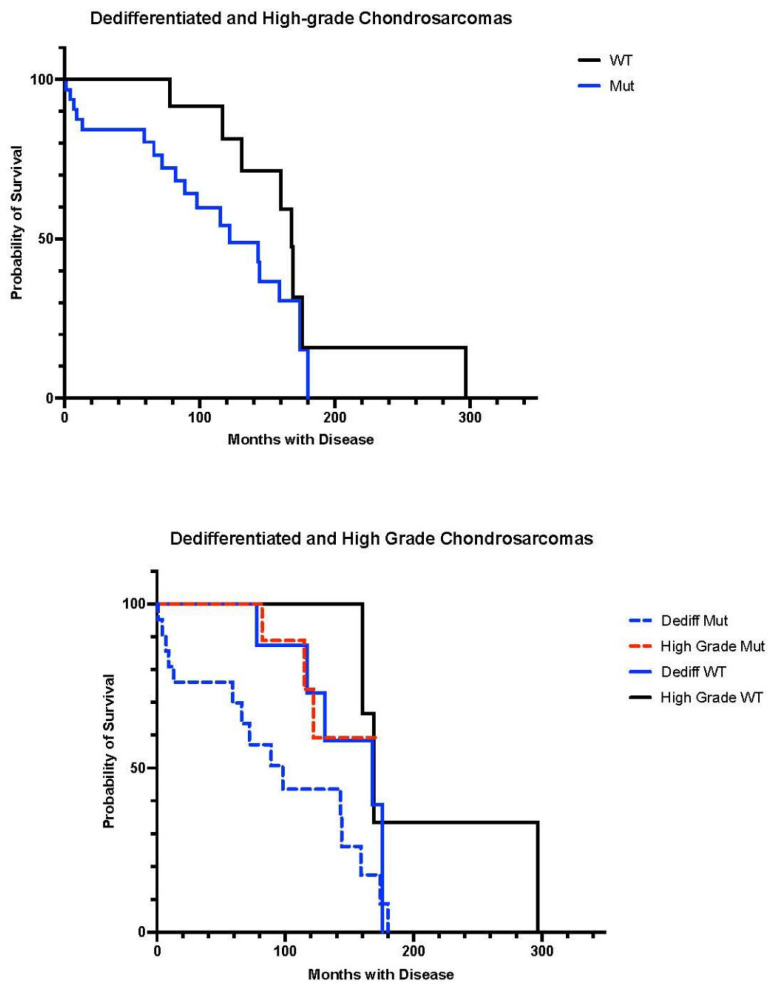
Outcome analysis was performed and Kaplan–Meier plots were generated (Figure 4). The difference in survival between IDH mutant and WT tumors was statistically significant (p = 0.04). Statistically significant differences were also seen between dedifferentiated mutant and high-grade mutant (p = 0.03), and dedifferentiated mutant and high-grade WT tumors (p = 0.03) In general, wild-type tumors showed a survival advantage.
Figure 4.
Kaplan–Meier plots of IDH mutant (both IDH1 and IDH2) vs. WT overall survival in dedifferentiated and high-grade (2, 3) chondrosarcomas. Top plot shows WT vs. mutant tumors, while bottom plot compares dedifferentiated and high-grade conventional CS stratified by IDH status.
4. Discussion
Histologic grading of conventional chondrosarcoma is correlated with outcome, with high-grade tumors demonstrating greater metastatic potential and mortality than lower-grade disease [1,5]. Dedifferentiated chondrosarcomas (DDCS) are thought to derive from conventional chondrosarcoma, and rarely from enchondroma or osteochondroma [10], and represent a clinically aggressive form of chondrosarcoma. DDCS is associated with a 7–24% 5-year survival [1]. In the majority of cases, treatment is limited mainly to surgery, leaving those with advanced disease without effective therapeutic options. In order to develop new therapies for higher-grade and dedifferentiated CS, the genomic and transcriptomic landscape of these tumors must be understood.
It is known that IDH status plays important prognostic and therapeutic roles in several malignancies such as acute myeloid leukemia, cholangiocarcinoma, and glioma [18], among others. In the central nervous system, IDH mutant infiltrating gliomas (oligodendroglioma and IDH mutant astrocytoma) show significantly longer survival and better clinical outcomes than their IDH wild type counterparts (glioblastoma, IDH wild type) [19]. The clinical outcomes are so divergent that IDH mutational status now defines these diagnostic entities in the most recent WHO classifications [20]. It is interesting to note that in the current study, IDH mutational status has the opposite prognostic impact in chondrosarcoma. IDH mutant gliomas additionally have distinct morphology and co-occurring molecular alterations; 1p/19q-codeletion and TERT promoter mutation in oligodendroglioma and p53/ATRX mutations in IDH mutant astrocytoma. By contrast, IDH mutant and WT chondrosarcomas are histologically identical, and at present are treated as a single disease entity.
IDH mutations affect 38–86% of conventional chondrosarcomas [21]. IDH status appeared to inform the prognosis of higher-grade CS and DDCS in our series. IDH WT cases demonstrated prolonged survival in comparison to IDH mutants. That IDH mutations in chondrosarcoma confer a worse outcome was also seen in Nakegawa’s study of 38 cases in 2022 [2]. In our study, IDH1 and IDH2 mutant cases were combined for outcome analysis due to a small number of IDH2 mutant chondrosarcomas in our series. This is commensurate with the literature, as it is known that chondrosarcomas are affected more often by IDH1 mutations than IDH2 [5]. Differentially expressed genes derived from RNA sequencing delineate differences between the IDH mutant and WT chondrosarcoma cohorts. They suggest that the molecular pathways utilized for tumor growth, maintenance, and malignant phenotype could be different between the two groups. In a study of 350 cases of chondrosarcoma by Cross et al., it was proposed that IDH mutant and wild type tumors utilized different molecular pathways and, furthermore, that IDH2 mutant and high-grade chondrosarcomas were more often associated with TERT mutations [5]. TERT mutations were also identified in approximately 35% of dedifferentiated chondrosarcomas in Nacev’s study [22]. We did not assess for TERT promoter mutations in our cohort.
Mutations in IDH1/2 are thought to be oncogenic through the aberrant production of D2-hydroxyglutarate (D2-HG) [7]. D2-HG is an oncometabolite that leads to DNA and histone hypermethylation and associated genome-wide alterations in gene expression [23]. Hypermethylation of IDH mutant chondrosarcomas activates proliferation and glycolysis [24], and is associated with higher histologic grade [25]. Amary et al. reported that D2-HG levels were elevated in patients with IDH mutated chondrosarcomas arising in the setting of Ollier and Maffucci syndromes [26], and Mohammad et al. showed increased 2-HG levels due to IDH1 mutations in chondrosarcoma [27].
IDH mutant chondrosarcoma demonstrated increased expression of collagen 10 alpha 1 (COL10A1) and matrix metalloproteinase 13 (MMP13) in our study. The elevated MMP13 expression suggested that extracellular matrix breakdown may promote aggressive behavior [28]. Vascular invasion may also be facilitated by MMP13 [29]. Furthermore, COL10A1 and MMP13 are involved in endochondral ossification, a process conserved in matrix-producing chondrosarcoma, and downregulated in dedifferentiated chondrosarcoma [24]. The Hedgehog pathway was also implicated in our study, by higher expression of HHIP (Table 2). Hedgehog signaling was noted in a study of chondrosarcomas by Isenlys et al., and Tiet et al. showed increased expression of PTCH1 and GlI1, target genes of the Hedgehog pathway [30,31]. TP53 showed increased expression in the mutant tumors. TP53 overexpression has been previously reported in chondrosarcoma by several authors [2,5,6,22,32].
Additionally, WRAP53 was overexpressed in the IDH-mutant tumors, suggesting that the tumor cells may achieve immortality by telomere lengthening [33].
Furthermore, fewer genes showing increased expression were seen in the IDH mutant cohort, suggesting loss of genetic material and/or increased chromosomal instability as compared to the WT group. Special AT-rich sequence-binding protein 2 (SATB2) was expressed only in the IDH mutants by immunohistochemistry, corroborating its presence in the GSEA, and possibly representing post-translational mechanisms for increased expression. Confirmation of MMP13, COL10A1 and SATB2 was attempted by RT-qPCR in the IDH mutant samples, and expression of these three genes was detected in all samples tested (n = 5). Furthermore, higher expression of MMP13, COL10A1 and SATB2 was found in two IDH mutant samples, both demonstrating an IDH1 R132C mutation, suggesting that a specific IDH1 mutation involving R132C may play a role in the expression of these genes. However, the case numbers were too small to form a conclusion. This finding would require a multi-center large chondrosarcoma cohort to study.
With regard to the IDH WT tumors, more differentially expressed developmental genes and pathways appeared to play roles in their genesis than seen in the mutants. These included SRY-box 2 (SOX2), Bone morphogenetic protein 7 (BMP7), Homeobox A2 (HOXA2), Gremlin 1 (GREM1), Forkhead box protein A1 (FOXA1), Transmembrane protein 52 (TMEM52), Insulin-like growth factor 2 (IGF2), melanoma antigen (MAGEA1), and ALX1 and ALX3 Homeobox genes. Braveheart (BvHT) was implicated by the IPA analysis. These suggested the involvement of long non-coding RNAs in tumor development, as well as in promotion of tumor cell survival and proliferation. Several of these genes participate in normal embryonic development [34,35,36]. Long non-coding RNAs are involved in cardiovascular disease and many cancers [37]. That MAGEA1 is expressed in the WT tumors and not in the mutants suggests that MAGEA1 is epigenetically silenced in the mutants, possibly by DNA methyltransferase 1 (DNMT1) and histone deacetylation [38]. MAGEA1 expression may contribute to malignancy in the WT tumors. WT1 and AR expression were identified in the WT cases and were used as immunohistochemical validation markers.
Interestingly, IDH WT tumors were identified more often than IDH mutant tumors in the chest wall and scapula, a finding also reported by Cross et al. [5]. This finding may be related to reduced endochondral ossification involved in chest wall development as compared to the long bones. This hypothesis requires further investigation.
Incorporating differential gene expression in IDH WT and mutant chondrosarcomas to inform present and future treatment strategies represents a substantial and potentially exciting challenge. With respect to current systemic therapy, while dedifferentiated chondrosarcoma is often treated along an osteosarcoma paradigm using cytotoxic drugs [12,13], chemotherapy in conventional chondrosarcoma has poor activity [14]. However, tyrosine kinase inhibition by dasatinib [15] or pazopanib [16] may offer benefit in terms of disease control and occasional responses in conventional chondrosarcoma. A phase I trial demonstrated the promising activity of an IDH1 inhibitor, ivosidenib, particularly in conventional chondrosarcoma, where the progression-free survival was greater than in dedifferentiated chondrosarcoma, although the study was not powered to specifically compare those groups [11]. Ivosidenib is currently listed on the National Comprehensive Cancer Network (NCCN) guidelines for conventional and dedifferentiated chondrosarcoma patients with susceptible mutations [12].
Based on the RNA-seq data generated as part of this study, anti-methylation drugs such as Decitabine could be explored pre-clinically in IDH mutant tumors, as well as inhibitors of IDH1 [11,18,21] or IDH2 [8]. ARL4C expression in the IDH mutants (Supplementary Materials Data S4) could represent a target for ASO-1316 [39]. For IDH WT tumors, Isotretinoin targeting SOX2 may be considered. Chemotherapeutic regimens including Doxorubicin, Carboplatin, Cyclophosphamide or Doxetaxel could be used to target WT1 [40]. Most likely, due to the complexity of pathways and number of genes involved, combination therapy should be tested preclinically.
Our findings are not without limitations because of a small sample size; however, our study uncovered an intriguing survival advantage of IDH wild type chondrosarcomas. The analysis of larger patient cohorts may uncover unique characteristics of IDH2 mutant tumors that our analysis was not able to capture. In addition, work in the future may be improved by better methods of extracting high-quality RNA from FFPE samples with chondroid matrix present. These methods may decrease the sample extraction batch effect that impacted our bioinformatic analysis.
5. Conclusions
In summary, dedifferentiated and higher-grade chondrosarcomas demonstrate genetic and probable epigenetic changes attributed in part to IDH status. The mutant and wild-type tumors utilize different molecular pathways which likely correlate with malignant behavior. In our admittedly small series, clinical outcomes are significantly different between IDH wild type and mutant groups. The combination of IDH mutated and dedifferentiated chondrosarcomas demonstrates the worst prognosis. However, dedifferentiated wild type tumors confer a better prognosis, which could be used in counseling patients. Future studies should explore whether targeting specific IDH mutations can be used to effectively inform therapeutic strategies for this aggressive disease with few options besides surgery.
Acknowledgments
The authors thank Benjamin Nacev of UPMC Department of Medicine for his thoughtful comments on the manuscript.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers16020247/s1, Data S1: Upregulated IDH Differentially Expressed Genes; Data S2: WT Upregulated Differentially Expressed Genes; Data S3: Heatmap of IDH mutant and WT chondrosarcomas with SATB2; Data S4: Heatmap of IDH mutant and WT CS with ARL4C; Webpage Details for gene set CTCTATG_MIR368 (C3 GSEA with SATB2); Webpage Details for gene set KAZMIN PBM (C7 GSEA with ARL4C).
Author Contributions
Conceptualization: K.S., K.W. and A.D.; Methodology: K.W., T.H., R.B., S.B. and L.P.; Software U.C. and A.C.; Validation: U.C., A.C. and K.S.; Formal Analysis: U.C., A.C., K.W., K.S., T.H. and A.S.-V.; Resources: K.W., K.S. and U.C.; Data Curation U.C., K.W. and K.S.; Writing—Original draft K.S.; Writing—Review and editing: A.D., K.W., I.L., H.S.-R. and D.M.; Supervision: K.W.; Funding Acquisition: K.S. and K.W. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study used archival material only and was approved by the Institutional Review Board of the University of Pittsburgh (IRB Study 20050109, accessed on 19 May 2020).
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Conflicts of Interest
The authors have no conflicts of interest to disclose.
Funding Statement
Pittsburgh Cure Sarcoma AWD00004925, Gibsonia, PA, USA. Gift through University of Pittsburgh Physicians to Dr. Weiss.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.WHO Classification of Tumours Editorial Board . WHO Classification of Tumours Series. 5th ed. Volume 3 IARC; Lyon, France: 2020. Soft tissue and bone tumours. [Google Scholar]
- 2.Nakagawa M., Sekimizu M., Endo M., Kobayashi E., Iwata S., Fukushima S., Yoshida A., Kitabayashi I., Ichikawa H., Kawai A., et al. Prognostic impact of IDH mutations in chondrosarcoma. J. Orthop. Sci. 2022;27:1315–1322. doi: 10.1016/j.jos.2021.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Micaily I., Roche M., Ibrahim M., Martinez-Outschoorn U., Mallick A. Metabolic pathways and targets in chondrosarcoma. Front. Oncol. 2021;11:772263. doi: 10.3389/fonc.2021.772263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H., Puviindran V., Puviindran N., Ding X., Shen L., Tang Y., Tsushima H., Yahara Y., Ban G., Zhang G. Distinct roles of glutamine metabolism in benign and malignant cartilage tumors with IDH mutations. J. Bone Miner. Res. 2022;37:983–996. doi: 10.1002/jbmr.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cross W., Lyskjaer I., Lesluyes T., Hargreaves S., Strobl A., Davies C., Waise S., Hames-Fathi S., Oukrif D., Ye H., et al. A genetic model for central chondrosarcoma evolution correlates with patient outcomes. Genome Med. 2022;14:99. doi: 10.1186/s13073-022-01084-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miwa S., Yamamoto N., Hayashi K., Takeuchi A., Igarashi K., Tsuchiya H. Therapeutic targets and emerging treatments in advanced chondrosarcoma. Int. J. Mol. Sci. 2022;23:1096. doi: 10.3390/ijms23031096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cairns R., Mak T. Oncogenic isocitrate dehydrogenase mutations: Mechanisms, models and clinical opportunities. Cancer Discov. 2013;3:730–741. doi: 10.1158/2159-8290.CD-13-0083. [DOI] [PubMed] [Google Scholar]
- 8.Molenaar R., Wilmink J. IDH1/2 mutations in cancer stem cells and their implications for differentiation therapy. J. Histochem. Cytochem. 2022;70:83–97. doi: 10.1369/00221554211062499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bovee J., Cleton-Jansen A., Rosenberg C., Taminiau A., Cornelisse C.J., Hogendoorn P. Molecular genetic characterization of both components of a dedifferentiated chondrosarcoma, with implications for its histogenesis. J. Pathol. 1999;189:454–462. doi: 10.1002/(SICI)1096-9896(199912)189:4<454::AID-PATH467>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 10.Nielsen G., Rosenberg A. Diagnostic Pathology Bone. 2nd ed. Elsevier; Philadelphia, PA, USA: 2017. [Google Scholar]
- 11.Tap W., Villalobos V., Cole G., Burris H., Janku F., Mir O., Beeram M., Wagner A., Jiang L., Wu B., et al. Phase 1 study of the mutant IDH1 inhibitor Ivosidenib: Safety and clinical activity in patients with advanced chondrosarcoma. J. Clin. Oncol. 2020;38:1693–1701. doi: 10.1200/JCO.19.02492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.National Comprehensive Cancer Network . NCCN Clinical Practice Guidelines in Oncology, Bone Cancer, Version 1.2024. National Comprehensive Cancer Network; Plymouth Meeting, PA, USA: 2023. [Google Scholar]
- 13.Mitchell A., Ayoub K., Mangham D., Grimer R., Carter S., Tillman R. Experience in the treatment of dedifferentiated chondrosarcoma. J. Bone Jt. Surg. Br. 2000;82:55–61. doi: 10.1302/0301-620X.82B1.0820055. [DOI] [PubMed] [Google Scholar]
- 14.Italiano A., Mir O., Cioffi A., Palmerini E., Piperno-Neumann S., Perrin C., Chaigneau L., Penel N., Duffaud F., Kurtz J., et al. Advanced chondrosarcomas: Role of chemotherapy and survival. Ann. Oncol. 2013;24:2916–2922. doi: 10.1093/annonc/mdt374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuetze S., Bolejack V., Choy E., Ganjoo K., Staddon A., Chow W., Tawbi H., Samuels B., Patel S., von Mehren M., et al. Phase 2 study of dasatinib in patients with alveolar soft part sarcoma, chondrosarcoma, chordoma, epithelioid sarcoma, or solitary fibrous tumor. Cancer. 2017;123:90–97. doi: 10.1002/cncr.30379. [DOI] [PubMed] [Google Scholar]
- 16.Chow W., Frankel P., Ruel C., Araujo D., Milhem M., Okuno S., Hartner L., Undevia S., Staddon A. Results of a prospective phase 2 study of pazopanib in patients with surgically unresectable or metastatic chondrosarcoma. Cancer. 2020;126:105–111. doi: 10.1002/cncr.32515. [DOI] [PubMed] [Google Scholar]
- 17.Horbinski C., Kofler J., Kelly L., Murdoch G., Nikiforova M. Diagnostic use of IDH1/2 mutation analysis in routine clinical testing of formalin fixed, paraffin embedded glioma tissues. J. Neuropathol. Exp. Neurol. 2009;68:1319–1325. doi: 10.1097/NEN.0b013e3181c391be. [DOI] [PubMed] [Google Scholar]
- 18.Pirozzi C., Yan H. The implications of IDH mutations for cancer development and therapy. Nat. Rev. Clin. Oncol. 2021;18:645–661. doi: 10.1038/s41571-021-00521-0. [DOI] [PubMed] [Google Scholar]
- 19.Han S., Liu Y., Cai S., Qian M., Ding J., Larion M., Gilbert M., Yang C. IDH mutation in glioma: Molecular mechanisms and potential therapeutic targets. Br. J. Cancer. 2020;122:1580–1589. doi: 10.1038/s41416-020-0814-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.WHO Classification of Tumours Editorial Board . WHO Classification of Tumours Series. 5th ed. Volume 6 IARC; Lyon, France: 2021. Central Nervous System Tumours. [Google Scholar]
- 21.Tian W., Zhang W., Wang Y., Jin R., Wang Y., Guo H., Tang Y., Yao X. Recent advances in IDH1 mutant inhibitor in cancer therapy. Front. Pharmacol. 2022;13:982424. doi: 10.3389/fphar.2022.982424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nacev B., Sanchez-Vega F., Smith S., Antonescu B., Rosenbaum E., Shi H., Tang C., Socci N., Rana S., Gularte-Merida R., et al. Clinical sequencing of soft tissue and bone sarcomas delineates diverse genomic landscapes and potential therapeutic targets. Nat. Commun. 2022;13:3405. doi: 10.1038/s41467-022-30453-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo C., Pirozzi C., Lopez G., Yan H. Isocitrate dehydrogenase mutations in gliomas: Mechanisms, biomarkers and therapeutic target. Curr. Opin. Neurol. 2011;24:648–652. doi: 10.1097/WCO.0b013e32834cd415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicolle R., Ayadi M., Gomez-Brouchet A., Armenoult L., Banneau G., Elarouci N., Tallegas M., Decouvelaere A., Aubert S., Redini F., et al. Integrated molecular characterization of chondrosarcoma reveals critical determinants of disease progression. Nat. Commun. 2019;10:4622. doi: 10.1038/s41467-019-12525-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venneker S., Kruisselbrink A., Baranski Z., Palubeckaite I., Briaire-de Bruijn I., Oosting J., French P., Danen E., Bovee J. Beyond the influence of IDH mutations: Exploring epigenetic vulnerabilities in chondrosarcoma. Cancers. 2020;12:3589. doi: 10.3390/cancers12123589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amary M., Bacsi K., Maggiani F., Damato S., Halai D., Berisha F., Pollock R., O’Donnell P., Grigoriadis A., Diss T., et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J. Pathol. 2011;224:334–343. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 27.Mohammad N., Wong D., Lum A., Lin J., Ho J., Lee C., Yip S. Characterization of isocitrate dehydrogenase 1/isocitrate dehydrogenase 2 gene mutation and the d-2-hydroxyglutarate oncometabolite level in dedifferentiated chondrosarcoma. Histopathology. 2020;76:722–730. doi: 10.1111/his.14018. [DOI] [PubMed] [Google Scholar]
- 28.Stoeckl S., Lindner G., Li S., Schuster P., Haferkamp S., Wagner F., Prodinger P., Multhoff G., Boxberg M., Hillman A., et al. SOX9 knockout induces polyploidy and changes sensitivity to tumor treatment strategies in a chondrosarcoma cell line. Int. J. Mol. Sci. 2020;21:7627. doi: 10.3390/ijms21207627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blumer M. Bone tissue and histological and molecular events during development of the long bones. Ann. Anat. 2021;235:151704. doi: 10.1016/j.aanat.2021.151704. [DOI] [PubMed] [Google Scholar]
- 30.Iseulys R., Gomez-Brouchet A., Bouvier C., Du Bouexic G., Karanian M., Blay J., Dutour A. The immune landscape of chondrosarcoma reveals an immunosuppressive environment in the dedifferentiated subtypes and exposes CSFR1 + macrophages as a promising therapeutic target. J. Bone Oncol. 2020;20:100271. doi: 10.1016/j.jbo.2019.100271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tiet T., Hopyan S., Nadesan P., Gokgoz N., Poon R., Lin A., Yan T., Andrulis I., Alman B., Wunder J. Constitutive hedgehog signaling in chondrosarcoma up-regulates tumor cell proliferation. Am. J. Pathol. 2006;168:321–330. doi: 10.2353/ajpath.2006.050001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meijer D., de Jong D., Pansuriya T.C., van den Akker B., Picci P., Szuhai K., Bovee J. Genetic characterization of mesenchymal, clear cell and dedifferentiated chondrosarcoma. Genes Chromosomes Cancer. 2012;51:899–909. doi: 10.1002/gcc.21974. [DOI] [PubMed] [Google Scholar]
- 33.Gadelha R., Machado C., de Pinho Pessoa F., Pantoja L., Barreto I., Ribeiro R., de Moraes Filho M., de Moraes M., Khayat A., Moreira-Nunes C. The role of WRAP53 in cell homeostasis and carcinogenesis onset. Curr. Issues Mol. Biol. 2022;44:5498–5514. doi: 10.3390/cimb44110372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 35.Liu Z., Shen F., Wang H., Li A., Wang J., Du L., Liu B., Zhang B., Lian X., Pang B., et al. Abnormally high expression of HOXA2 as an independent factor for poor prognosis in glioma patients. Cell Cycle. 2020;19:1632. doi: 10.1080/15384101.2020.1762038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Pagter-Holthuizen P., Jansen M., van der Kammen R., van Schaik F., Sussenback J. Differential expression of the human insulin-like growth factor II gene. Characterization of the IGF-II mRNAs and an mRNA encoding a putative IGF-II associated protein. Biochem. Biophys. Acta. 1988;950:282–295. doi: 10.1016/0167-4781(88)90124-8. [DOI] [PubMed] [Google Scholar]
- 37.Palmini G., Marini F., Brandi M. What is new in the miRNA world regarding osteosarcoma and chondrosarcoma? Molecules. 2017;22:417. doi: 10.3390/molecules22030417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Florke Gee R., Chen H., Lee A., Daly C., Wilander B., Tacer K., Potts P. Emerging roles of the MAGE protein family in stress response pathways. J. Biol. Chem. 2020;295:16121–16155. doi: 10.1074/jbc.REV120.008029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kimura K., Matsumoto S., Harada T., Morii E., Nagatomo I., Shintani Y., Kikuchi A. ARL4C is associated with initiation and progression of lung adenocarcinoma and represents a therapeutic target. Cancer Sci. 2020;111:951–961. doi: 10.1111/cas.14303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. [(accessed on 1 September 2023)]. Available online: www.genecards.org.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are contained within the article and Supplementary Materials.