Abstract
We describe a case of Clostridium intestinale bacteremia in a previously healthy adolescent female presenting with fever and abdominal pain. The bacterium was definitively identified via 16S rRNA gene sequencing. This is the first report, in the world literature, of human infection caused by this microorganism.
CASE REPORT
A 17-year-old girl in previously good health presented to hospital with a 1-day history of severe sharp, nonradiating right lower quadrant (RLQ) abdominal pain associated with a 3-day history of fever, anorexia, nausea, vomiting, diarrhea, and dizziness. The pain was worse with activity. There was no history of trauma. Her last normal menstrual period began 10 days prior to the onset of her complaints. She denied any gynecologic symptoms and had no other active health problems. She was not taking any medications other than an oral contraceptive. There was no history of recent foreign travel. On examination, she appeared to be in some distress. Her temperature was 38.0°C. Her abdomen was soft but markedly tender in the right lower quadrant on deep palpation. Bowel sounds were present, and there was no abdominal guarding or rebound tenderness. The rest of the physical examination, including a gynecologic exam, was unremarkable. Laboratory investigations revealed a normal complete blood count (hemoglobin, 145 g/liter; white blood cells, 7.4 × 109/liter; platelets, 344 × 109/liter), normal electrolytes, and an unremarkable abdominal ultrasound exam, although the appendix could not be easily visualized. The clinical diagnosis remained uncertain, although acute appendicitis was considered. Tests for sexually transmitted diseases were not performed. She was admitted to hospital without a defined etiology of her abdominal pain. Two sets of BacT/Alert PF Pediatric FAN (Biomerieux Inc., Durham, N.C.) blood cultures were collected upon admission. She was managed conservatively with intravenous fluids and anti-inflammatory agents.
Within 48 h, both sets of blood cultures were positive for large, gram-positive, boxcar-like, rod-shaped bacteria resembling Clostridium spp., after which empirical therapy with intravenous ampicillin was begun. After subculture of the blood culture bottles, growth was observed aerobically on 5% sheep blood agar medium and anaerobically on brucella blood agar medium (PML Microbiologicals, Wilsonville, Oreg.). After 48 h of anaerobic incubation on brucella blood agar medium, colonial growth was scant. Gram staining revealed the presence of gram-positive bacilli, with terminal endospores seen occasionally. The organism was motile but nonhemolytic. Growth was not observed on kanamycin-vancomycin laked blood or Bacteroides bile esculin agars (PML Microbiologicals). Sensitivity to vancomycin (5 μg) and kanamycin (1,00 μg) and resistance to colistin (10 μg) special-potency identification disks were demonstrated. Tests for catalase, lecithinase, and lipase and reverse Christie-Atkins-Munch-Peterson test results were negative. The organism was identified as an aerotolerant Clostridium sp. on the basis of its phenotypic characteristics and its ability to grow in an aerobic atmosphere with 5% carbon dioxide. However, it was further characterized by partial 16S rRNA gene sequencing using MicroSeq 500 kits and an ABI Prism 3100 genetic analyzer (Applied Biosystems, Foster City, Calif.). Full-length sequencing of the 16S rRNA gene was subsequently performed for more definitive identification. A GenBank BLAST search revealed a match of greater than 99% of the full-length 16S rRNA gene profile with that of a previously characterized strain of Clostridium intestinale (ATCC 49213; GenBank accession no. X76740). Further phylogenetic analysis indicated that our sequence (GenBank accession no. AY781385) clustered tightly with the 16S rRNA sequence of the single C. intestinale strain in the GenBank database and helped confirm the species identification of the present isolate. The organism was susceptible to penicillin G (MIC = 0.5 μg/ml), clindamycin (MIC = 1.5 μg/ml), and metronidazole (MIC = 0.094 μg/ml) by E-test methodology. The patient's fever, abdominal pain, and other symptoms gradually resolved over the course of her hospital stay. After receiving intravenous ampicillin for 48 h, she was discharged home on a 7-day course of oral amoxicillin therapy.
The genus Clostridium is one of the largest and most phylogenetically diverse genera of bacteria, being comprised of more than 150 unique species of obligately anaerobic or aerotolerant endospore-forming, usually gram-positive, rod-shaped bacteria (1, 3, 9). Clostridia are widely distributed in nature, existing primarily as apparently harmless soil saprophytes or as part of the permanent or transient normal gastrointestinal and vaginal bacterial flora of humans and other animals (1). They are also one of the most commonly encountered groups of anaerobic bacteria recovered from human clinical specimens (1, 8, 12). Several members of this genus, most notably C. perfringens, have been implicated in a wide array of infectious (usually arising from colonizing strains) and/or toxin-mediated (usually arising from exogenously acquired strains) illnesses in humans, ranging from conditions such as food poisoning and uncomplicated skin and soft-tissue infections to life-threatening illnesses such as myonecrosis, septicemia, central nervous system infections, tetanus, and botulism (1, 9, 11). While the majority of disease has been caused by a small proportion of the Clostridium species described to date, a number of other seemingly saprophytic species of Clostridium have only recently been implicated in human illness (4, 5).
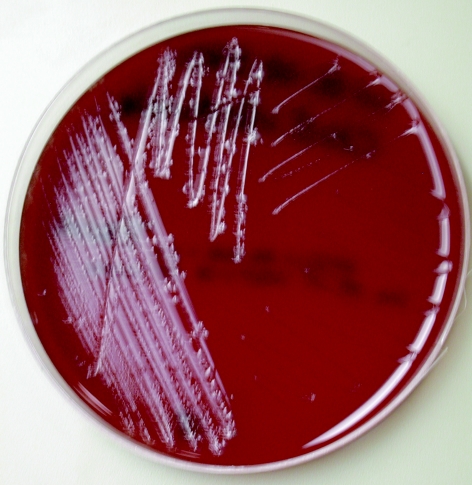
Clostridium intestinale (formerly Clostridium intestinalis) is a newly recognized species that was first described by Lee and coworkers in 1989 (6, 7). These investigators reported the recovery of five aerotolerant, gram-positive, endospore-forming, motile, rod-shaped bacterial strains from the feces of healthy cattle and pigs during quantitative and qualitative studies of the fecal flora of these animals (7). DNA homology and biochemical, and physiological studies served to distinguish these strains from other known Clostridium species (7). Microscopically, cells of C. intestinale are gram positive, straight or slightly curved motile rods that occur singly, in pairs, or occasionally in short chains (7). Endospores may be visible, being large, slightly oval, and located terminally (7). On nonselective blood agar medium, colonies of C. intestinale are approximately 2 to 4 mm in diameter, beta-hemolytic, grayish-white, translucent, raised, undulate, and rough after anaerobic incubation for 48 h (7), in similarity to our observations (Fig. 1). Isolates are typically saccharolytic and ferment a variety of carbohydrate compounds and hydrolyze esculin but do not reduce nitrate, fail to hydrolyze gelatin, and do not produce lecithinase, lipase, or indole (7).
FIG. 1.
Colonial morphology of Clostridium intestinale on brucella blood agar medium after 48 h of anaerobic incubation.
The ability of C. intestinale to ferment sorbitol may help distinguish this organism from other aerotolerant clostridia (7). However, detailed phenotypic identification is generally not practical for most clinical microbiology laboratories due to time and labor considerations. An increasing number of laboratories are resorting to the use of more-rapid and -robust microbial identification methods, such as those that are performed on the basis of sequencing of microbial genes. The 16S rRNA gene has served as the prototypic target of sequence-based bacterial identification algorithms for phenotypically difficult-to-identify bacteria (13). This gene is present in all eubacteria, with virtually all unique bacterial species having a distinctive 16S rRNA gene sequence (approximately 1,500 bp in size). While 16S rRNA gene sequence analysis of our patient's Clostridium isolate supported an identification of C. intestinale, a similar degree of 16S rRNA gene sequence identity was observed with several unspeciated nitrogen-fixing strains of Clostridium spp. (Clostridium spp. Kas301-1, P303, P301, Kas107-1, Kas401-3, Kas104-4, UsIt102-1, Kas303, Kas 404-1, and Kas 203-1, with GenBank accession numbers AB114242, AB114247, AB114246, AB114241, AB114240, AB114239, AB114237, AB114244, AB114243, and AB114238, respectively) recovered from a variety of nonleguminous plants from Japan and the Philippines (10). Strains Kas107-1, Kas410-3, Kas104-4, and UsIt102-2 demonstrated 100% 16S rRNA gene sequence homology with our strain (as determined on the basis of their complete GenBank sequences, which span from base 70 to 1367 of the 16S rRNA gene of our isolate), suggesting they belong to the same species. Hence, C. intestinale may be ubiquitous in the environment.
However, there have been no previous reports, in the world literature, of human colonization with or infection caused by Clostridium intestinale. Since most episodes of clostridial bacteremia arise endogenously (i.e., usually from the colon), we postulate that our patient's gastrointestinal tract was colonized with C. intestinale prior to the onset of her bacteremic episode. Risk factors for clostridial bacteremia include malignancy, chemotherapy- or radiation-induced tissue damage, hypoxia, trauma, recent surgery, diabetes mellitus, alcoholism, and bowel perforation, although clinically insignificant transient bacteremia is believed to account for over half of bacteremic cases (1, 9, 11). In a recent review of 65 cases of clostridial bacteremia in noninfant pediatric patients, C. septicum and C. perfringens were the species most frequently encountered (2). Most of those cases were associated with underlying immunocompromise, gastrointestinal tract disease, and/or trauma, although a few otherwise healthy individuals developed bacteremia and acute abdominal pain, with or without fever (2). Although a definitive diagnosis of our patient's fever and abdominal pain could not be made, the recovery of C. intestinale from two separate blood culture sets, combined with our patient's clinical presentation, lends credence to the purported clinical significance of this C. intestinale, although animal pathogenicity studies are essential to prove this supposition. Clostridium intestinale displays the closest phylogenetic relationships with a number of other Clostridium spp. of clinical importance (1), particularly C. fallax and C. cadaveris. Our case report highlights the importance of C. intestinale as a potential human pathogen.
REFERENCES
- 1.Allen, S. D., C. L. Emery, and D. M. Lyerly. 2003. Clostridium, p. 835-856. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. American Society for Microbiology, Washington, D.C.
- 2.Caya, J. G., and A. L. Truant. 1999. Clostridial bacteremia in the non-infant pediatric population: a report of two cases and review of the literature. Pediatr. Infect. Dis. J. 18:291-298. [DOI] [PubMed] [Google Scholar]
- 3.Collins, M. D., P. A. Lawson, A. Willems, J. J. Cordoba, J. Fernandez-Garayzabal, P. Garcia, J. Cai, H. Hippe, and J. A. E. Farrow. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812-826. [DOI] [PubMed] [Google Scholar]
- 4.Elsayed, S., and K. Zhang. 2004. Bacteremia caused by Clostridium symbiosum. J. Clin. Microbiol. 42:4390-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elsayed, S., and K. Zhang. 2004. Human infection caused by Clostridium hathewayi. Emerg. Infect. Dis. 10:1950-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Euzeby, J. P. 1998. Taxonomic note: necessary correction of specific and subspecific epithets according to rules 12c and 13b of the International Code of Nomenclature of Bacteria (1990 Revision). Int. J. Syst. Bacteriol. 48:1073-1075. [DOI] [PubMed] [Google Scholar]
- 7.Lee, W.-K., T. Fujisawa, S. Kawamura, I. Kikuji, and T. Mitsuoka. 1989. Clostridium intestinalis sp. nov., an aerotolerant species isolated from the feces of cattle and pigs. Int. J. Syst. Bacteriol. 39:334-336. [Google Scholar]
- 8.Lombardi, D. P., and N. C. Engleberg. 1992. Anaerobic bacteremia: incidence, patient characteristics, and clinical significance. Am. J. Med. 92:53-60. [DOI] [PubMed] [Google Scholar]
- 9.Lorber, B. 2000. Gas gangrene and other Clostridium-associated diseases, p. 2549-2561. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases, 5th ed. Churchill Livingstone, Philadelphia, Pa.
- 10.Minamisawa, K., K. Nishioka, T. Miyaki, B. Ye, T. Miyamoto, M. You, A. Saito, M. Saito, W. L. Barraquio, N. Teaumroong, T. Sein, and T. Sato. 2004. Anaerobic nitrogen-fixing consortia consisting of clostridia isolated from gramineous plants. Appl. Environ. Microbiol. 70:3096-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rechner, P. M., W. A. Agger, K. Mruz, and T. H. Cogbill. 2001. Clinical features of clostridial bacteremia: a review from a rural area. Clin. Infect. Dis. 33:349-353. [DOI] [PubMed] [Google Scholar]
- 12.Saito, T., K. Senda, S. Takakura, N. Fujihara, T. Kudo, Y. Linuma, N. Fujita, T. Komori, N. Baba, T. Horii, K. Matsuoka, M. Tanimoto, and S. Ichiyama. 2003. Anaerobic bacteremia: the yield of positive anaerobic blood cultures: patient characteristics and potential risk factors. Clin. Chem. Lab. Med. 42:293-297. [DOI] [PubMed] [Google Scholar]
- 13.Woo, P. C. Y., K. H. L. Ng, S. K. P. Lau, K.-T. Yip, A. M. Y. Fung, K.-W. Leung, D. M. W. Tam, T.-L. Que, and K.-Y. Yuen. 2003. Usefulness of the MicroSeq 500 16S ribosomal DNA-based bacterial identification system for identification of clinically significant bacterial isolates with ambiguous biochemical profiles. J. Clin. Microbiol. 41:1996-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]