Abstract
The genomic diversity of Helicobacter pylori from the vast Indian subcontinent is largely unknown. We compared the genomes of 10 H. pylori strains from Ladakh, North India. Molecular analysis was carried out to identify rearrangements within and outside the cag pathogenicity island (cag PAI) and DNA sequence divergence in candidate genes. Analyses of virulence genes (such as the cag PAI as a whole, cagA, vacA, iceA, oipA, babB, and the plasticity cluster) revealed that H. pylori strains from Ladakh are genetically distinct and possibly less virulent than the isolates from East Asian countries, such as China and Japan. Phylogenetic analyses based on the cagA-glr motifs, enterobacterial repetitive intergenic consensus patterns, repetitive extragenic palindromic signatures, the glmM gene mutations, and several genomic markers representing fluorescent amplified fragment length polymorphisms revealed that Ladakhi strains share features of the Indo-European, as well as the East Asian, gene pools. However, the contribution of genetic features from the Indo-European gene pool was more prominent.
Helicobacter pylori infection (1) is very common in India, where millions of adults and children are at risk for developing gastric inflammation, ulcers, and carcinoma (2). Distinct H. pylori genotypes have been observed in the Indian subcontinent, although their dissemination dynamics and routes of infection have not been uncovered completely (2, 7, 19, 25). Also, H. pylori from India has been resistant to commonly prescribed drugs, like metronidazole (19). Due to an extremely high rate of colonization, the incidence of gastroduodenal diseases linked to H. pylori, such as gastric-duodenal ulcers and stomach cancer, is thought to be very high, but these incidence rates have been grossly underestimated as low to negligible (22). Genetic analyses have been carried out recently on isolates recovered from different Indian populations (2, 7, 15, 19). However, the information needed to link particular genotypes to disease outcome or progression is not sufficient to build up a scenario to predict the effects of H. pylori on the occurrence of gastroduodenal diseases in different parts of India. This also appears to be a difficult exercise due to the fact that the affected population is extremely diverse, with close-knit communities, each consisting of peoples of different ethnicities, linguistic groups, and social status, often living quite close to one another. Although H. pylori populations from major cities, like Kolkata (15) and Hyderabad (2), have been characterized at the molecular level, such information is not available from geographically isolated and tribal areas. This lack of scientific data is especially evident in the case of the sparsely populated region of Ladakh in North India, where people of different ethnic backgrounds and genetic makeups inhabit a desert that has very typical high-altitude weather and an extremely difficult climate for life, with exceptionally low population density. The prevalence of H. pylori infection in Ladakh is reflected by a high seroprevalence of the cagA antigen, which is recorded to be up to 95% (27).
Profiling of the H. pylori gene pool serves as a surrogate marker (1) for population migration and demographic studies, thus constituting the so-called “geographic genomics” approach in microbiology. Ladakhi Buddhist people are genetically very close to Ladakhi Muslims, and Ladakhis in general have a genetic affinity to Mongolians and Chinese (21). Moreover, since Ladakh lies on the crossroads of India, China, and Pakistan (formerly Baltistan), H. pylori gene pools from Ladakhis have become very interesting in revealing peopling and migration patterns and the effects of religion and societal patterns on the host population and the pathogen (29). However, there has been a frustrating lack of genetic information on informative disease-linked loci and several other loci of phylogenetic relevance in such an important population of H. pylori. The unavailability of this vital data on Ladakhi strains made it almost impossible to understand the prevalent genotypes in the context of the development of gastroduodenal pathology in Ladakhis. Our study attempted to analyze the genomes of the 10 Ladakhi strains (29) to gain insights into the status of important genetic landmarks associated with pathogenesis and those important in defining the evolutionary history and genetic identity of H. pylori in Ladakh.
MATERIALS AND METHODS
Molecular genotyping.
Purified DNA preparations from Ladakhi strains were obtained from Mark Achtman (Department of Molecular Biology, Max-Planck Institut für Infektionsbiologie, Berlin, Germany) as a gift. These isolates were cultured from dyspeptic-gastritis patients, among whom five were of Buddhist ethnicity and the rest were Muslims. Additional details of the isolates used in the study are available in the Multi Locus Sequence Typing database (http://pubmlst.org/helicobacter/projects/ladakh/). These DNAs were used for PCRs for amplification and sequencing of genes, such as glmM, babB, and oipA, as described previously (23, 24, 31). The amplified products of the ureC fragment (glmM) and oipA (HP0638) and babB genes were gel eluted and purified with a QIAquick Gel Extraction kit (QIAGEN, Hilden, Germany). Sequencing was performed with both forward and reverse primers, using an ABI Prism 3100 DNA sequencer (Applied Biosystems). PCR and direct sequencing were performed at least twice to determine and confirm the DNA sequences for each strain. Consensus sequences for each of the samples were generated using Genedoc (version 2.6.002). Multiple alignments of sequenced nucleotides were carried out using Clustal X (version 1.81). Phylogenetic trees were developed using Treeview (version 1.6.6). The frame status for the oipA gene was analyzed using the DNA Star package.
The cag pathogenicity island (cag PAI) status was evaluated by PCR using eight sets of primers (Table 1) spanning the cagA gene, its promoter region, the cagE and cagT genes, and the left end of the PAI (LEC), as mentioned elsewhere (14, 16). The presence of the cagA gene and rearrangement analysis of the right end of the cag PAI (17), vacA genotyping (6, 17), iceA allele status (25), babA-babB (11, 32) gene status, and sequencing were carried out by molecular genotyping methods as described previously (6, 11, 17, 25, 32). The presence or absence of the plasticity region open reading frames (ORFs) JHP947, HP986, JHP912, JHP926, JHP931, JHP933, JHP944, and JHP945 was detected based on PCR amplifications employing target sequences and reaction parameters as described by Occhialini et al. (20).
TABLE 1.
Details of the PCR primers used in the study
| Locus | Primer name | Primer sequence (5′-3′) | Annealing temp (°C) | PCR product size (bp) | Reference |
|---|---|---|---|---|---|
| cag PAI | CagA F1 | AACAGGACAAGTAGCTAGCC | 52 | 701 | 14, 16 |
| CagA R1 | TATTAATGCGTGTGTGGCTG | ||||
| CagA F2 | GATAACAGGCAAGCTTTTGA | 52 | 349 | 14, 16 | |
| CagA R2 | TCTGCCAAACAATCTTTTGCAG | ||||
| CagAP-F1 | GTGGGTAAAAATGTGAATCG | 52 | 730 | 14, 16 | |
| CagA R2 | TCTGCCAAACAATCTTTTGCAG | ||||
| CagAP-F2 | CTACTTGTCCCAACCATTTT | 52 | 1181 | 14, 16 | |
| CagA R2 | TCTGCCAAACAATCTTTTGCAG | ||||
| CagE F1 | GCGATTGTTATTGTGCTTGTAG | 52 | 329 | 14, 16 | |
| CagE R1 | GAAGTGGTTAAAAAATCAATGCCCC | ||||
| CagT F1 | CCATGTTTATACGCCTGTGT | 52 | 301 | 14, 16 | |
| CagT R1 | CCATGTTTATACGCCTGTGT | ||||
| Lec F1 | ACATTTTGGCTAAATAAACGCTG | 52 | 384 | 14, 16 | |
| Lec R1 | TCTCCATGTTGCCATTATGCT | ||||
| Lec F2 | ATAGCGTTTTGTGCATAGAA | 52 | 877 | 14, 16 | |
| Lec R2 | ATCTTTAGTCTCTTTAGCTT | ||||
| cagA-glr motif typing (cag RJa) | cagF4584 (1) | GTTAATACAAAAGGTGGTTTCCAAAAATC | 52 | 1000/800 | 17 |
| cagR5280 (3) | GGTTGCACGCATTTTCCCTTAATC | ||||
| cagF4584 (1) | GTTAATACAAAAGGTGGTTTCCAAAAATC | 52 | 400 | 17 | |
| miniIS605.R (8) | CCGCTAAAGACGATTGGGCTT | ||||
| fcn unk (6) | TGGATTAAATCTTAATGAATTATCG | 52 | 350 | 17 | |
| cagR5280 (3) | GGTTGCACGCATTTTCCCTTAATC | ||||
| fcn unk (6a) | ACTCTATTTTGCTTGCAGTGCTTTTGG | 52 | 350 | 17 | |
| cagR5280 (3) | GGTTGCACGCATTTTCCCTTAATC | ||||
| cagF4856 (4) | GCGATGAGAAGAATATCTTTAGCG | 52 | 350 | 17 | |
| cagR5280 (3) | GGTTGCACGCATTTTCCCTTAATC | ||||
| IS606-1692 (5) | CTAACAATTTGCCATTATGCTGT | 52 | 2000 | 17 | |
| cagR5280 (3) | GGTTGCACGCATTTTCCCTTAATC | ||||
| cagF4584 (1) | GTTAATACAAAAGGTGGTTTCCAAAAATC | 52 | 400 | 17 | |
| Xins.R (7) | CGCTCTCTAATTGTTCTAGGA | ||||
| vacA signal region | VAIF | ATGAAAAAAACCCTTTTAC | 54 | 259 (s1) | 6 |
| VAIXR | CGAATTGCAAGTGATGGT | 286 (s2) | |||
| vacA middle region | VA3-F | GGTCAAAATGCGGTCATGG | 54 | 300 (m1a) | 17 |
| VA3-R | CCATTGGTACCTGTAGAAAC | ||||
| VAm-F3 | GGCCCCAATGCAGTCATGGAT | 54 | 300 (m1b) | 17 | |
| VAm-R3 | GCTGTTAGTGCCTAAAGAAGCAT | ||||
| VA4-F | GGAGCCCCAGGAAACATTG | 54 | 400 (m2) | 17 | |
| VA4-R | CATAACTAGCGCCTTGCAC | ||||
| glmM | GlmM1-R | GCTTACTTTCTAACACTAACGCGC | 52 | 296 | 21 |
| GlmM2-F | GGATAAGCTTTTAGGGGTGTTAGGGG | ||||
| oipA | HP0638-F | GTTTTTGATGCATGGGATTT | 52 | 401 | 28 |
| HP0638-R | GTGCATCTCTTATGGCTTT | ||||
| babA | BABA2F | AATCCAAAAAGGAGAAAAAGTATGAAA | 61 | 832/601 | 22 |
| BABA2R | TGTTAGTGATTTCGGTGTAGGACA | 55 | |||
| BABA2R607 | CTTTGAGCGCGGGTAAGC | ||||
| babB | Bab B F | ATGAAAAAAACCCTTTTAC | 40 | 496 | 22 |
| Bab B R | CGAATTGCAAGTGATGGT | ||||
| iceA | iceA1F | TATTTCTGGAACTTGCGCAACCTGAT | 52 | 696 (A1) | 23 |
| M.Hpy1R | GGCCTACAACCGCATGGATAT | 608 (A2) | |||
| CycSF | CGGCTGTAGGCACTAAAGCTA | ||||
| IceA2R | TCAATCCTATGTGAAACAATGATCGTT | ||||
| Plasticity region ORFs | JHP912F | CAATAGCCTTGCTCACGCTTC | 59 | 624 | 19 |
| JHP912R | GTTAAATGGTGAGAGCCTACG | ||||
| JHP931F | GTATTAGCGAAGTGCAATCAC | 57 | 1,133 | 19 | |
| JHP931R | GCTAATTTGTTTAGGCGTAGC | ||||
| JHP944 F | CTATGAGTGAAGAATTAACGC | 59 | 358 | 19 | |
| JHP944 R | CGCTCCATTCCAATATCTTTG | ||||
| JHP945 F | CAATGCGACTAACAGCATAG | 66 | 1,028 | 19 | |
| JHP945 R | CGCATTTGCTGTCATCTTTG | ||||
| JHP986 F | GCATGTCCCAAATCGTAGG | 58 | 566 | 19 | |
| JHP986 R | TGCATTTCGCATTGGCTCC | ||||
| JHP947 F | GATAATCCTACGCAGAACG | 60 | 611 | 19 | |
| JHP947 R | GCTAAAGTCATTTGGCTGTC | ||||
| JHP926 F | GATGAGCAAATCAATGGCATG | 59 | 991 | 19 | |
| JHP926 R | ACCTTTCAATACCGCTAGAAG | ||||
| JHP933 F | GAGTGAGTTTAAGCGAAC | 58 | 708 | 19 | |
| JHP933 R | CTTGTTGCTCTTGCAAGG | ||||
| REP typing | REPAF1- Dt | IIIGCGACGGCATCAGGC | 49 | Multilocus | 25 |
| REPAR2- Dt | ACGGCTTATCGGGCCTAC | ||||
| ERIC typing | ERIC1R | ATGTAAGCTCCTGGGGATTCAC | 49 | Multilocus | 13 |
| ERIC2R | AAGTAAGTGACTGGGGTGAGCG | ||||
| FAFLP typing | EcoRI + A/MseI + 0 | GTAGACTGCGTACCAATTCA | Touchdown | Multilocus | 2, 5 |
| GACGATGAGTCCTGAGTAA | From 66 to 56 | ||||
| EcoRI + G/MseI + 0 | GTAGACTGCGTACCAATTCG | Touchdown | Multilocus | 2, 5 | |
| GACGATGAGTCCTGAGTAA | From 66 to 56 |
RJ, right junction.
DNA fingerprinting.
The PCR methods for the enterobacterial repetitive intergenic consensus (ERIC) fingerprinting technique were employed exactly as explained earlier (13). The repetitive extragenic palindromic (REP) typing procedure involved primers for amplifying unique DNA sequences between the two REP signatures (28). All the gel images corresponding to ERIC and REP PCRs were analyzed using the Quantity 1.0 software in a gel documentation system (Bio-Rad). These images were then uploaded into a Diversity version 2.2.0 database (Bio-Rad). Band sizes, band attributes, and standard molecular weights were assigned alongside the molecular weight markers. Cluster analysis of DNA profiles was conducted on the basis of fingerprint characteristics. Based on the data for the presence or absence of 3 to 15 different DNA fragments in the fingerprints of strains of H. pylori, a binary data matrix was created. Overall similarity between the pairs of strains was calculated from the binary data matrix using the simple matching Dice coefficient. The resulting similarity matrix was used for cluster analysis by the unweighted pair group method with arithmetic averages to generate trees.
Whole-genome fingerprinting based on fluorescent amplified fragment length polymorphism (FAFLP) genotyping was done as described previously (2, 5). Briefly, the profiling of whole-genome microrestriction fingerprints with EcoRI/MseI enzymes using the fluorescence-tagged primer pairs EcoRI+A-MseI+0 and EcoRI+G or A-MseI+0 was performed for all 10 strains. The PCR-amplified fragments for each of the strains were then subjected to electrophoretic separation on a 5% acrylamide gel, and scoring of the fluorescent markers was done using an automated DNA analysis workstation (ABI Prism 3100 DNA sequencer).
Genomewide comparisons.
All the data obtained through DNA profiling were deposited in the AmpliBASE HP database (http://www.cdfd.org.in/amplibase/HP). The AmpliBASE HP server was queried for genomewide comparisons. The cag PAI rearrangement profiles and cagA-glr motif types were also compared to existing records in the database.
Nucleotide sequence accession numbers.
The nucleotide sequences of the gene loci of the Ladakhi strains were deposited in GenBank under the following accession numbers: AY843016 to AY843024 (glmM), AY845039 to AY845048 (oipA), and AY845049 to AY845051 (babB).
RESULTS
Rearrangements within and outside the cag PAI.
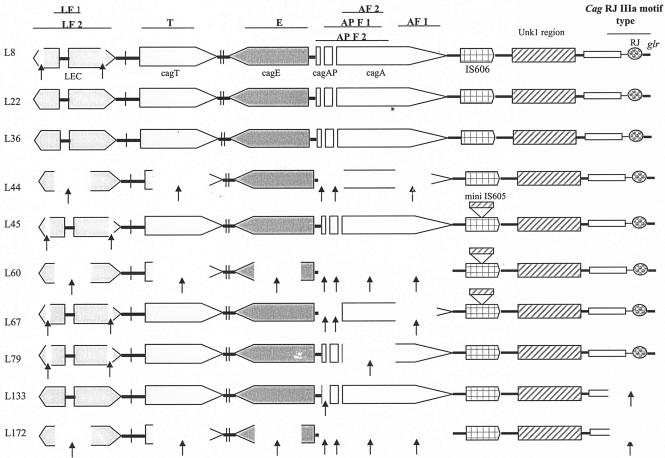
The cag PAI status was evaluated using eight sets of primers spanning the 3′ end, the middle region, and the 5′ end or left end of the cag PAI (Fig. 1). Based on this approach, we found only two strains with an intact cag PAI, while the rest had a rearranged PAI with frequent deletions in one or the other gene (Fig. 1 and Table 2). Furthermore, rearrangements were more frequent in the promoter region of cagA and the LEC. The cagE gene was also observed to be present in eight of the strains, while there was no significant difference between the PCR amplification percentages of the cagT and cagA genes. Analysis of the rearrangement activities on the extreme right end of the cag PAI and within the 3′ end of the glutamate racemase (cagA-glr) gene revealed a type III motif (comprised of a canonical IS606, IS606-IS605 chimeric sequences, and a 200-bp unknown sequence replacing the entire hel gene [17]) for most of the Ladakhi strains (Fig. 1). This motif type is particularly common in Indian strains from Kolkata and from Bangladesh (17, 19). Two of the strains did not reveal any motif types.
FIG. 1.
Genetic rearrangements in Ladakhi strains corresponding to the cag PAI and the region from the extreme right end of the cagA gene to the start of the glutamate racemase (glr) gene. Precise locations of rearrangements have been marked with arrows. LF1, left end of cag PCR product 1; LF2, left end of cag PCR product 2; T, cagT gene; E, cagE gene; APF1, cagA promoter PCR product 1; APF2, cagA promoter PCR product 2; AF1, cagA gene PCR product 1; AF2, cagA gene PCR product 2; Unk1, unknown region 1 as described by Kersulyte et al. (17); RJ, right junction of cagA; glr, glutamate racemase gene.
TABLE 2.
Schematic overview of H. pylori genotypes in 10 Ladakhi strains recovered from dyspeptic patients with gastritisa
| Sample | Ethnicityb | Clinical statusc | glmM | oipA frame status | oipA no. of CT repeats | babA | babB | Plasticity region cluster
|
iceA allele
|
vac alleles |
cag PAI PCR status
|
IS605 | cag RJ motif | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| JHP912 | JHP926 | JHP931 | JHP933 | JHP944 | JHP945 | JHP947 | HP986 | iceA1 | iceA2 | AF1 | AF2 | AP1 | AP2 | E | T | LF1 | LF2 | |||||||||||
| L8 | Bud | G | + | Off | 2 + 2 | − | + | + | + | + | − | + | − | − | − | + | − | s2 | + | + | + | + | + | + | + | − | − | IIIa |
| L22 | Bud | G | + | On | 3 | − | − | + | − | + | + | − | + | − | + | + | − | m2 | + | + | + | + | + | + | + | + | − | III |
| L36 | Mus | G | + | On | 2 + 2 | − | + | + | − | + | + | − | + | − | + | − | + | s2m1a | + | + | + | + | + | + | + | + | − | III |
| L44 | Mus | G | + | On | 2 + 1 | − | + | + | − | + | + | − | + | − | − | − | + | m2 | − | + | − | − | + | − | − | + | − | IIIa |
| L45 | Mus | G | + | On | 2 + 1 | + | + | + | − | + | − | − | + | − | + | − | + | s2m1b | + | + | + | + | + | + | + | − | + | III |
| L60 | Mus | G | + | Off | 7 | − | − | − | − | + | + | − | + | − | − | − | − | m2 | − | − | − | − | − | − | − | + | + | III |
| L67 | Mus | G | + | On | 2 + 3 | + | + | − | − | + | + | + | + | − | − | + | − | s2m1a | − | + | − | − | + | + | + | − | + | IIIa & I |
| L79 | Bud | G | + | On | 3 | − | + | + | − | + | − | + | − | − | − | + | − | m1b | + | − | + | + | + | + | + | − | − | IIIa |
| L133 | Bud | G | + | On | 2 + 1 | + | + | + | − | + | + | − | − | − | − | + | − | s2 | + | + | + | − | + | + | + | + | − | − |
| L172 | Bud | G | + | On | 3 | − | + | − | − | − | − | + | − | − | − | − | − | s2 | − | − | − | − | − | − | − | + | − | − |
Distribution (+, present; −, absent) of the genes glmM (PCR product, 296 bp), oipA (401 bp), babA (babA2F + babA2R, 832 bp; babA2F + BabA2R607, 607 bp; babB, 496 bp); ORFs specific for gastritis, JHP986 (566 bp), and gastric cancer, JHP947 (611 bp); the iceA (A1, 700 bp; A2, 700 bp) and vacA (s2, 286 bp; m2, 400 bp; m1a, 300 bp; m1b, 300 bp) allele combinations; the cag PAI status (AF1, 349 bp; AF2, 701 bp; APF1, 730 bp; APF2, 1,181 bp; E, 329 bp; T, 301 bp; LF1, 384 bp; LF2, 877 bp) with the motif type (III, 350 bp; IIIa, 350 bp; I, 350 bp) flanking the right end of the cagA gene and the presence of the insertion element IS605 (400 bp) have been represented. See the legend to Fig. 1 for abbreviations.
Bud, Buddhist; Mus, Muslim.
G, gastritis.
vacA and iceA allele status.
In our analyses, vacA genotypes were obtained by PCR of the signal and middle regions of the gene. The s2 genotype was found to be the most common (60% of strains). This is contrary to an earlier report suggesting that the frequency of the s1 allele was greater in gastritis isolates (82.9%) (10). The vacA middle region was amplifiable in 7 of the 10 strains in which m1a-m1b and m2 genotypes were observed with equal frequencies. None of the Ladakhi strains showed the s1m1 genotype. A particular genotype, s2m1a, not reported earlier (4, 12), was observed in two strains. The results of vacA genotyping and the analyses of other gene loci are summarized in Table 2. In our study, the iceA1 allele was observed in 5 out of 10 strains, while the iceA2 allele was present in 3 strains. This gene could not be amplified in two strains (L60 and L172), and none of the strains were positive for both iceA1 and iceA2 alleles.
OipA polymorphisms and frame status.
Sequence analysis of the 401-bp fragment of the oipA gene, which encodes a proinflammatory OipA protein, was analyzed for the presence of CT repeats. Variations in the CT repeat number and pattern determine the frame status of this gene. The oipA gene was found to be in frame for 8 of the 10 strains with a mixed CT repeat pattern, although two CT repeats followed by one TT with a single, double, or triple CT was more common (6 strains). The strains with three CT repeats were all from the patients of Buddhist ancestry. The CT repeat numbers of ≤5 are characteristic of the East Asian strains (3). In one of the Ladakh strains (L60), the oipA gene with seven CT repeats was out of frame, and thus the gene status was off (Table 2). This result was in good agreement with previous reports suggesting that a CT repeat number of five or seven put the frame out, and the gene was off (31). Differences in the number of CT repeats in genes coding for the outer membrane proteins introduce a stop codon in the coding region, thereby interrupting translation of the gene into a functional protein. It has recently been confirmed that the functional status of the OipA protein is in agreement with sequence-based genotyping (18).
Other informative loci (glmM, babA-babB, and plasticity region ORFs).
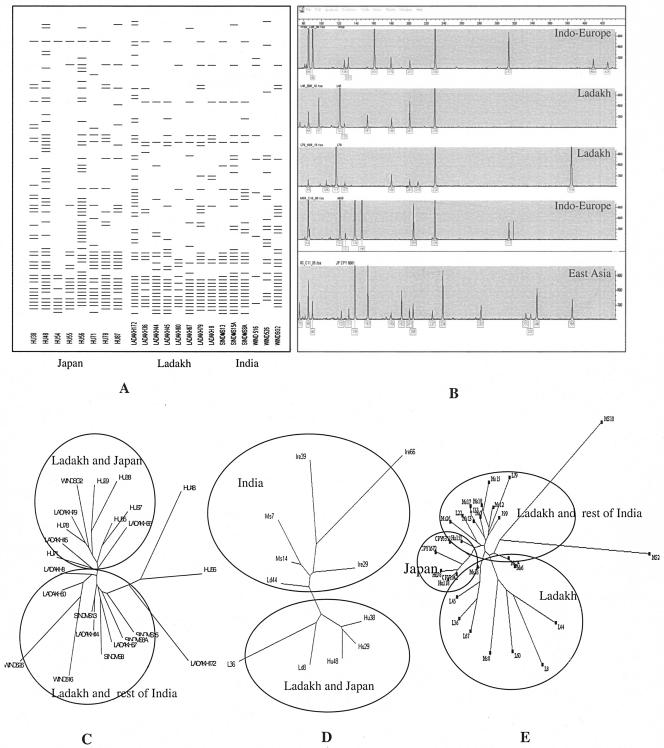
The glmM (phosphoglucosamine mutase) gene (26) was PCR amplified in all the strains. Sequence analysis of the 296-bp fragment of this gene and alignment with sequences from other regions, including Hyderabad (South India) and East Asia (Japan), revealed two clusters (Fig. 2). Most of the Ladakhi strains grouped with the South Indian strains, while only two strains, L22 and L133, clustered with the Japanese strains (Fig. 2). These two strains originated from the Buddhist patients. The babA2 gene was amplified in three strains, whereas babB was present in all of the strains except L22 and L60. The babB gene has also been observed to be more conserved than the babA2 gene in most H. pylori strains (24). Sequence analysis of the 496-bp fragment of the babB gene in strains from Ladakh and a few representative strains from South India, Ireland, and Japan revealed an Indo-European cluster. This cluster was mainly populated by isolates representing Ladakh, Ireland, and South India. In addition, there was a separate Asian cluster, with two Ladakhi strains (L8 and L36) and the Japanese strains (Fig. 2). In the Ladakhi strains, the plasticity region ORF HP0986 was detected in three strains, while none of the strains were positive for the cancer-associated gene JHP947 (20). We also attempted to obtain profiles of other important plasticity region ORFs, among which JHP931 and JHP912 were present in 9 and 6 of the 10 Ladakhi strains, respectively. Six strains were positive for both JHP933 and JH0945, while JHP944 was observed in four strains. The ORF JHP926 was detected in only one strain.
FIG. 2.
Genomic diversity of H. pylori from Ladakh as revealed by FAFLP-based genotyping (A, B, and C) and polymorphisms within the glmM and babB genes. Part A corresponds to the two-dimensional gel image depicting the number of polymorphic loci that were compared before genetic affinities among Ladakhi, Indian, and Japanese strains were deduced in the form of a phylogenetic tree (C). The Genotyper plot (B) corresponding to FAFLP analysis was developed by the Genescan and Genotyper software (Applied Biosystems). It compares FAFLP allelic profiles of Ladakhi strains with Indo-European and East Asian gene pools. Phylogenetic trees were also deduced based on DNA sequence divergence analyses corresponding to glmM (D) and babB (E) genes.
DNA-profiling studies.
ERIC and REP fingerprinting profiles from Ladakhi strains were compared with those of isolates from other regions, including Europe (Ireland and England), South India, and East Asia (Japan). All 10 strains studied were found to be analyzable using ERIC- and REP-based PCR techniques. The number of bands observed in each strain varied from six to nine in the size range of 0.25 to 3 kb in ERIC and four to nine in the size range of 0.3 to 7.0 kb in REP fingerprinting methods. None of the profiles were found to be identical. Most prominent among these were bands corresponding to 2, 2.5, and 3 kb in ERIC genotyping and 1.25 and 2.5 kb with REP PCR. Although ERIC- and REP-based PCRs revealed equal numbers of bands, the ERIC PCR revealed more distinct and prominent bands. Results from both the methods were reproducible. No specific ERIC profiles distinguished the strains from Buddhists from those obtained from Muslims. Comparison of REP profiles obtained from Ladakhi strains with those from Ireland and England gave two clusters, one comprised of the Irish and English strains, while only Ladakhi strains, along with an English strain, formed the other cluster (data not shown). This indicates that the Indian strains are distinct from the European strains based on REP signatures. Based on ERIC profiles, the Ladakhi strains clustered with Japanese or Irish strains (Indo-European), and a few others shared similar profiles with South Indian strains (data not shown). FAFLP analysis using primers MseI+0-EcoRI+A and MseI+0-EcoRI+G revealed a number of polymorphic alleles. All the strains were individualized based on unique traces of alleles (fluorescence-tagged amplicons). Phylogenetic analysis revealed a final evolutionary scenario in which genotypes of the Ladakhi strains were frequently found to share alleles specific to either the East Asian or the Indo-European gene pool. However, the contribution of alleles from the Indian gene pool was more prominent, as the Ladakhi strains clustered more closely with the Indian strains than with the European strains (Fig. 2).
DISCUSSION
Genetic variation within bacterial populations can provide information relating to their evolution. However, it is very rare that this variation can provide a window into their hosts' evolution. Coevolution between host and pathogen has been verified only if pathogens do not move horizontally between eukaryotic hosts (9). On this basis, a strict phylogenetic and evolutionary parallel of host and pathogen genomes could be envisaged. Unfortunately, for most bacterial pathogens, frequent transmission between hosts separates the evolution of the bacterium from that of the host. For H. pylori, however, transmission is faithfully restricted to families within specific communities. Recently, this phenomenon has provided evidence regarding patterns of human migration (8, 11, 17, 29, 30). In this context, the strains from geographical areas with conflicting yet interesting human histories, such as Ladakh, received immediate attention (8, 29). Bounded by two of the world's mightiest mountain ranges, the Himalayan and the Karakoram, Ladakh lies at altitudes ranging from ∼9,000 feet (2,750 m) at Kargil to 25,170 feet (7,672 m) at Saser Kangri in the Karakoram. It is an isolated trans-Himalayan region with low population density. Due to typical geoclimatic positioning and interesting past events linked to the peopling of Ladakh, H. pylori strains from this region may constitute an important model for host-pathogen coevolution and human migration, in addition to representing some of the precious bacterial gene pools in the hands of evolutionary microbiologists. A recent landmark study described the genetic descent and phylogeography of these strains (29). In this description of human history and peopling through H. pylori genetics, the major issue of pathogenic potential and epidemiology of H. pylori in Ladakh has probably not been addressed in full. This lack of information was the impetus for our study, in which for the first time we attempted to describe the molecular fine structure of important genetic landmarks in these strains to gain insights into their virulence potential and microevolution.
Our study surveyed important informative loci other than those already studied in an anthropological context (29). Interestingly, the cag PAI was found to be rearranged or split in 8 of the 10 strains studied. Although these analyses were based on the presence or absence of specific amplicons, we also used nested primers (LEC region) and sequencing in some cases to confirm complete or partial deletion or rearrangements. This PCR-based method was recently tested in 335 isolates from around the world and was found to generate reproducible results (16). All of the strains (those with an intact cag PAI and those with a partial cag PAI) were recovered from the same clinical outcome (gastritis), indicating that the intactness or rearrangement of the cag PAI alone may not be the sole determinant of the outcome of infection in these cases. This was also independent of the status of other potentially proinflammatory proteins, such as OipA and IceA. cagA-glr motif typing of the 10 Ladakhi strains revealed type III motifs when we looked for insertion-deletion and substitution motifs on the right end of the cag PAI. It is noteworthy that this motif type has been exclusively defined for strains of Indian origin. The insertion element mini-IS605 was detected in three strains, but there was no correlation between the intactness of the cag PAI and the presence of this mobile element, since irrespective of its presence or absence, most of the strains had a rearranged cag PAI. Also, we observed that the Ladakhi strains, in addition to their genetic relatedness with the Indo-European gene pool, showed affinity with East Asian strains based on the glmM and babB sequences and on ERIC profiles. Interestingly, the vacA genotypes from Ladakh did not represent the potentially toxigenic s1m1 genotype. A particular genotype, s2m1a, not reported earlier was observed in two strains. Also, the cancer-associated ORF JHP947 was missing in all the strains. The disruption of the cag PAI and the absence of the s1m1 alleles and JPH947, as well as the fact that all the strains were recovered from patients with dyspepsia and gastritis, hint that the virulence of these strains is mild. We observed no correlation between the cagA status and the frame status of oipA and the vacA genotype, contrary to an earlier study (3) that described strains from the United States that had the oipA gene in frame and carried the cagA gene and the vacA s1 genotype. This observation, however, needs to be validated with a large number of isolates.
Phylogenetic reconstruction based on multiple loci appears to be a good approach for this pathogen. We found the H. pylori genome from Ladakh to be a mosaic of genetic elements contributed by Asian and European gene pools. These results are in agreement with recent findings based on polymorphisms in housekeeping genes (8, 29). However, from our study, which employed multiple typing approaches, it appears that the genetic affinity of these strains is more toward Indo-European strains than East Asian strains. This was evident even from the analyses of virulence-linked loci, such as the cag PAI, which is largely intact in ∼60% of Japanese strains (16) but was rearranged in the majority of Ladakhi strains. Although obtaining a large number of strains from remote and difficult areas of Ladakh and nearby Kashmir will be a challenge, we suggest that our studies may be extended further to a large number of isolates from these places, especially those linked to severe pathological outcomes.
However, the fullest possible molecular dissection of the present strains may be viewed as an opportunity to understand the diversity of H. pylori in India, as they might constitute missing pieces of the large biological jigsaw puzzle.
In conclusion, we have compared important genomic landmarks in the context of the pathological and evolutionary behavior of the strains from Ladakh. Such a molecular dissection of genotypes is likely to contribute to our understanding of the role of H. pylori in gastroduodenal pathology in India. Given that the two genomes sequenced to date are each from H. pylori strains isolated from European patients, genomic comparisons modeled on these two genomes should facilitate the identification of novel loci from strains, especially from the understudied Asian populations. The identification and characterization of such loci, which are more abundant in the Asian gene pool, may lead to newer insights into the mechanisms of H. pylori colonization, carriage, and virulence in the countries of Asia, which are under more serious threat from H. pylori.
Acknowledgments
We are very grateful to Seyed E. Hasnain for his support. We thank Mark Achtman for making available the DNAs of Ladakhi strains. Our thanks are also due to Ian M. Carroll and Cyril J. Smyth for some of the Irish isolates used here. We are thankful to John Atherton and Douglas E. Berg for the English and Japanese strains, respectively.
N.A. is a staff scientist and currently leads the pathogen evolution program at CDFD.
REFERENCES
- 1.Achtman, M., T. Azuma, D. E. Berg, Y. Ito, G. Morelli, Z. J. Pan, S. Suerbaum, S. Thompson, A. van der Ende, and L. J. van Doorn. 1999. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32:459-470. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed, N., A. A. Khan, A. Alvi, S. Tiwari, C. S. Jyothirmayee, F. Kauser, M. Ali, and C. M. Habibullah. 2003. Genomic analysis of Helicobacter pylori from Andhra Pradesh, South India: molecular evidence for three major genetic clusters. Curr. Sci. 85:101-108. [Google Scholar]
- 3.Ando, T., R. M. Peek, D. Pride, S. M. Levine, T. Takata, Y. C. Lee, K. Kusugami, A. van der Ende, E. J. Kuipers, J. G. Kusters, and M. J. Blaser. 2002. Polymorphisms of Helicobacter pylori HP0638 reflect geographic origin and correlate with cagA status. J. Clin. Microbiol. 40:239-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atherton, J. C., P. Cao, R. M. J. Peek, M. K. Tummuru, M. J. Blaser, and T. L. Cover. 1995. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 270:17771-17777. [DOI] [PubMed] [Google Scholar]
- 5.Carroll, I. M., N. Ahmed, S. M. Beesley, A. A. Khan, S. Ghousunnissa, C. A. O'Morain, and C. J. Smyth. 2003. Fine-structure molecular typing of Irish Helicobacter pylori isolates and their genetic relatedness to strains from four different continents. J. Clin. Microbiol. 41:5755-5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll, I. M., N. Ahmed, S. M. Beesley, A. A. Khan, S. Ghousunnissa, C. A. Moráin, C. M. Habibullah, and Cyril J. Smyth. 2004. Microevolution between paired antral and paired antrum and corpus Helicobacter pylori isolates recovered from individual patients. J. Med. Microbiol. 53:1-9. [DOI] [PubMed] [Google Scholar]
- 7.Datta, S., S. Chattopadhyay, G. B. Nair, A. K. Mukhopadhyay, J. Hembram, D. E. Berg, D. R. Saha, A. Khan, A. Santra, S. K. Bhattacharya, and A. Chowdhury. 2003. Virulence genes and neutral DNA markers of Helicobacter pylori isolates from different ethnic communities of West Bengal, India. J. Clin. Microbiol. 41:3737-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falush, D., T. Wirth, B. Linz, J. K. Pritchard, M. Stephens, M. Kidd, M. J. Blaser, D. Y. Graham, S. Vacher, G. I. Perez-Perez, Y. Yamaoka, F. Mégraud, K. Otto, U. Reichard, E. Katzowitsch, X. Wang, M. Achtman, and S. Suerbaum. 2003. Traces of human migrations in Helicobacter pylori populations. Science 299:1582-1585. [DOI] [PubMed] [Google Scholar]
- 9.Funk, D. J., L. Helbling, J. J. Wernegreen, and N. A. Moran. 2000. Intraspecific phylogenetic congruence among multiple symbiont genomes. Proc. R. Soc. Lond. Ser. B 267:2517-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerhard, M. N. L., N. Neumayer, T. Boren, R. Rad, W. Schepp, S. Miehlke, M. Classen, and C. Prinz. 1999. Clinical relevance of the Helicobacter pylori gene for blood-group antigen-binding adhesin. Proc. Natl. Acad. Sci. USA 96:12778-12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghose, C., G. I. Perez-Perez, M. G. D. Bello, D. T. Pride, C. M. Bravi, and M. J. Blaser. 2002. East Asian genotypes of Helicobacter pylori strains in Amerindians provide evidence for its ancient human carriage. Proc. Natl. Acad. Sci. USA 99:15107-15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han, S.-R., H.-J. Schreiber, S. Bhakdi, M. Loos, and M. J. Maeurer. 1998. vacA genotypes and genetic diversity in clinical isolates of Helicobacter pylori. Clin. Diagn. Lab. Immunol. 5:139-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hussain, M. A., F. Kauser, A. A. Khan, S. Tiwari, C. M. Habibullah, and N. Ahmed. 2004. Implications of molecular genotyping of Helicobacter pylori isolates from different human populations by genomic fingerprinting of enterobacterial repetitive intergenic consensus regions for strain identification and geographic evolution. J. Clin. Microbiol. 42:2372-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikenoue, T., S. Maeda, K. O. Gura, M. Akanuma, Y. Mitsuno, Y. Imai, H. Yoshida, Y. Shiratori, and M. Omata. 2001. Determination of Helicobacter pylori virulence by simple gene analysis of the cag pathogenicity island. Clin. Diagn. Lab. Immunol. 8:181-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katelaris, P. H., G. H. Tippett, P. Norbu, D. G. Lowe, R. Brennan, and M. J. Farthing. Dyspepsia, Helicobacter pylori and peptic ulcer in a randomly selected population in India. Gut 33:1462-1466. [DOI] [PMC free article] [PubMed]
- 16.Kauser, F., A. A. Khan, M. A. Hussain, I. M. Carroll, N. Ahmad, S. Tiwari, Y. Shouche, B. Das, M. Alam, S. M. Ali, C. M. Habibullah, R. Sierra, F. Megraud, L. A. Sechi, and N. Ahmed. 2004. The cag pathogenicity island of Helicobacter pylori is disrupted in a majority of patient isolates from different human populations. J. Clin. Microbiol. 42:5302-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kersulyte, D., A. K. Mukhopadhyay, B. Velapatino, W. W. Su, Z. J. Pan, C. Garcia, V. Hernandez, Y. Valdez, R. S. Mistry, R. H. Gilman, Y. Yuan, H. Gao, T. Alarcón, M. López-Brea, G. B. Nair, A. Chowdhury, S. Datta, M. Shirai, T. Nakazawa, R. Ally, I. Segal, B. C. Y. Wong, S. K. Lam, F. Olfat, T. Borén, L. Engstrand, O. Torres, R. Schneider, J. E. Thomas, S. Czinn, and D. E. Berg. 2000. Differences in genotypes of Helicobacter pylori from different human populations. J. Bacteriol. 182:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kudo, T., Z. Z. Nurgalieva, M. E. Conner, S. Crawford, S. Odenbreit, R. Haas, D. Y. Graham, and Y. Yamaoka. 2004. Correlation between Helicobacter pylori OipA protein expression and oipA gene switch status. J. Clin. Microbiol. 42:2279-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukhopadhyay, A. K., D. Kersulyte, J. Y. Jeong, S. Datta, Y. Ito, A. Chowdhury, S. Chowdhury, A. Santra, S. K. Bhattacharya, T. Azuma, G. B. Nair, and D. E. Berg. 2000. Distinctiveness of genotypes of Helicobacter pylori in Calcutta, India. J. Bacteriol. 182:3219-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Occhialini, A., A. Marais, R. Alm, F. Garcia, R. Sierra, and F. Mégraud. 2000. Distribution of open reading frames of plasticity region of strain J99 in Helicobacter pylori strains isolated from gastric carcinoma and gastritis patients in Costa Rica. Infect. Immun. 68:6240-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oota, H., N. Saitou, and S. Ueda. 2002. A large-scale analysis of human mitochondrial DNA sequences with special reference to the population history of East Eurasia. Anthropol. Sci. 110:293-312. [Google Scholar]
- 22.Pavithran, K., D. C. Doval, and K. K. Pandey. 2002. Gastric cancer in India. Gastric Cancer 5:240-243. [DOI] [PubMed] [Google Scholar]
- 23.Pride, D. T., and M. J. Blaser. 2002. Concerted evolution between duplicated genetic elements in Helicobacter pylori. J. Mol. Biol. 316:629-642. [DOI] [PubMed] [Google Scholar]
- 24.Pride, D. T., R. J. Meinersmann, and M. J. Blaser. 2001. Allelic variation within Helicobacter pylori BabA and BabB. Infect. Immun. 69:1160-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rahman, M., A. K. Mukhopadhyay, S. Nahar, S. Datta, M. M. Ahmad, S. Sarker, I. M. Masud, L. Engstrand, M. J. Albert, G. B. Nair, and D. E. Berg. 2003. DNA-level characterization of Helicobacter pylori strains from patients with overt disease and with benign infections in Bangladesh. J. Clin. Microbiol. 41:2008-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reuse, H. D., A. Labigne, and D. M. Lecreulx. 1997. The Helicobacter pylori ureC gene codes for a phosphoglucosamine mutase. J. Bacteriol. 179:3488-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romero-Gallo, J., G. I. Pérez-Pérez, R. P. Novick, P. Kamath, T. Norbu, and M. J. Blaser. 2002. Responses of endoscopy patients in Ladakh, India, to Helicobacter pylori whole-cell and CagA antigens. Clin. Diagn. Lab. Immunol. 9:1313-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veralovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in Eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wirth, T., X. Wang, B. Linz, R. P. Novick, J. K. Lum, M. Blaser, G. Morelli, D. Falush, and M. Achtman. 2004. Distinguishing human ethnic groups by means of sequences from Helicobacter pylori: lessons from Ladakh. Proc. Natl. Acad. Sci. USA 101:4746-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamaoka, Y., E. Orito, M. Mizokami, O. Gutierrez, N. Saitou, T. Kodama, M. S. Osato, J. G. Kim, F. C. Ramirez, V. Mahachai, and D. Y. Graham. 2002. Helicobacter pylori in North and South America before Columbus. FEBS Lett. 517:180-184. [DOI] [PubMed] [Google Scholar]
- 31.Yamaoka, Y., D. Kwon, and D. Graham. 2000. A Mr 34,000 proinflammatory outer membrane protein (Oip) of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 97:7533-7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zambon, C. F., F. Navaglia, D. Basso, M. Rugge, and M. Plebani. 2003. Helicobacter pylori babA2, cagA, and s1 vacA genes work synergistically in causing intestinal metaplasia. J. Clin. Pathol. 56:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]