Abstract
The ability of Legionella pneumophila to cause Legionnaires’ disease is dependent on its capacity to survive in the intracellular environment of its host cells. Furthermore, outbreaks of this disease have been associated with contaminated water sources where L. pneumophila survives as a parasite of protozoa. In this study, we determined the effect of nutritional auxotrophy on the ability of L. pneumophila to survive in the intracellular environment of its host cells. We generated a diaminopimelic acid (DAP) auxotroph (AA400) of L. pneumophila by disruption of the aspartate-β-semialdehyde (asd) gene. The ability of AA400 to survive within macrophages and protozoa was found to be defective. This defect was due solely to the asd disruption since complementation of the mutant with the wild-type asd gene restored its capacity for intracellular survival. Furthermore, the defect was not completely complemented by DAP supplementation to the culture media. Thus, our results suggest that disruption of the asd gene may prove to be useful in the design of attenuated vaccines against Legionnaires’ disease.
Legionella pneumophila, a gram-negative facultative intracellular bacterium, is a major cause of community-acquired and nosocomial pneumonia and is associated with a high level of mortality, particularly among immunocompromised individuals (14, 17, 32). The ability of L. pneumophila to cause disease is dependent on its ability to survive and replicate within a specialized membrane-bound phagosome in human macrophages and epithelial cells (29, 34). At the ultrastructural and molecular levels, the life cycle of L. pneumophila within human phagocytic cells is very similar to its life cycle in its environmental host, the protozoa (1, 13, 21). We have recently described the expression of type IV pili by L. pneumophila and their role in adherence to both mammalian and protozoan cells (41). Although some of the attachment and uptake mechanisms of the two hosts are similar, there are also distinct differences (4, 13, 21–23, 25, 31, 45).
Protection against bacterial pathogens has become increasingly urgent due in part to the alarming rise in drug resistance among these pathogens (43). Interestingly, it has been shown that L. pneumophila becomes 1,000-fold more resistant to antibiotics after intracellular growth (11). This rise in antibiotic resistance may be partly due to the expression of unique bacterial genes in the intracellular environment (2, 3, 5, 6, 42). Furthermore, treatment of patients infected with L. pneumophila with the commonly used antibiotics often fails to eradicate the disease (35). Thus, alternative methods for the treatment of Legionnaires’ disease may be necessary.
The most effective means to generate a protective immune response against intracellular bacterial pathogens is by live attenuated bacteria that deliver particulate antigens to antigen-processing cells (27, 30). One promising method of bacterial attenuation is the generation of auxotrophic strains that are unable to survive intracellularly due to the absence of the essential nutrient that they require within the host cell. Examples of these are the aromatic amino acid auxotrophs of Salmonella typhimurium and the leucine and diaminopimelic acid (DAP) auxotrophs of mycobacteria (10, 20, 38).
As in most gram-negative bacteria, a major constituent of the peptidoglycan layer of L. pneumophila is DAP (8, 24). DAP is involved in the peptide cross-linking of the peptidoglycan. Since mammalian cells do not utilize or synthesize DAP, intracellular bacterial pathogens that are auxotrophic for DAP would not be able to survive in the mammalian host. Therefore, it may be possible to develop chemotherapeutic agents that target enzymes of the DAP biosynthetic pathway. Such an auxotroph may also be a potential live attenuated vaccine candidate or a vehicle for the delivery of immunogenic antigens of other intracellular pathogens. To identify a possible target of chemotherapeutic agents and to determine the feasibility of using L. pneumophila as a particulate antigen delivery system, we cloned and characterized the aspartate-β-semialdehyde dehydrogenase (asd) gene of L. pneumophila and examined the phenotype of a null mutant.
MATERIALS AND METHODS
Bacterial strains and vectors.
The virulent AA100 strain of L. pneumophila has been described previously (3). Escherichia coli DH5α (Bethesda Research Laboratories [BRL], Gaithersburg, Md.) was used for the majority of cloning experiments. E. coli K-12 derivative strain χ2981 carries an asd insertion mutation that renders it auxotrophic for DAP (5, 7). Plasmid pUC-4K was purchased from Pharmacia (Piscataway, N.J.) and was the source of the kanamycin (kan) resistance gene used as a probe for Southern hybridization. Plasmids pBC-SK+ and pBlueScript were purchased from Stratagene (La Jolla, Calif.). Plasmid pBOC20 is a chloramphenicol-resistant plasmid that contains oriT and the sacB gene from Bacillus subtilis (6). The sacB gene encodes levansucrase and is lethal to L. pneumophila grown in the presence of sucrose (16). The conjugative plasmid pRK212.1 was used as a helper plasmid to mobilize plasmid pBOC20 by conjugation as described previously (6).
DNA manipulations.
Chromosomal DNA preparations, transfection, restriction enzyme digestion, and DNA ligation were performed as described elsewhere (39) unless specified otherwise. Restriction enzymes and T4 DNA ligase were obtained from BRL.
Plasmid DNA preparations were performed by using a Qiagen (Chatsworth, Calif.) plasmid kit according to the manufacturer’s recommendations. Transformations were carried out by electroporation using a Gene Pulser as recommended by the manufacturer (Bio-Rad, Hercules, Calif.). Purification of DNA fragments from agarose gels for subcloning or labeling for Southern hybridization was done by using a Qiaex kit as recommended by the manufacturer (Qiagen). Transfer of DNA from agarose gels into membranes, fluorescein labeling of DNA probes, hybridizations, and detection were performed as previously described (5).
Tissue culture and protozoan culture.
Macrophage-like U937 cells were maintained at 37°C and 5% CO2 in RPMI 1640 tissue culture medium supplemented with 10% heat-inactivated fetal calf serum (Sigma Chemical Co., St. Louis, Mo.). Prior to infection, cells were differentiated with phorbol 12-myristate 13-acetate (PMA) for 48 h as described previously (5). Differentiated cells are nonreplicative adherent macrophage-like cells. Monolayers were washed three times with the tissue culture medium prior to infection. For infection of monolayers, L. pneumophila grown for 48 h at 37°C on buffered charcoal-yeast extract (BCYE) agar plates was resuspended in RPMI 1640. The infection was carried out as described for each experiment.
Hartmannella vermiformis CDC-19 (ATCC 50237) has been cloned and grown in axenic culture as a model for the study of the pathogenesis of L. pneumophila (19). The amoebae were maintained in ATCC culture medium 1034, and infection with L. pneumophila was done in assay medium as previously described (4).
Gene cloning and amplification of the L. pneumophila asd gene.
To generate an L. pneumophila chromosomal DNA library, AA100 DNA was partially digested with Sau3A and sucrose gradient size-selected fragments of 35 to 50 kb were ligated into a BamHI site in cosmid pLTP6, a derivative of pLTP5 (9). The ligation mixture was packaged into λ phage with the use of a λ packaging mix (Stratagene). The packaged DNA was transfected into E. coli HB101. To isolate clones that carry the asd gene of L. pneumophila, the cosmid DNA library was conjugated from HB101 into the E. coli DAP auxotroph χ2981. Clones that complemented the χ2981 defect were selected. A common 4.3-kb HindIII fragment from these clones was found to complement the asd defect in χ2981. The 4.3-kb fragment was subcloned into pBlueScript (pOH1) and into pBOC20 (pOHBOC1).
To amplify the L. pneumophila asd gene, two primers complementary to sequence 200 bp upstream (BAM-1) and downstream (X-1) (5′-GCGGGATCCCTGCGTGGG-3′ and 5′-GCGCTCGAGAAGTAAGACA-3′, respectively) of the asd gene were used for PCR. BAM-1 was designed to generate a BamHI restriction site and X-1 was designed to generate an XhoI restriction site to facilitate subcloning. A 1.4-kb fragment was isolated and subcloned into pBOC20 (pOHBOC2).
DNA sequencing.
To locate and partially sequence the asd gene of L. pneumophila, unidirectional deletion with exonuclease III was used as described elsewhere (26, 37). The dideoxy-chain termination method of Sanger et al. (40) was employed, using a Sequenase kit (United States Biochemical, Cleveland, Ohio). The T3 promoter primer of the vector was used for sequencing of the exonuclease III-digested DNA. Once homology to other known asd sequences was found, oligonucleotides were synthesized (BRL) and sequencing of both strands of the gene was performed. The DNA sequences were compared to other sequences at the National Center for Biotechnology Information databases by using blastX and blastP programs.
Transposon mutagenesis and allelic exchange into the L. pneumophila chromosome.
A 4.3-kb fragment containing the asd gene of L. pneumophila was subcloned into pBOC20, designated pOHBOC1, and transformed into the E. coli χ2981. Mini-Tn10::kan mutagenesis was performed exactly as described previously (6). An insertion within the asd gene in pOHBOC1 was conjugated into L. pneumophila, and allelic exchange was performed exactly as described previously (6). Confirmation of the chromosomal insertion was obtained by Southern hybridization analysis.
Cytopathogenicity of L. pneumophila mutants to U937 cells.
Infection of PMA-differentiated U937 monolayers was performed in triplicate wells of 96-well plates containing 5 × 104 CFU/well at a multiplicity of infection of 1. The infected monolayers were incubated at 37°C for 24, 48, or 72 h. For measurements of the number of remaining viable cells in the monolayer, the monolayers were treated with alamar blue dye as described previously (5). The relative degree of cytopathogenicity of L. pneumophila to the monolayers was expressed as the optical density of the reduced alamar blue as an indicator of the remaining viable cells that reduced the dye.
Growth kinetics of L. pneumophila mutants in U937 cells and in H. vermiformis.
Differentiated 5 × 104 U937 cells in 96-well plates were infected with L. pneumophila in triplicate at a multiplicity of infection of 10 for 30 min at 37°C. Wells were then washed three times with tissue culture medium and incubated in the presence of 50 μg of gentamicin per ml at 37°C for 1 h to kill extracellular bacteria. Wells were washed three times with tissue culture medium to remove gentamicin. Time zero was harvested at this point by hypotonic lysis of the monolayers and plating dilutions onto BCYE plates for colony enumeration. The rest of the monolayers were incubated at 37°C for several time intervals. At the end of each time interval, the supernatant of the monolayer was transferred to a sterile well and monolayers were lysed hypotonically. The lysate was combined with the supernatant, and aliquots were plated onto BCYE agar plates for enumeration of bacteria.
To determine the infectivity of the L. pneumophila strain to H. vermiformis, 105 amoebae/ml were infected with 103 CFU of L. pneumophila/ml in 5 ml of culture in triplicate as described previously (4). At several time points, aliquots were plated on BCYE agar plates for enumeration of bacteria.
Nucleotide sequence accession number.
The sequence shown in Fig. 2 has been assigned GenBank accession no. AF034213.
FIG. 2.
DNA sequence of the L. pneumophila asd gene. The start codon is indicated in bold.
RESULTS
Cloning of the L. pneumophila asd gene.
The Asd enzyme plays a central role in the biosynthesis of several amino acids in addition to DAP (44). Since the asd gene is conserved across several species, our strategy to clone the asd gene of L. pneumophila was to complement an asd defect in E. coli χ2981 with an L. pneumophila cosmid library for growth without DAP supplementation. Twelve clones that grew without a supplement of DAP were isolated. The cosmid DNA from these clones was purified, and a common 4.3-kb HindIII fragment was purified, subcloned into pBlueScript, and designated pOH1. pOH1 complemented the asd mutation of χ2981 for growth without DAP. Thus, pOH1 contained the asd gene of L. pneumophila.
Sequencing and analysis of the L. pneumophila asd gene.
Restriction mapping of pOH1 was used to further localize the L. pneumophila asd gene. Several digestion products were subcloned into pBlueScript and tested for the ability to complement the asd defect in χ2981. The L. pneumophila asd gene was localized to a 2.6-kb region of the 4.3-kb fragment between the PstI and SacI sites (Fig. 1A). Sequencing of the fragment revealed the presence of a potential 1,041-nucleotide open reading frame (Fig. 2).
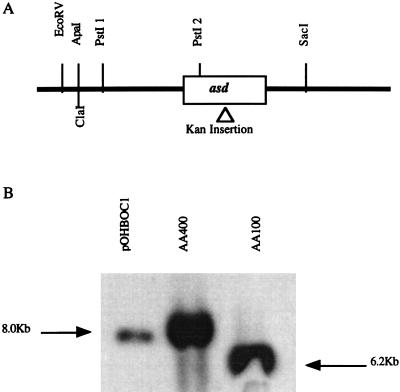
FIG. 1.
(A) Restriction map of the L. pneumophila 4.3-kb fragment containing the asd gene. The relative location of a mini-Tn10::kan insertion in the asd gene is indicated. (B) Southern hybridization of DNA from wild-type L. pneumophila (AA100), Δasd L. pneumophila (AA400), and plasmid pOHBOC1K harboring the 4.3-kb L. pneumophila fragment with the mini-Tn10::kan insertion. All DNA samples were digested with PstI and probed with pOH1 harboring the 4.3-kb L. pneumophila fragment. The 1.8-kb increase in the size of the band in pOHBOC1K and AA400 corresponds to the size of the kan cassette insertion.
Alignment of the predicted protein sequence of the open reading frame to other protein sequences in the databases revealed that it was most similar to the Asd of Vibrio cholerae, with 61% identity and 80% similarity. Several conserved amino acids among all known asd sequences were also present in the predicted L. pneumophila asd gene product. Most notably were glycines 11, 14, and 17 involved in coenzyme binding, cysteine 132 and glutamine 159 in the active subunit of the enzyme, and arginine 238 important for substrate binding (36).
Generation of an asd insertion mutant of L. pneumophila.
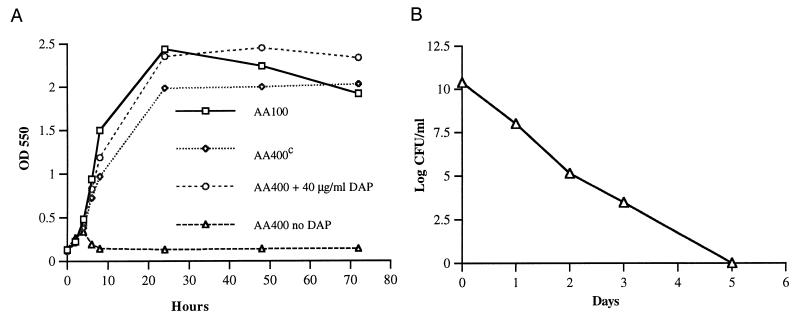
To facilitate allelic exchange into the L. pneumophila chromosome, the 4.3-kb fragment harboring the L. pneumophila asd gene was subcloned into pBOC20, designated pOHBOC1, and subjected to mini-Tn10::kan mutagenesis. Selection for clones that did not complement the χ2981 asd defect was used to screen for potential asd mini-Tn10::kan insertion mutants. One of the mini-Tn10::kan asd insertion mutants, designated pOHBOC1K, was mobilized into L. pneumophila AA100 by conjugation, and an asd mutant designated AA400 was isolated (Materials and Methods) (Fig. 1A). Allelic exchange into the L. pneumophila chromosome was confirmed by Southern hybridization, which showed a 1.8-kb increase in the size of the fragment that contained the asd gene (Fig. 1B). AA400 was auxotrophic for DAP and exhibited DAP dose-dependent growth in culture medium (data not shown). A DAP concentration of 40 μg/ml was the lowest concentration sufficient for optimal in vitro growth of AA400 (Fig. 3A). Growth of AA400 in BYE medium without a supplement of DAP was restored by introduction of pOHBOC1 into AA400 (AA400c [AA400 complemented with wild-type asd]; Fig. 3A). AA400 did not grow in the absence of DAP supplement, and viable bacteria were absent from the culture 6 days after incubation without DAP (Fig. 3B).
FIG. 3.
(A) In vitro growth kinetics of AA400 in BYE medium. Cell density was determined at an optical density of 550 nm (OD 550). (B) AA400 grown in BYE medium in the absence of DAP. Error bars cannot be seen due to their small values.
Cytopathogenicity of AA400 to U937 macrophage-like cells.
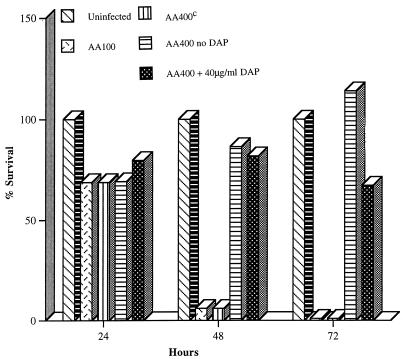
AA100 was completely cytopathogenic to U937 cells 48 and 72 h postinfection (Fig. 4). In contrast, AA400 was completely defective in cytopathogenicity to U937 cells (Fig. 4). A 40-μg/ml supplement of DAP to the infection with AA400 only partially restored the cytopathogenicity defect of AA400 (Fig. 4). Increasing the concentration of DAP to 200 μg/ml did not enhance the cytopathogenicity of AA400 (data not shown). AA400c exhibited wild-type cytopathogenicity to U937 cells (Fig. 4). A supplement of DAP had no effect on the cytopathogenicity of AA100 (data not shown). Thus, DAP auxotrophy in L. pneumophila rendered it defective for cytopathogenicity to U937 macrophages, and a supplement of DAP was unable to completely rescue the mutant.
FIG. 4.
Cytopathogenicity of L. pneumophila AA400 (Δasd), AA400c, and AA100 (wild type) to U937 macrophage-like cells. Error bars cannot be seen due to their small values.
In vivo growth of AA400 in U937 macrophage-like cells.
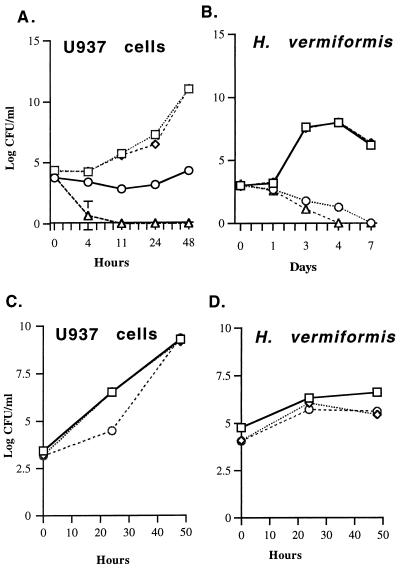
The ability of AA400 to survive and replicate in U937 macrophage-like cells was determined. In the absence of DAP, AA400 did not replicate and reached undetectable levels at 11 h postinfection in U937 cells (Fig. 5A). A supplement of DAP to U937 cells infected with AA400 resulted in a constant number of bacterial counts over the time course of the experiment, although a slight increase in bacterial numbers was observed 48 h postinfection (Fig. 5A). Furthermore, preincubation of the U937 cells with DAP prior to infection for up to 24 h or increasing the concentration of DAP to 200 μg/ml did not enhance the viability of AA400 (data not shown). AA400c exhibited a wild-type growth pattern in U937 cells (Fig. 5A).
FIG. 5.
(A and B) Intracellular growth kinetics of L. pneumophila AA400 (Δasd), AA400c, and AA100 (wild type). ○, AA400 plus 40 μg of DAP per ml; □, AA100; ◊, AA400c; ▵, AA400, no DAP. (C and D) Comparison of intracellular survival of AA400 complemented with the PCR-generated asd (AA400cc) to that of AA100 and AA400c. CFU levels were determined by plating dilutions from each time point on BCYE agar. Some error bars cannot be seen due to their small values. □, AA100; ◊, AA400cc; ○, AA400c.
In vivo growth of AA400 in protozoa.
Since protozoa are the environmental hosts of L. pneumophila, we determined the ability of AA400 to survive within the protozoan H. vermiformis. In the absence of DAP, AA400 did not replicate within H. vermiformis and was undetectable 4 days postinfection (Fig. 5B). The viability of AA400 was slightly enhanced in the presence of DAP, although bacteria were undetectable 7 days postinfection (Fig. 5B). AA400c exhibited a growth pattern similar to that of AA100 (Fig. 5B). Preincubation of H. vermiformis or another protozoan host (Acanthamoeba polyphaga) with DAP for 24 h prior to infection had no effect on the viability of AA400 (data not shown).
Does the insertion in asd have a polar effect?
Since DAP did not rescue the mutant in U937 macrophages or H. vermiformis, it is possible that the mini-Tn10::kan insertion in AA400 has polar effects on genes downstream of asd which may be important for intracellular survival of L. pneumophila. Thus, to determine if the asd defect of AA400 is the sole contributor to its inability to survive intracellularly, we complemented this strain with a PCR-generated asd gene. A 1.4-kb region from pOH1 harboring the L. pneumophila asd gene alone was amplified by PCR. This fragment was subcloned into pBOC20 and designated pOHBOC2. pOHBOC2 was found to complement the asd defect of χ2981. pOHBOC2 was mobilized into AA400, and colonies were selected for kanamycin resistance and the ability to grow without DAP supplement.
The ability of one of the asd-complemented clones of AA400, designated AA400cc, to survive and replicate within H. vermiformis, A. polyphaga, and U937 cells was compared to that of AA100 and AA400c. pOHBOC2 complemented AA400 for intracellular survival and growth in all cell lines tested (Fig. 5C and D and data not shown). Thus, the inability of AA400 to grow intracellularly was due solely to the lack of a functional asd gene.
DISCUSSION
Nutritional auxotrophs of several facultative intracellular pathogens have been generated and shown to be defective in intracellular survival and avirulent in animal models (12, 20, 28). Furthermore, targeting bacterium-specific metabolic pathways with antibiotics may prove to be an effective means of interrupting a pathogen’s intracellular life cycle. To determine the effect of nutritional auxotrophy on the intracellular life cycle of L. pneumophila and the feasibility of using the Asd enzyme as a potential target for treatment, we generated an aspartate-β-semialdehyde dehydrogenase (asd) mutant of L. pneumophila.
The asd mutant (AA400) was found to be completely defective in cytopathogenicity to U937 cells. In addition, AA400 was defective in intracellular growth within U937 macrophages, H. vermiformis, and A. polyphaga. In contrast to the thymidine and tryptophan auxotrophs of L. pneumophila that can be rescued by the respective nutrient, AA400 was not rescued by DAP (33). The inability of a DAP supplement to complement the intracellular growth of AA400 may be explained by several possibilities. First, in U937 cells the bacterial counts remained constant in the presence of DAP, which may indicate that some DAP reached the L. pneumophila phagosome, thereby prolonging the intracellular survival of AA400, or that the rate of replication of AA400 in the presence of DAP is equal to its rate of death in the intracellular environment. Second, although eukaryotic cells do not utilize DAP, it is possible that DAP is degraded by the cell lines examined in this study before it reaches the L. pneumophila phagosome. Third, it is possible that DAP is unable to cross the cell membrane of the cell lines tested or, if it does, may not reach the L. pneumophila phagosome in sufficient quantities, especially since 40 μg of DAP per ml was required for optimal in vitro growth. However, recently an asd mutant of S. typhimurium was shown to be capable of taking up DAP in the intracellular environment of Int-407 cells (15). This may indicate a difference between the phagosomal compartments of L. pneumophila and S. typhimurium, the lack of a DAP transport mechanism in the tested cell lines, or both.
It is possible that the insertion mutation in the L. pneumophila asd gene resulted in a polar effect, thereby altering the expression of downstream genes that may be necessary for intracellular survival. However, complementation of AA400 with the wild-type asd gene restored its ability to grow within all the tested cell lines. This finding indicates that the asd defect alone in AA400 is sufficient to disrupt its intracellular growth.
The ability of L. pneumophila to cause disease in humans has been closely associated with its ability to survive within protozoa in the contaminating water sources (18). The inability of AA400 to survive within H. vermiformis and A. polyphaga even in the presence of DAP in the culture media may indicate the possibility of targeting bacterium-specific metabolic pathways with antimicrobial agents in water systems. However, it should be cautioned that the inability of AA400 to survive intracellularly in tissue culture conditions may not reflect its ability for intracellular survival in environmental water reservoirs.
In summary, a DAP auxotroph of L. pneumophila was generated and found to be unable to survive in the intracellular environment of mammalian and protozoan cells. Our data indicate that the DAP biosynthetic pathway is a potential target for antibiotic therapy, and its disruption may be useful in the design of attenuated L. pneumophila vaccines. Furthermore, the inability of this mutant to survive in its environmental host may extend the idea of antimicrobial treatment of infected individuals to the treatment of water supplies that are associated with Legionnaires’ disease outbreaks.
ACKNOWLEDGMENTS
We thank Gopi Shankar, Chandrasekar Venkataraman, Lian-Yong Gao, and Tayfun Carli for critical review of the manuscript.
Y.A. was supported by Public Health Service grant R29AI38410. O.S.H. was supported by NIH training grant 5T32CA09509.
REFERENCES
- 1.Abu Kwaik Y. The phagosome containing Legionella pneumophila within the protozoan Hartmannella vermiformis is surrounded by the rough endoplasmic reticulum. Appl Environ Microbiol. 1996;62:2022–2028. doi: 10.1128/aem.62.6.2022-2028.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu Kwaik Y. Induced expression of a 20-kDa encoding gene of Legionella pneumophila during intracellular infection. Infect Immun. 1998;66:203–212. doi: 10.1128/iai.66.1.203-212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abu Kwaik Y, Eisenstein B I, Engleberg N C. Phenotypic modulation by Legionella pneumophila upon infection of macrophages. Infect Immun. 1993;61:1320–1329. doi: 10.1128/iai.61.4.1320-1329.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abu Kwaik Y, Fields B S, Engleberg N C. Protein expression by the protozoan Hartmannella vermiformis upon contact with its bacterial parasite Legionella pneumophila. Infect Immun. 1994;62:1860–1866. doi: 10.1128/iai.62.5.1860-1866.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abu Kwaik Y, Gao L-Y, Harb O S, Stone B J. Transcriptional regulation of the macrophage-induced gene (gspA) of Legionella pneumophila and phenotypic characterization of a null mutant. Mol Microbiol. 1997;24:629–642. doi: 10.1046/j.1365-2958.1997.3661739.x. [DOI] [PubMed] [Google Scholar]
- 6.Abu Kwaik Y, Pederson L L. The use of differential display-PCR to isolate and characterize a Legionella pneumophila locus induced during the intracellular infection of macrophages. Mol Microbiol. 1996;21:543–556. doi: 10.1111/j.1365-2958.1996.tb02563.x. [DOI] [PubMed] [Google Scholar]
- 7.Albano A A, Arroyo J, Eisenstein B I, Engleberg N C. PhoA gene fusions in Legionella pneumophila generated in vivo using a new transposon, MudphoA. Mol Microbiol. 1992;6:1829–1839. doi: 10.1111/j.1365-2958.1992.tb01355.x. [DOI] [PubMed] [Google Scholar]
- 8.Amano K-I, Williams J C. Peptidoglycan of Legionella pneumophila: apparent resistance to lysozyme hydrolysis correlates with a high degree of peptide cross-linking. J Bacteriol. 1983;153:520–526. doi: 10.1128/jb.153.1.520-526.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arroyo J, Hurley M C, Wolf M, McClain M S, Eisenstein B I, Engleberg N C. Shuttle mutagenesis of Legionella pneumophila: identification of a gene associated with host cell cytopathicity. Infect Immun. 1994;62:4075–4080. doi: 10.1128/iai.62.9.4075-4080.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bange F, Brown A M, Jacobs W R., Jr Leucine auxotrophy restricts the growth of Mycobacterium bovis BCG in macrophages. Infect Immun. 1996;64:1794–1799. doi: 10.1128/iai.64.5.1794-1799.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barker J, Scaife H, Brown M R W. Intraphagocytic growth induces an antibiotic-resistant phenotype of Legionella pneumophila. Antimicrob Agents Chemother. 1995;39:2684–2688. doi: 10.1128/aac.39.12.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biellmann J, Eid P, Hirth C, Jornvall H. Aspartate-β-semialdehyde dehydrogenase from Escherichia coli purification and general properties. Eur J Biochem. 1980;104:53–58. doi: 10.1111/j.1432-1033.1980.tb04398.x. [DOI] [PubMed] [Google Scholar]
- 13.Bozue J A, Johnson W. Interaction of Legionella pneumophila with Acanthamoeba castellanii: uptake by coiling phagocytosis and inhibition of phagosome-lysosome fusion. Infect Immun. 1996;64:668–673. doi: 10.1128/iai.64.2.668-673.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bozzoni M, Radice L, Frosi A, Vezzoli S, Cuboni A, Vezzoli F. Prevalence of pneumonia due to Legionella pneumophila and Mycoplasma pneumoniae in a population admitted to a department of internal medicine. Respiration. 1995;62:331–335. doi: 10.1159/000196475. [DOI] [PubMed] [Google Scholar]
- 15.Burns-Keliher L L, Portteus A, Curtiss R., III Specific detection of Salmonella typhimurium proteins synthesized intracellularly. J Bacteriol. 1997;179:3604–3612. doi: 10.1128/jb.179.11.3604-3612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cianciotto N P, Long R, Eisenstein B I, Engleberg N C. Site-specific mutagenesis in Legionella pneumophila by allelic exchange using counterselectable ColE1 vectors. FEMS Microbiol Lett. 1988;56:203–208. [Google Scholar]
- 17.Edelstein P H. Legionnaires’ disease. Clin Infect Dis. 1993;16:741–749. doi: 10.1093/clind/16.6.741. [DOI] [PubMed] [Google Scholar]
- 18.Fields B S. The molecular ecology of legionellae. Trends Microbiol. 1996;4:286–290. doi: 10.1016/0966-842x(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 19.Fields B S, Nerad T A, Sawyer T K, King C H, Barbaree J M, Martin W T, Morrill W E, Sanden G N. Characterization of an axenic strain of Hartmannella vermiformis obtained from an investigation of nosocomial legionellosis. J Protozool. 1990;37:581–583. doi: 10.1111/j.1550-7408.1990.tb01269.x. [DOI] [PubMed] [Google Scholar]
- 20.Fields P I, Swanson R V, Haidaris C G, Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao L-Y, Harb O S, Abu Kwaik Y. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant hosts, mammalian and protozoan cells. Infect Immun. 1997;65:4738–4746. doi: 10.1128/iai.65.11.4738-4746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao L-Y, Harb O S, Abu Kwaik Y. Identification of macrophage-specific infectivity loci (mil) of Legionella pneumophila that are not required for infectivity of protozoa. Infect Immun. 1998;66:883–892. doi: 10.1128/iai.66.3.883-892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao, L.-Y., B. J. Stone, M. Guzman, J. K. Brieland, and Y. Abu Kwaik. Submitted for publication.
- 24.Guerrant G O, Lambart M S, Moss C W. Identification of diaminopimelic acid in the Legionnaires’ disease bacterium. J Clin Microbiol. 1979;10:815–818. doi: 10.1128/jcm.10.6.815-818.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harb O S, Venkataraman C, Haack B J, Gao L-Y, Abu Kwaik Y. Heterogeneity in the attachment and uptake mechanisms of the Legionnaires’ disease bacterium, Legionella pneumophila, by protozoan hosts. Appl Environ Microbiol. 1998;64:126–132. doi: 10.1128/aem.64.1.126-132.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 27.Hess J, Kaufmann H E. Vaccination strategies against intracellular microbes. FEMS Immunol Med Microbiol. 1993;7:95–104. doi: 10.1111/j.1574-695X.1993.tb00387.x. [DOI] [PubMed] [Google Scholar]
- 28.Hoiseth S K, Stocker B A D. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 29.Horwitz M A. Formation of a novel phagosome by the Legionnaires’ disease bacterium (Legionella pneumophila) in human monocytes. J Exp Med. 1983;158:1319–1331. doi: 10.1084/jem.158.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaufmann S H E. Immunity to intracellular bacteria. Annu Rev Immunol. 1993;11:129–163. doi: 10.1146/annurev.iy.11.040193.001021. [DOI] [PubMed] [Google Scholar]
- 31.King C H, Fields B S, Shotts E B, Jr, White E H. Effects of cytochalasin D and methylamine on intracellular growth of Legionella pneumophila in amoebae and human monocyte-like cells. Infect Immun. 1991;59:758–763. doi: 10.1128/iai.59.3.758-763.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marston B J. Epidemiology of community-acquired pneumonia. Infect Dis Clin Pract. 1995;4:S232–S239. [Google Scholar]
- 33.Mintz C S, Jianxing C, Shuman H A. Isolation and characterization of auxotrophic mutants of Legionella pneumophila that fail to multiply in human monocytes. Infect Immun. 1988;56:1449–1455. doi: 10.1128/iai.56.6.1449-1455.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oldham L J, Rodgers F G. Adhesion, penetration and intracellular replication of Legionella pneumophila: an in vitro model of pathogenesis. J Gen Microbiol. 1985;131:697–706. doi: 10.1099/00221287-131-4-697. [DOI] [PubMed] [Google Scholar]
- 35.Onody C, Matsiota-Bernard P, Nauciel C. Lack of resistance to erythromycin, rifampicin and ciprofloxacin in 98 clinical isolates of Legionella pneumophila. J Antimicrob Chemother. 1997;39:815–816. doi: 10.1093/jac/39.6.815. [DOI] [PubMed] [Google Scholar]
- 36.Ouyang J, Viola R E. Use of structural comparisons to select mutagenic targets in aspartate-β-semialdehyde dehydrogenase. Biochemistry. 1995;34:6394–6399. doi: 10.1021/bi00019a019. [DOI] [PubMed] [Google Scholar]
- 37.Ozkaynak E, Putney S D. A unidirectional deletion technique for the generation of clones for sequencing. BioTechniques. 1987;5:770–773. [Google Scholar]
- 38.Pavelka M S, Jr, Jacobs W R., Jr Biosynthesis of diaminopimelate, the precursor of lysine and a component of peptidoglycan, is an essential function of Mycobacterium smegmatis. J Bacteriol. 1996;178:6496–6507. doi: 10.1128/jb.178.22.6496-6507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stone B J, Abu Kwaik Y. Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infect Immun. 1998;66:1768–1775. doi: 10.1128/iai.66.4.1768-1775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Susa M, Hacker J, Marre R. De novo synthesis of Legionella pneumophila antigens during intracellular growth in phagocytic cells. Infect Immun. 1996;64:1679–1684. doi: 10.1128/iai.64.5.1679-1684.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tenover F C. Emerging problems in antimicrobial resistance. J Intraven Nurs. 1995;18:297–300. [PubMed] [Google Scholar]
- 44.Umbarger H E. Amino acid biosynthesis and its regulation. Annu Rev Biochem. 1978;47:533–606. doi: 10.1146/annurev.bi.47.070178.002533. [DOI] [PubMed] [Google Scholar]
- 45.Venkataraman C, Haack B J, Bondada S, Abu Kwaik Y. Identification of a Gal/GalNAc lectin in the protozoan Hartmanella vermiformis as a potential receptor for attachment and invasion by the Legionnaires’ disease bacterium, Legionella pneumophila. J Exp Med. 1997;186:537–547. doi: 10.1084/jem.186.4.537. [DOI] [PMC free article] [PubMed] [Google Scholar]